Transcriptomic Analysis Identifies GhSACPD-Mediated Fatty Acid Regulation in the Cotton Boll Abscission
Abstract
1. Introduction
2. Materials and Methods
2.1. Investigation of Plant Materials and Squares-Bolls Abscission Rate
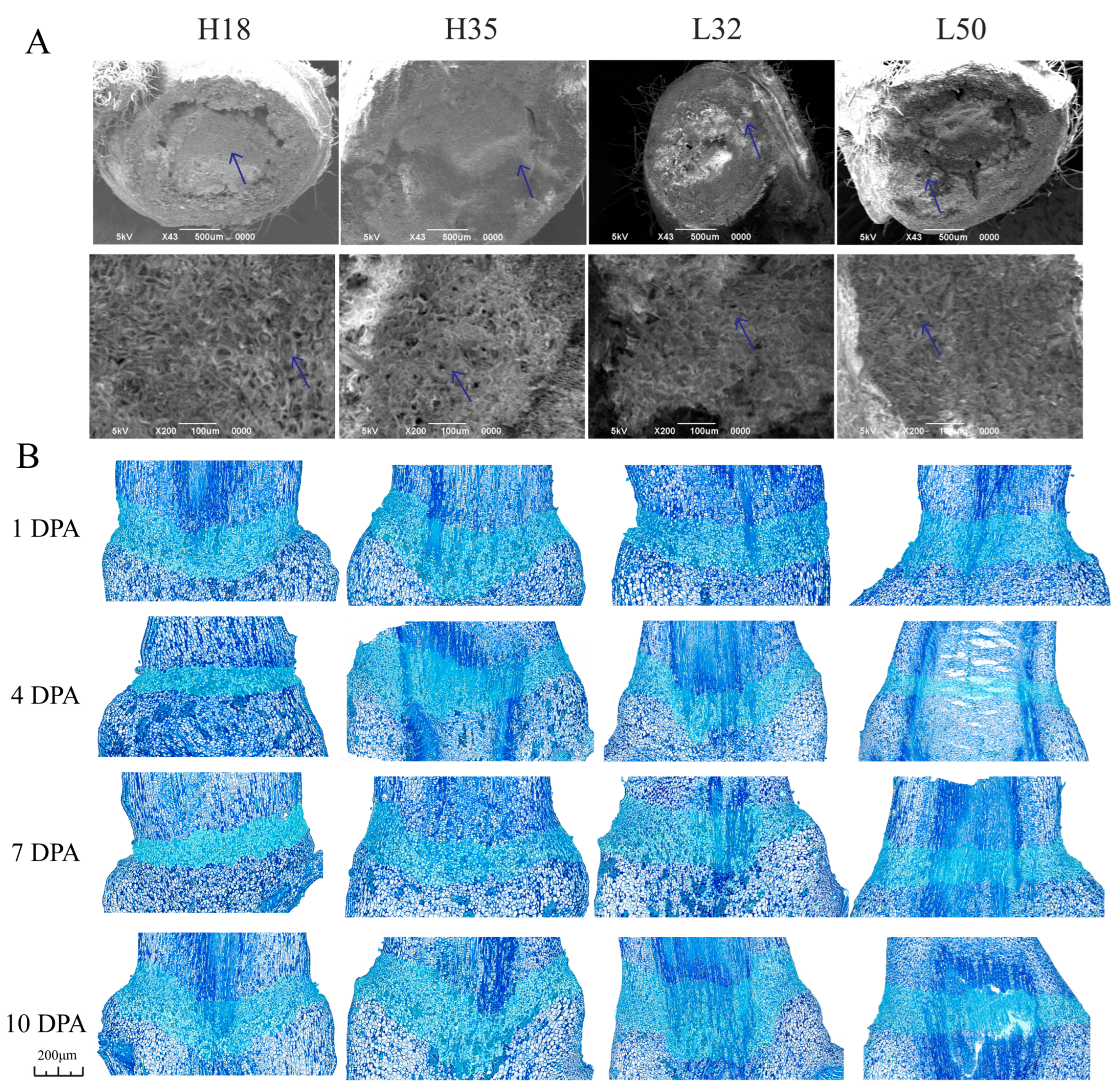
2.2. Cellular Structure Observation of Boll Pedicel Abscission Zone
2.3. RNA-seq Profiling and Computational Analysis of Boll Pedicel Abscission Zone Transcriptomes
2.4. Identification and Characterization of SACPD Gene Family Members
2.5. Phylogeny and Synteny Analysis Reveal Evolutionary Patterns of the SACPD Gene Family
2.6. Expression Pattern Analysis of SACPD Genes Across Cotton Tissues
3. Results
3.1. Identification of the Bolls Abscission Phenotype and Anatomical Characterization
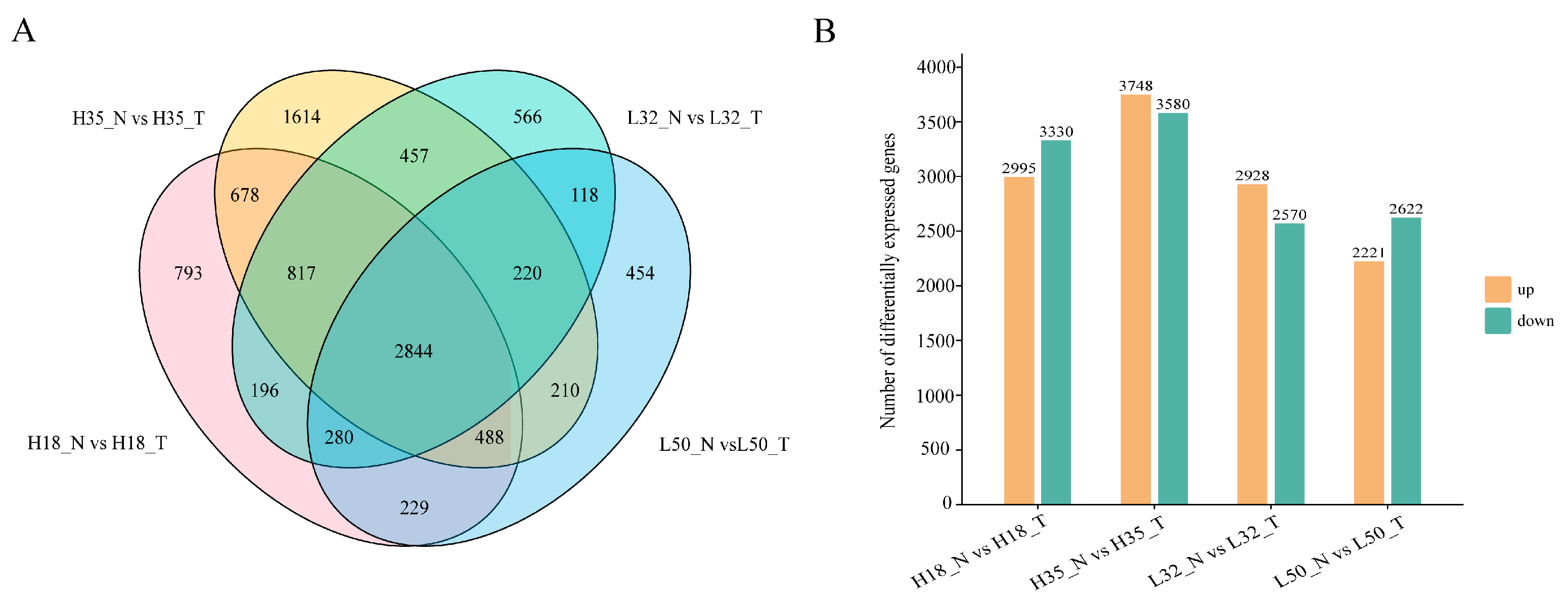
3.2. Comparative Transcriptome Analysis of AZ Tissues Between Abscission-Sensitive and Abscission-Resistant Cotton Lines
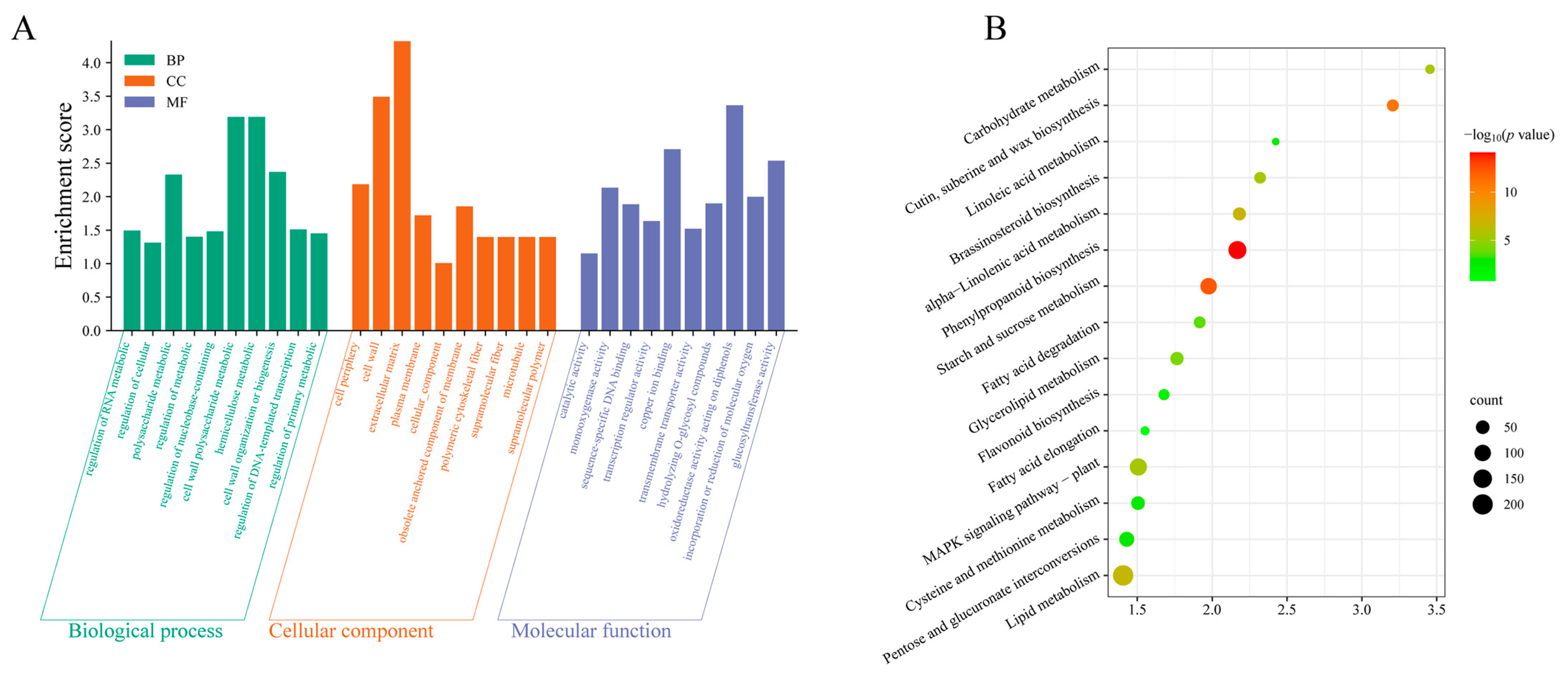
3.3. Lipid Metabolism Is a Key Metabolic Pathway in Cotton Boll Abscission as Revealed by GO and KEGG Analysis
3.4. The SACPD Gene as the Key Hub Gene Identified by WGCNA-Based Construction of Gene Co-Expression Networks
3.5. Genome-Wide Identification and Characterization of Stearoyl-Acyl Carrier Protein Desaturase Genes in Cotton Species
3.6. Structure and Motif Analysis of SACPD Family Genes
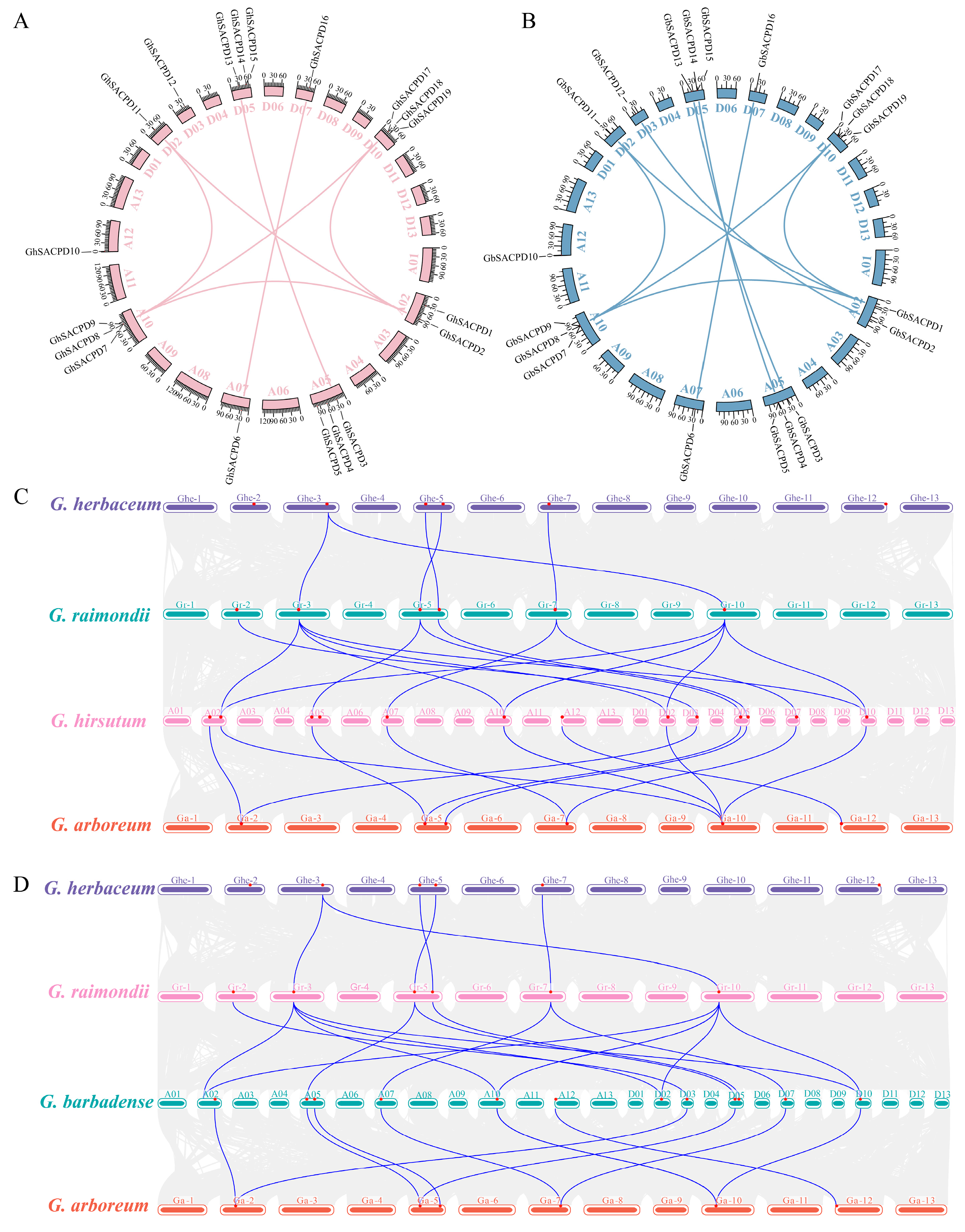
3.7. Characterization of Chromosome Distribution and Gene Duplication in the SACPD Gene Family
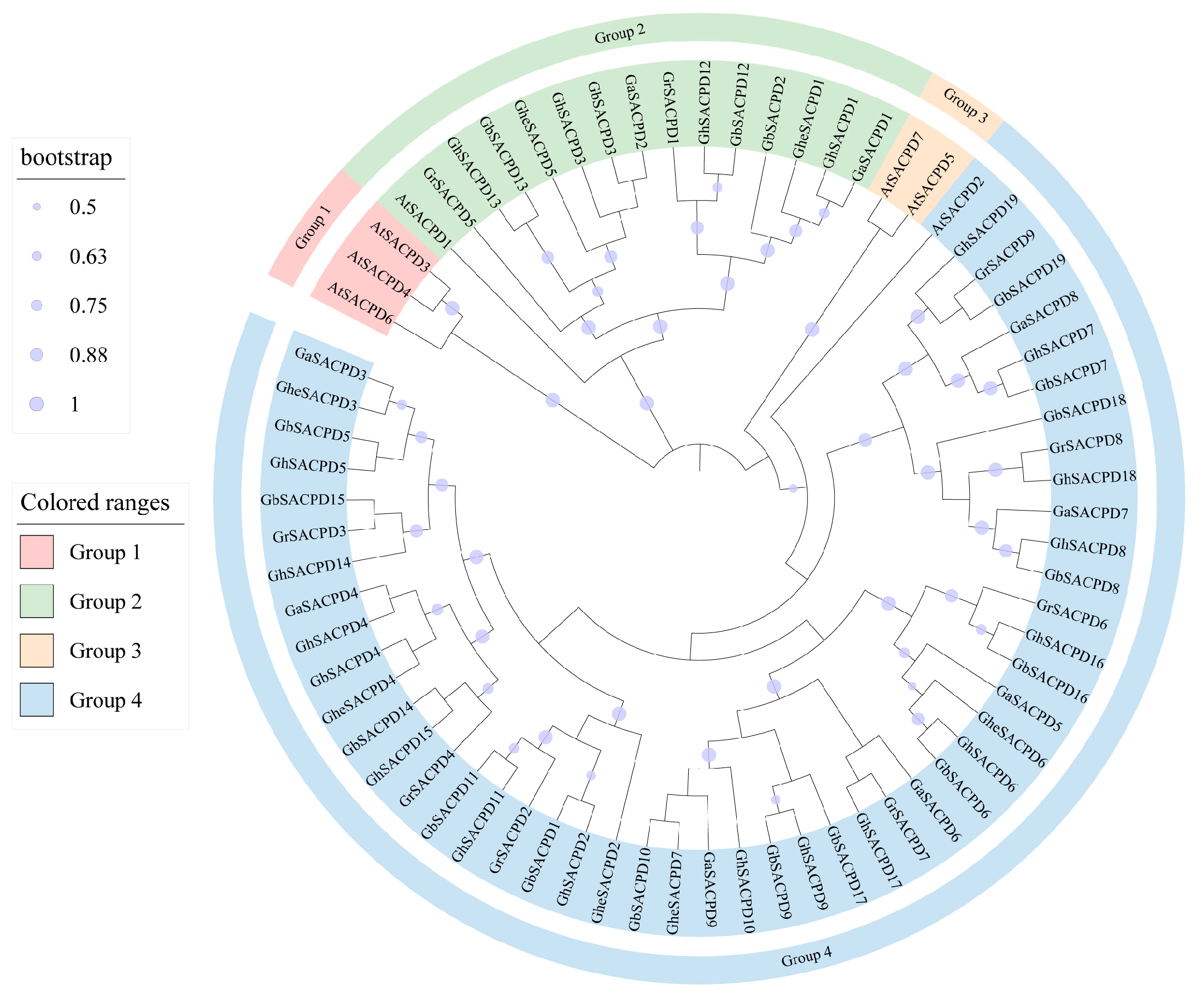
3.8. Genomic Collinearity and Homologous Evolution Analysis of the SACPD Gene Family in Cotton
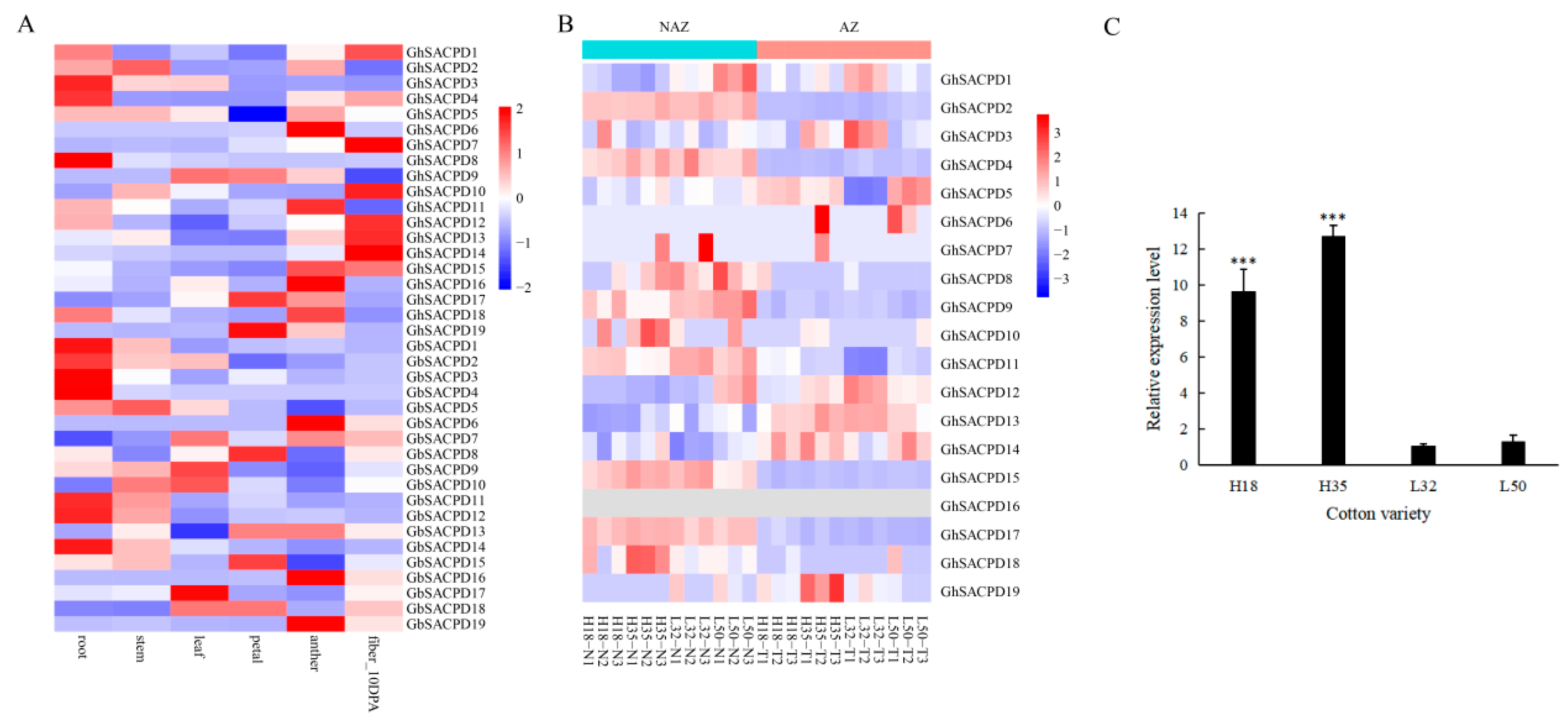
3.9. Expression Profiling of the SACPD Gene Family in Cotton
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- He, S.; Sun, G.; Geng, X.; Gong, W.; Dai, P.; Jia, Y.; Shi, W.; Pan, Z.; Wang, J.; Wang, L.; et al. The genomic basis of geographic differentiation and fiber improvement in cultivated cotton. Nat. Genet. 2021, 53, 916–924. [Google Scholar] [CrossRef]
- Huang, X.; Hu, Y.; Li, Z.; Jiao, B.; Ma, X.; Guo, Q.; Wang, Q. Dephenolization methods, quality characteristics, applications, and advancements of dephenolized cottonseed protein: Review. Foods 2025, 14, 628. [Google Scholar] [CrossRef]
- Hu, G.; Wang, Z.; Tian, Z.; Wang, K.; Ji, G.; Wang, X.; Zhang, X.; Yang, Z.; Liu, X.; Niu, R.; et al. A telomere-to-telomere genome assembly of cotton provides insights into centromere evolution and short-season adaptation. Nat. Genet. 2025, 57, 1031–1043. [Google Scholar] [CrossRef] [PubMed]
- He, Z.; Ma, X.; Wang, F.; Li, J.; Zhao, M. LcERF10 functions as a positive regulator of litchi fruitlet abscission. Int. J. Biol. Macromol. 2023, 250, 126264. [Google Scholar] [CrossRef]
- Wang, Y.; Ye, H.; Bai, J.; Ren, F. The regulatory framework of developmentally programmed cell death in floral organs: A review. Plant Physiol. Biochem. 2021, 158, 103–112. [Google Scholar] [CrossRef] [PubMed]
- Parra-Lobato, M.C.; Paredes, M.A.; Labrador, J.; Saucedo-García, M.; Gavilanes-Ruiz, M.; Gomez-Jimenez, M.C. Localization of sphingolipid enriched plasma membrane regions and long-chain base composition during mature-fruit abscission in olive. Front. Plant Sci. 2017, 8, 1138. [Google Scholar] [CrossRef] [PubMed]
- Chang, L.; Liu, Z.; Ying, X.; Kalandarov, B.; Ergashev, M.; Tong, X.; Zhang, J.; Jin, J.; Ying, J. Molecular basis of lipid metabolism in Oryza sativa L. Plants 2024, 13, 3263. [Google Scholar] [CrossRef]
- Lee, S.B.; Suh, M.C. Advances in the understanding of cuticular waxes in Arabidopsis thaliana and crop species. Plant Cell Rep. 2015, 34, 557–572. [Google Scholar] [CrossRef]
- Grifé-Ruiz, M.; Hierrezuelo-León, J.; de Vicente, A.; Pérez-García, A.; Romero, D. Diversification of lipopeptide analogues drives versatility in biological activities. J. Agric. Food Chem. 2025, 73, 1403–1416. [Google Scholar] [CrossRef]
- Zhang, Y.; Liu, X.; Sun, Y.; Liu, Y.; Zhang, Y.; Ding, T.; Chen, J. Salivary protein cyclin-dependent kinase-like from Grain aphid Sitobion avenae suppresses wheat defense response and enhances aphid adaptation. Int. J. Mol. Sci. 2024, 25, 4579. [Google Scholar] [CrossRef]
- Cao, Y.; Xian, M.; Yang, J.; Xu, X.; Liu, W.; Li, L. Heterologous expression of stearoyl-acyl carrier protein desaturase (S-ACP-DES) from Arabidopsis thaliana in Escherichia coli. Protein Expr. Purif. 2010, 69, 209–214. [Google Scholar] [CrossRef]
- Kachroo, P.; Shanklin, J.; Shah, J.; Whittle, E.J.; Klessig, D.F. A fatty acid desaturase modulates the activation of defense signaling pathways in plants. Proc. Natl. Acad. Sci. USA 2001, 98, 9448–9453. [Google Scholar] [CrossRef]
- Kachroo, A.; Shanklin, J.; Whittle, E.; Lapchyk, L.; Hildebrand, D.; Kachroo, P. The Arabidopsis stearoyl-acyl carrier protein-desaturase family and the contribution of leaf isoforms to oleic acid synthesis. Plant Mol. Biol. 2007, 63, 257–271. [Google Scholar] [CrossRef]
- Adolf, J.E.; Place, A.R.; Stoecker, D.K.; Harding, L.W. Modulation of polyunsaturated fatty acids in mixotrophic Karlodinium Veneficum (Dinophyceae) and its prey, Storeatula major (Cryptophyceae). J. Phycol. 2007, 43, 1259–1270. [Google Scholar] [CrossRef]
- Zhang, L.; Yuan, Q.; Hu, C.; Sun, X.; Gong, Y.; Xu, N. Characterization of monogalactosyldiacylglycerol synthases in Gracilariopsis lemaneiformis and their potential roles in the fading of the thallus. J. Phycol. 2023, 59, 1258–1271. [Google Scholar] [CrossRef]
- Lightner, J.; Wu, J.; Browse, J. A mutant of arabidopsis with increased levels of stearic-acid. Plant Physiol. 1994, 106, 1443–1451. [Google Scholar] [CrossRef]
- Kachroo, P.; Kachroo, A.; Lapchyk, L.; Hildebrand, D.; Klessig, D.F.; Li, J.; Galla, A.; Avila, C.A.; Flattmann, K.; Vaughn, K.; et al. Restoration of defective cross talk in ssi2 mutants: Role of salicylic acid, jasmonic acid, and fatty acids in SSI2-mediated signaling. Mol. Plant-Microbe Interact. 2003, 16, 1022–1029. [Google Scholar] [CrossRef]
- Kachroo, P.; Venugopal, S.; Navarre, D.; Lapchyk, L.; Kachroo, A. Role of salicylic acid and fatty acid desaturation pathways in ssi2-mediated signaling. Plant Physiol. 2005, 139, 1717–1735. [Google Scholar] [CrossRef]
- Shah, J.; Kachroo, P.; Nandi, A.; Klessig, D.F. A recessive mutation in the Arabidopsis SSI2 gene confers SA- and NPR1-independent expression of PR genes and resistance against bacterial and oomycete pathogens. Plant J. 2001, 25, 563–574. [Google Scholar] [CrossRef]
- Mandal, M.K.; Chandra-Shekara, A.C.; Jeong, R.-D.; Yu, K.; Zhu, S.; Chanda, B.; Navarre, D.; Kachroo, A.; Kachroo, P. Oleic acid-dependent modulation of NITRIC OXIDE ASSOCIATED1 protein levels regulates nitric oxide-mediated defense signaling in Arabidopsis. Plant Cell 2012, 24, 1654–1674. [Google Scholar] [CrossRef]
- Wang, D.; Zhao, X.; Su, T.; Wang, W.; Xin, X.; Zhang, B.; Zhang, D.; Yu, Y.; Wang, Z.; Zhang, F.; et al. Exogenous gibberellin delays postharvest leaf senescence in pak choi by modulating transcriptomic and metabolomic profiles. Foods 2025, 14, 981. [Google Scholar] [CrossRef]
- Yang, B.; Tan, Z.; Yan, J.; Zhang, K.; Ouyang, Z.; Fan, R.; Lu, Y.; Zhang, Y.; Yao, X.; Zhao, H.; et al. Phospholipase-mediated phosphate recycling during plant leaf senescence. Genome Biol. 2024, 25, 199. [Google Scholar] [CrossRef]
- Briegas, B.; Camarero, M.C.; Corbacho, J.; Labrador, J.; Sanchez-Vera, V.; Gavilanes-Ruiz, M.; Saucedo-García, M.; Gomez-Jimenez, M.C. Sphingolipid long chain bases as mediators of cell death in olive fruit abscission. Physiol. Plant. 2025, 177, e70061. [Google Scholar] [CrossRef]
- Nandi, A.; Moeder, W.; Kachroo, P.; Klessig, D.F.; Shah, J. Arabidopsis ssi2-conferred susceptibility to Botrytis cinerea is dependent on EDS5 and PAD4. Mol. Plant-Microbe Interact. 2005, 18, 363–370. [Google Scholar] [CrossRef]
- Kućko, A.; Smoliński, D.; Wilmowicz, E.; Florkiewicz, A.; Alché, J.d.D. Spatio-temporal localization of LlBOP following early events of floral abscission in yellow lupine. Protoplasma 2019, 256, 1173–1183. [Google Scholar] [CrossRef]
- Li, J.; Liu, Z.; You, C.; Qi, Z.; You, J.; Grover, C.E.; Long, Y.; Huang, X.; Lu, S.; Wang, Y.; et al. Convergence and divergence of diploid and tetraploid cotton genomes. Nat. Genet. 2024, 56, 2562–2573. [Google Scholar] [CrossRef]
- Munkvold, J.D.; Laudencia-Chingcuanco, D.; Sorrells, M.E. Systems genetics of environmental response in the mature wheat embryo. Genetics 2013, 194, 265–277. [Google Scholar] [CrossRef]
- Zheng, J.; He, C.; Qin, Y.; Lin, G.; Park, W.D.; Sun, M.; Li, J.; Lu, X.; Zhang, C.; Yeh, C.; et al. Co-expression analysis aids in the identification of genes in the cuticular wax pathway in maize. Plant J. 2019, 97, 530–542. [Google Scholar] [CrossRef]
- Wang, X.; Bai, Y.; Zhang, L.; Jiang, G.; Zhang, P.; Liu, J.; Li, L.; Huang, L.; Qin, P. Identification and core gene-mining of weighted gene co-expression network Analysis-based co-expression modules related to flood resistance in quinoa seedlings. Bmc Genom. 2024, 25, 728. [Google Scholar] [CrossRef]
- Huang, Q.; Liu, L.; Li, H.; Wang, X.; Si, A.; He, L.; Yu, Y. Identification of co-expression modules of cotton plant height-related genes based on weighted gene co-expression network analysis. Agronomy 2025, 15, 196. [Google Scholar] [CrossRef]
- Chen, C.; Wu, Y.; Li, J.; Wang, X.; Zeng, Z.; Xu, J.; Liu, Y.; Feng, J.; Chen, H.; He, Y.; et al. TBtools-II: A “one for all, all for one”bioinformatics platform for biological big-data mining. Mol. Plant 2023, 16, 1733–1742. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive tree of life (iTOL) v5: An online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2021, 49, W293–W296. [Google Scholar] [CrossRef]
- Hu, Y.; Chen, J.; Fang, L.; Zhang, Z.; Ma, W.; Niu, Y.; Ju, L.; Deng, J.; Zhao, T.; Lian, J.; et al. Gossypium barbadense and Gossypium hirsutum genomes provide insights into the origin and evolution of allotetraploid cotton. Nat. Genet. 2019, 51, 739–748. [Google Scholar] [CrossRef]
- Mandal, S.; Ji, W.; McKnight, T.D. Candidate gene networks for acylsugar metabolism and plant defense in wild tomato Solanum pennellii. Plant Cell 2020, 32, 81–99. [Google Scholar] [CrossRef]
- Beno-Moualem, D.; Gusev, L.; Dvir, O.; Pesis, E.; Meir, S.; Lichter, A. The effects of ethylene, methyl jasmonate and 1-MCP on abscission of cherry tomatoes from the bunch and expression of endo-1,4-β-glucanases. Plant Sci. 2004, 167, 499–507. [Google Scholar] [CrossRef]
- Sexton, R.; Roberts, J. Cell biology of abscission. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1982, 33, 133–162. [Google Scholar] [CrossRef]
- Pautot, V.; Crick, J.; Hepworth, S.R. Abscission zones: Cellular interfaces for the programmed separation of organs. Ann. Bot. 2025, 136, 29–48. [Google Scholar] [CrossRef]
- Strom, N.I.; Gerring, Z.F.; Galimberti, M.; Yu, D.; Halvorsen, M.W.; Abdellaoui, A.; Rodriguez-Fontenla, C.; Sealock, J.M.; Bigdeli, T.; Coleman, J.R.; et al. Genome-wide analyses identify 30 loci associated with obsessive-compulsive disorder. Nat. Genet. 2025, 57, 1389–1401. [Google Scholar] [CrossRef]
- Qi, G.; Li, Y.; Zhang, W.; Han, Z.; Chen, J.; Zhang, Z.; Xuan, L.; Chen, R.; Fang, L.; Hu, Y.; et al. Reveal genomic insights into cotton domestication and improvement using gene level functional haplotype-based GWAS. Nat. Commun. 2025, 16, 4734. [Google Scholar] [CrossRef]
- Zhang, X.; Zhang, X.; Fu, Y.; Cui, Y.; Wu, N.; Li, Y.; Yang, Z.; Zhang, C.; Song, H.; He, G.; et al. HTT1, a stearoyl-acyl carrier protein desaturase involved in unsaturated fatty acid biosynthesis, affects rice heat tolerance. Plant Cell Environ. 2025, 48, 3391–3405. [Google Scholar] [CrossRef]
- Shilman, F.; Brand, Y.; Brand, A.; Hedvat, I.; Hovav, R. Identification and molecular characterization of homeologous Δ9-stearoyl acyl carrier protein desaturase 3 genes from the allotetraploid peanut (Arachis hypogaea). Plant Mol. Biol. Report. 2011, 29, 232–241. [Google Scholar] [CrossRef]
- Ding, M.; Zhou, D.; Ye, Y.; Wen, S.; Zhang, X.; Tian, Q.; Zhang, X.; Mou, W.; Dang, C.; Fang, Y.; et al. Genome-wide identification and expression analysis of the stearoyl-acyl carrier protein delta9 desaturase gene family under abiotic stress in barley. Int. J. Mol. Sci. 2023, 25, 113. [Google Scholar] [CrossRef]
- Liu, X.; Inoue, H.; Tang, X.; Tan, Y.; Xu, X.; Wang, C.; Jiang, C.-J. Rice OsAAA-ATPase1 is induced during blast infection in a salicylic acid-dependent manner, and promotes blast fungus resistance. Int. J. Mol. Sci. 2020, 21, 1443. [Google Scholar] [CrossRef]
- Mo, S.; Zhang, Y.; Wang, X.; Yang, J.; Sun, Z.; Zhang, D.; Chen, B.; Wang, G.; Ke, H.; Liu, Z.; et al. Cotton GhSSI2 isoforms from the stearoyl acyl carrier protein fatty acid desaturase family regulate Verticillium wilt resistance. Mol. Plant Pathol. 2021, 22, 1041–1056. [Google Scholar] [CrossRef]
- Wang, W.; Tan, Z.; Xu, Y.; Zhu, A.-A.; Li, Y.; Yao, J.; Tian, R.; Fang, X.-M.; Liu, X.-Y.; Tian, Y.-M.; et al. Chromosome structural variation of two cultivated tetraploid cottons and their ancestral diploid species based on a new high-density genetic map. Sci. Rep. 2017, 7, 7640. [Google Scholar] [CrossRef]
- Gao, Q.-M.; Venugopal, S.; Navarre, D.; Kachroo, A. Low oleic acid-derived repression of jasmonic acid-inducible defense responses requires the WRKY50 and WRKY51 proteins. Plant Physiol. 2011, 155, 464–476. [Google Scholar] [CrossRef]
- Yang, W.; Dong, R.; Liu, L.; Hu, Z.; Li, J.; Wang, Y.; Ding, X.; Chu, Z. A novel mutant allele of SSI2 confers a better balance between disease resistance and plant growth inhibition on Arabidopsis thaliana. BMC Plant Biol. 2016, 16, 208. [Google Scholar] [CrossRef]


| Accession ID | Defoliant Sensitivity | 2018-KEL-AR1 | 2018-SHZ-AR1 | 2019-KEL-AR1 | 2019-SHZ-AR1 | Mean |
|---|---|---|---|---|---|---|
| L32 | R | 8.91 | 2.06 | 22.24 | 3.57 | 9.20 |
| L50 | R | 2.15 | 1.60 | 37.60 | 3.40 | 11.19 |
| H35 | S | 12.72 | 36.58 | 53.82 | 20.77 | 30.97 |
| H18 | S | 25.74 | 38.72 | 51.81 | 43.51 | 39.95 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shui, G.; Chang, Z.; Han, P.; Zhang, Q.; Li, Z.; Lin, H.; Wang, X.; Wu, Y.; Nie, X. Transcriptomic Analysis Identifies GhSACPD-Mediated Fatty Acid Regulation in the Cotton Boll Abscission. Agriculture 2025, 15, 2166. https://doi.org/10.3390/agriculture15202166
Shui G, Chang Z, Han P, Zhang Q, Li Z, Lin H, Wang X, Wu Y, Nie X. Transcriptomic Analysis Identifies GhSACPD-Mediated Fatty Acid Regulation in the Cotton Boll Abscission. Agriculture. 2025; 15(20):2166. https://doi.org/10.3390/agriculture15202166
Chicago/Turabian StyleShui, Guangling, Zewei Chang, Peng Han, Qi Zhang, Zhibo Li, Hairong Lin, Xin Wang, Yuanlong Wu, and Xinhui Nie. 2025. "Transcriptomic Analysis Identifies GhSACPD-Mediated Fatty Acid Regulation in the Cotton Boll Abscission" Agriculture 15, no. 20: 2166. https://doi.org/10.3390/agriculture15202166
APA StyleShui, G., Chang, Z., Han, P., Zhang, Q., Li, Z., Lin, H., Wang, X., Wu, Y., & Nie, X. (2025). Transcriptomic Analysis Identifies GhSACPD-Mediated Fatty Acid Regulation in the Cotton Boll Abscission. Agriculture, 15(20), 2166. https://doi.org/10.3390/agriculture15202166






