Crop Disease Spore Detection Method Based on Au@Ag NRS
Abstract
1. Introduction
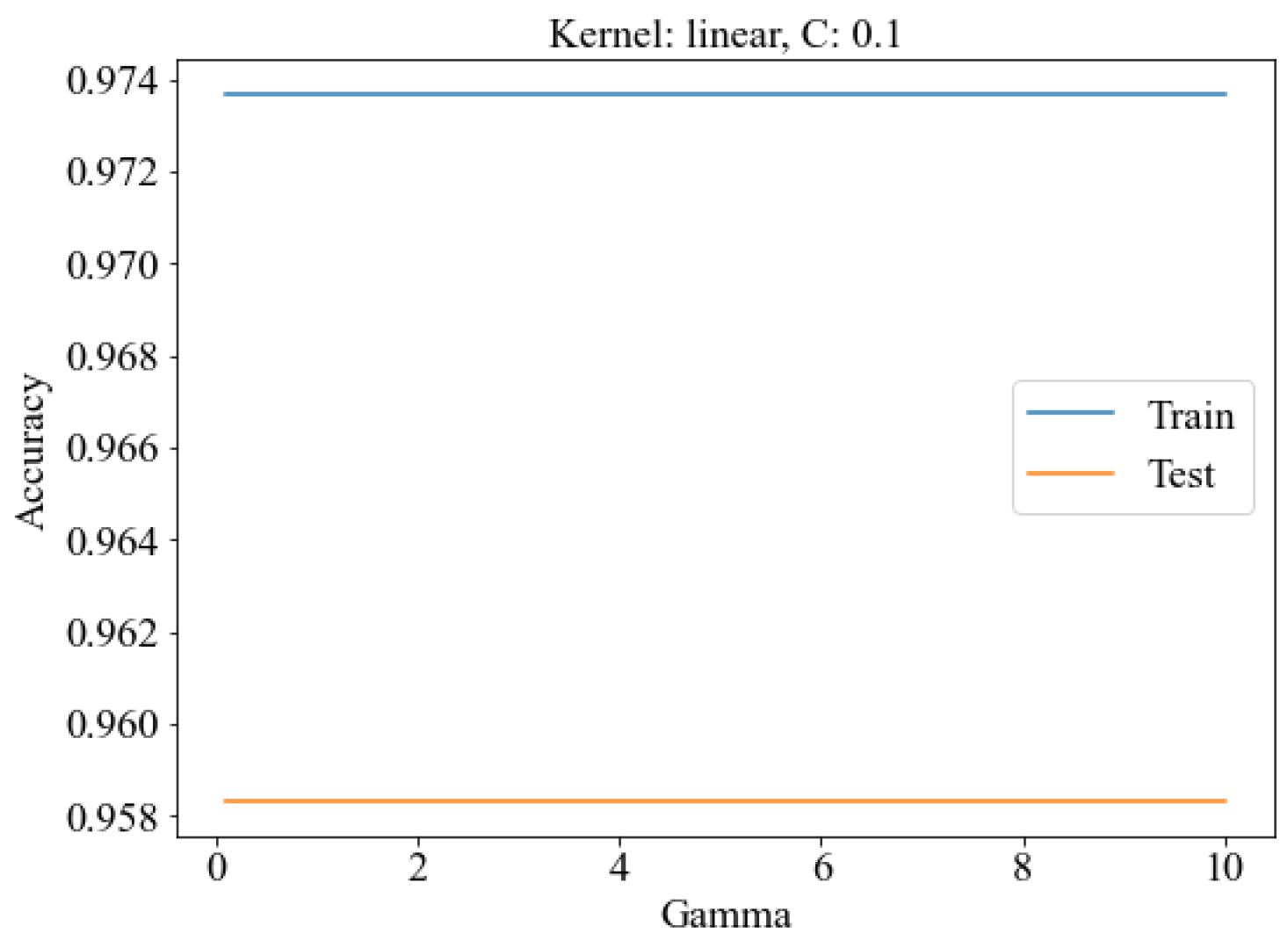
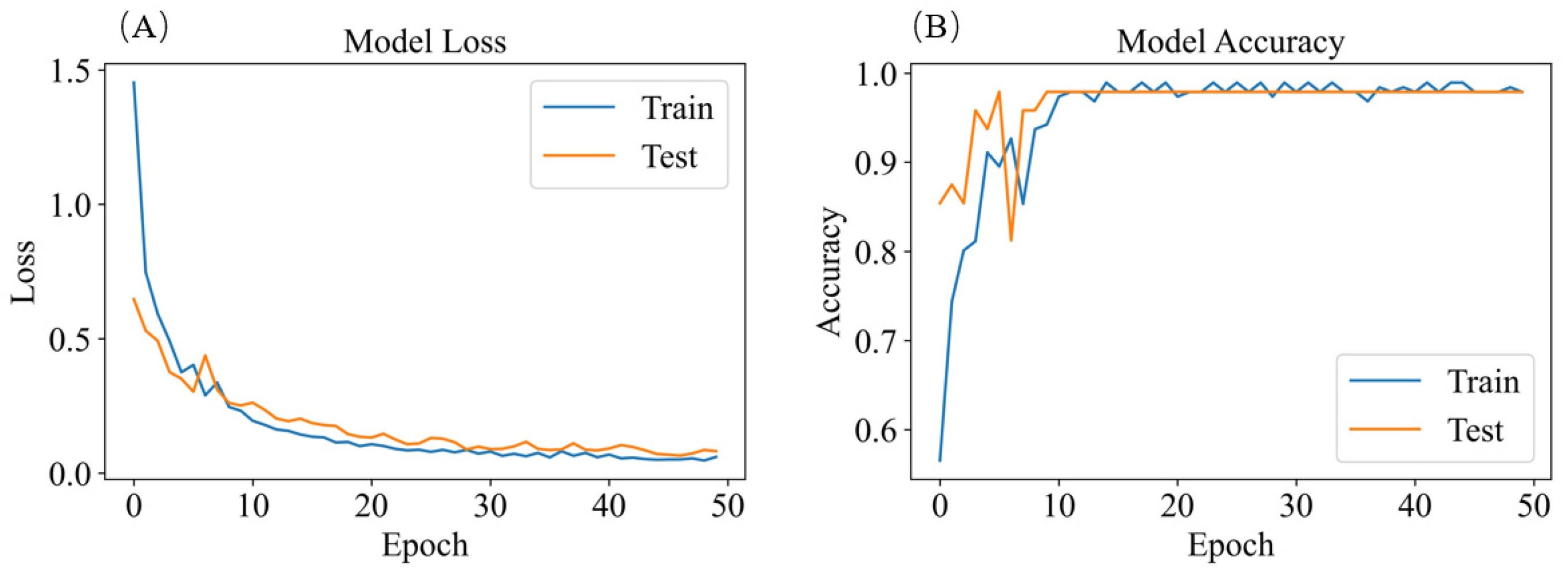
2. Materials and Methods
2.1. Laboratory Reagents
2.2. Synthesis Method of Au@Ag NRS
2.2.1. Synthesis of Au NRS
- Synthesis of Gold Seeds. In a constant-temperature water bath with magnetic stirring, 8 mL of 0.1 mol/L CTAB (Hexadecyl Trimethyl Ammonium Bromide) aqueous solution was added to a glass vial containing a magnetic stir bar. Under stirring at 640 r/min, 200 μL of 0.01 mol/L Chloroauric Acid (HAuCl4) aqueous solution was slowly added, resulting in a golden-yellow color. The stirring speed was then increased to 1200 r/min, and 480 μL of ice-cold 0.1 mol/L Sodium Borohydride (NaBH4) solution (stored at 0–4 °C) was added, turning the solution brownish yellow. After thorough mixing, the reaction mixture was statically incubated at 30 °C for 2 h in an oven, yielding a tea-brown gold seed solution.
- Preparation of Growth Solution. To 20 mL of 0.1 mol/L CTAB aqueous solution, the following reagents were sequentially added: ① 1 mL of 0.01 mol/L HAuCl4 aqueous solution, ② 250 μL of 0.01 mol/L Silver Nitrate (AgNO3) aqueous solution, ③ 34 μL of 38% Hydrochloric Acid (HCl) aqueous solution (shifting the color to khaki), ④ 160 μL of 0.1 mol/L Ascorbic Acid (AA) aqueous solution, ⑤ The mixture was stirred at 1200 r/min for 30 s, resulting in a colorless transparent growth solution.
- Growth of Au NRS. 10 μL of the gold seed solution was added to the growth solution and stirred at 700 r/min for 1 min. The mixture was then statically reacted at 30 °C for 12 h in an oven.
- Centrifugal Washing. After reaction completion, centrifugal washing was performed to remove excess CTAB and AgNO3 impurities: ① Centrifugation at 10,000 r/min for 10 min (with centrifuge tubes symmetrically placed), ② The supernatant was discarded, and the pellet was resuspended in 15 mL of ultrapure water, ③ The colloid was ultrasonicated for 5 min to ensure homogeneous dispersion, ④ This washing cycle was repeated multiple times with minimal solvent volume, ⑤ the colloid was concentrated to 5 mL and stored at 4 °C.
2.2.2. Synthesis of Au@Ag NRS
- Add 600 μL of 0.08 mol/L CTAC (Cetyltrimethylammonium Chloride) solution to a 10 mL centrifuge tube, followed by 200 μL of 0.01 mol/L Silver Nitrate (AgNO3) aqueous solution. Oscillate the mixture for homogenization and incubate at 30 °C for 10 min.
- Remove the mixture from the oven, add 500 μL of concentrated Au NRS solution and 2 mL of ultrapure water. Oscillate for homogenization and incubate at 30 °C for 5 min.
- Add 100 μL of 0.1 mol/L Ascorbic Acid (AA) solution to the mixture. Oscillate vigorously at 1000 r/min for 1 min, then react in a constant-temperature oscillating water bath at 30 °C for 4 h.
2.3. SERS-Based Detection Method for Crop Disease Spores
2.4. SERS Spectral Analysis of Crop Disease Spores
2.5. MLP Classification of Pathogenic Spores
3. Results and Discussion
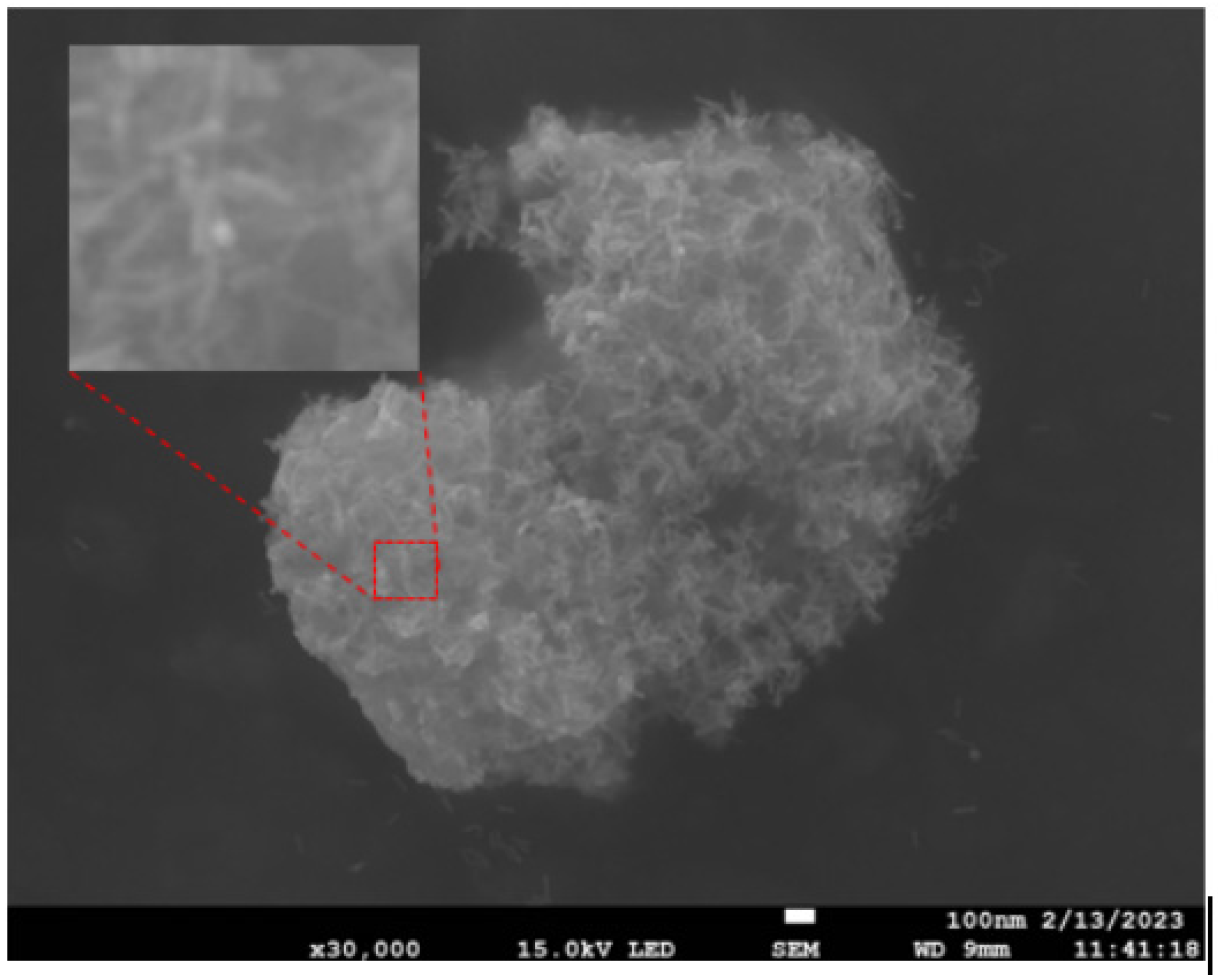
3.1. Analysis of Preparation Results for Au NRS and Au@Ag NRS
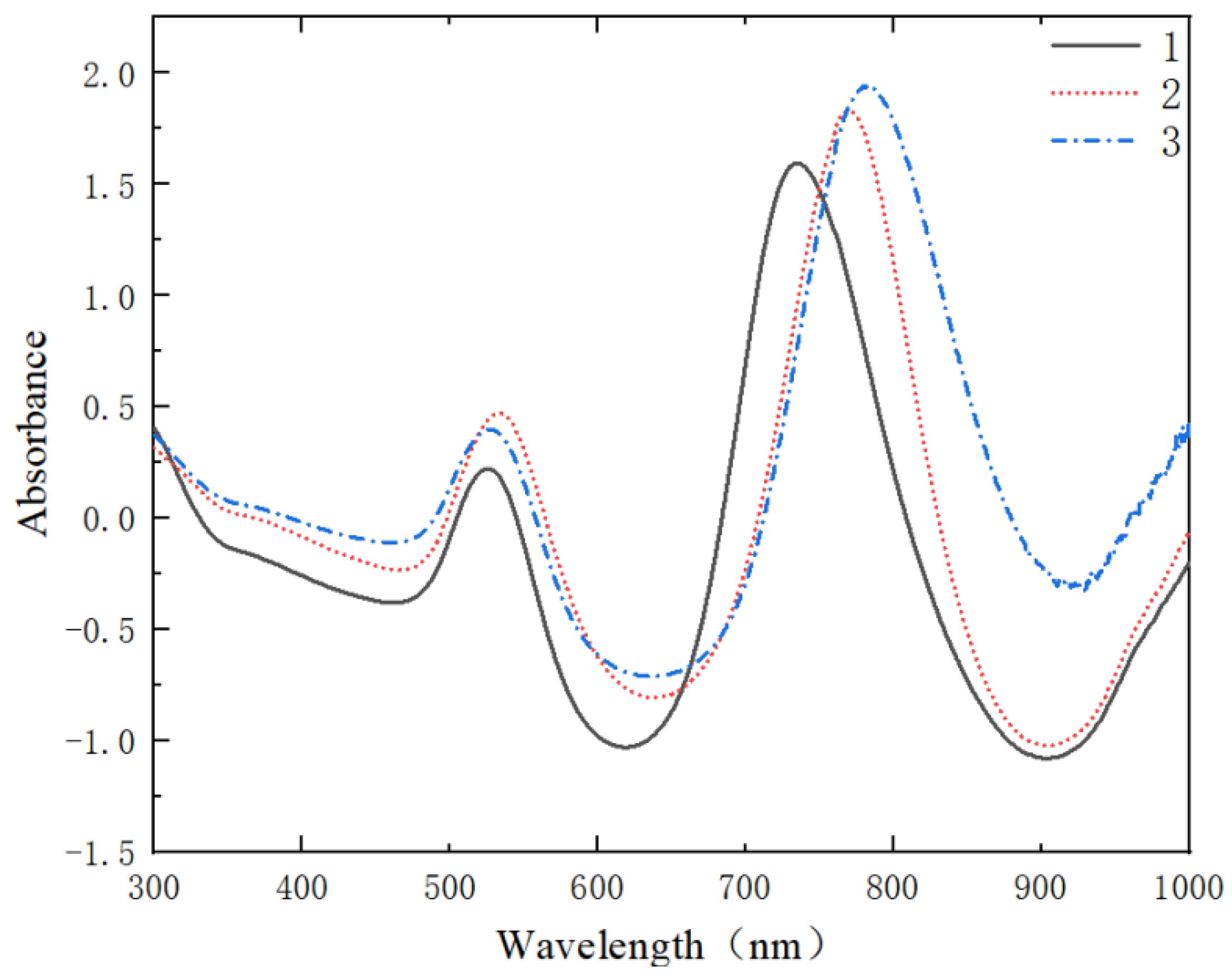
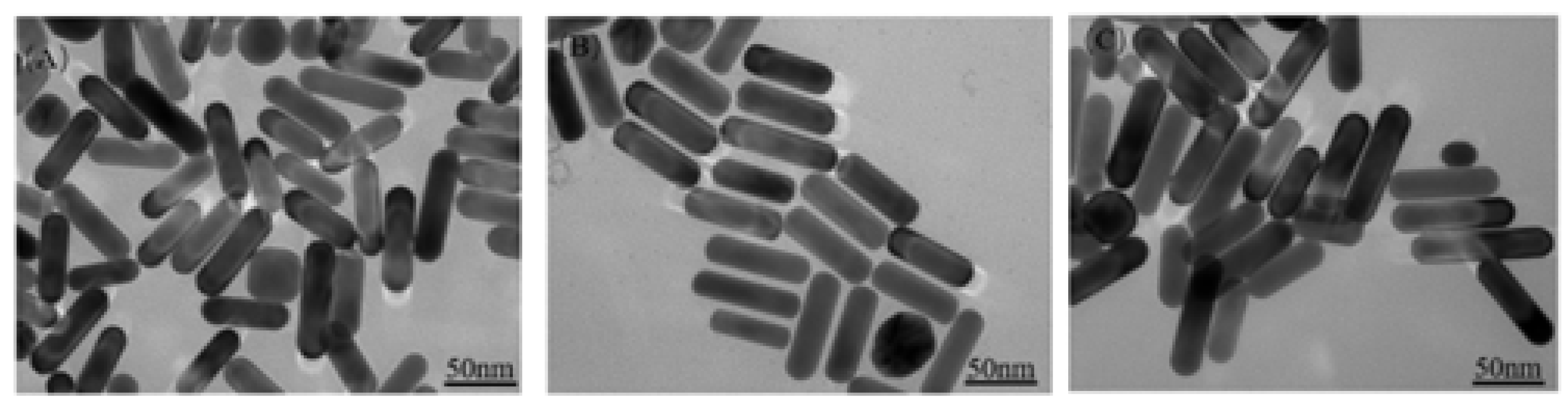
3.1.1. Effect of Different Concentrations on Au NRS Formation
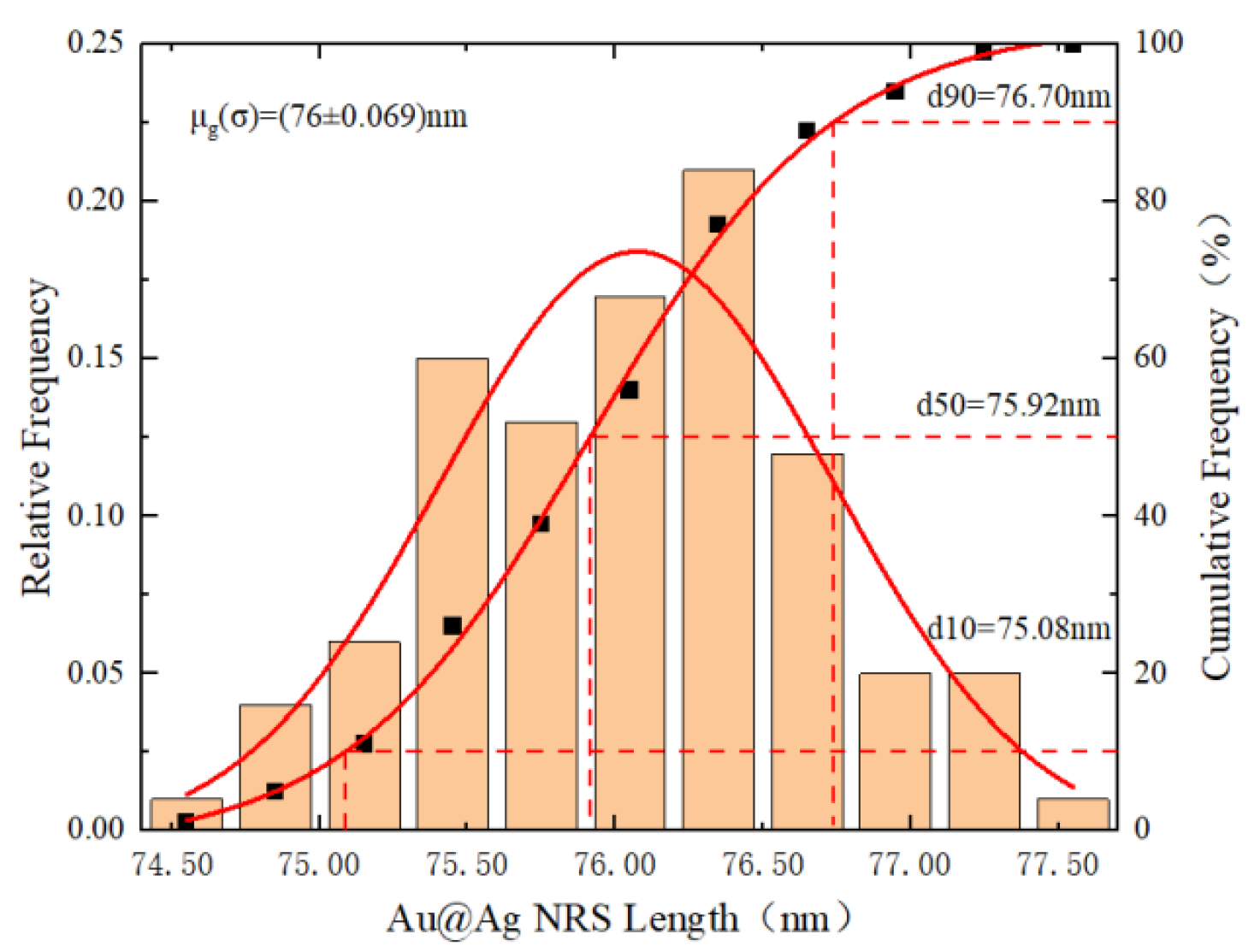
3.1.2. Synthesis Analysis of Au@Ag NRS
3.1.3. EDS Analysis of Au@Ag Core-Shell Structures
3.1.4. R6G Detection Using Au@Ag NRS
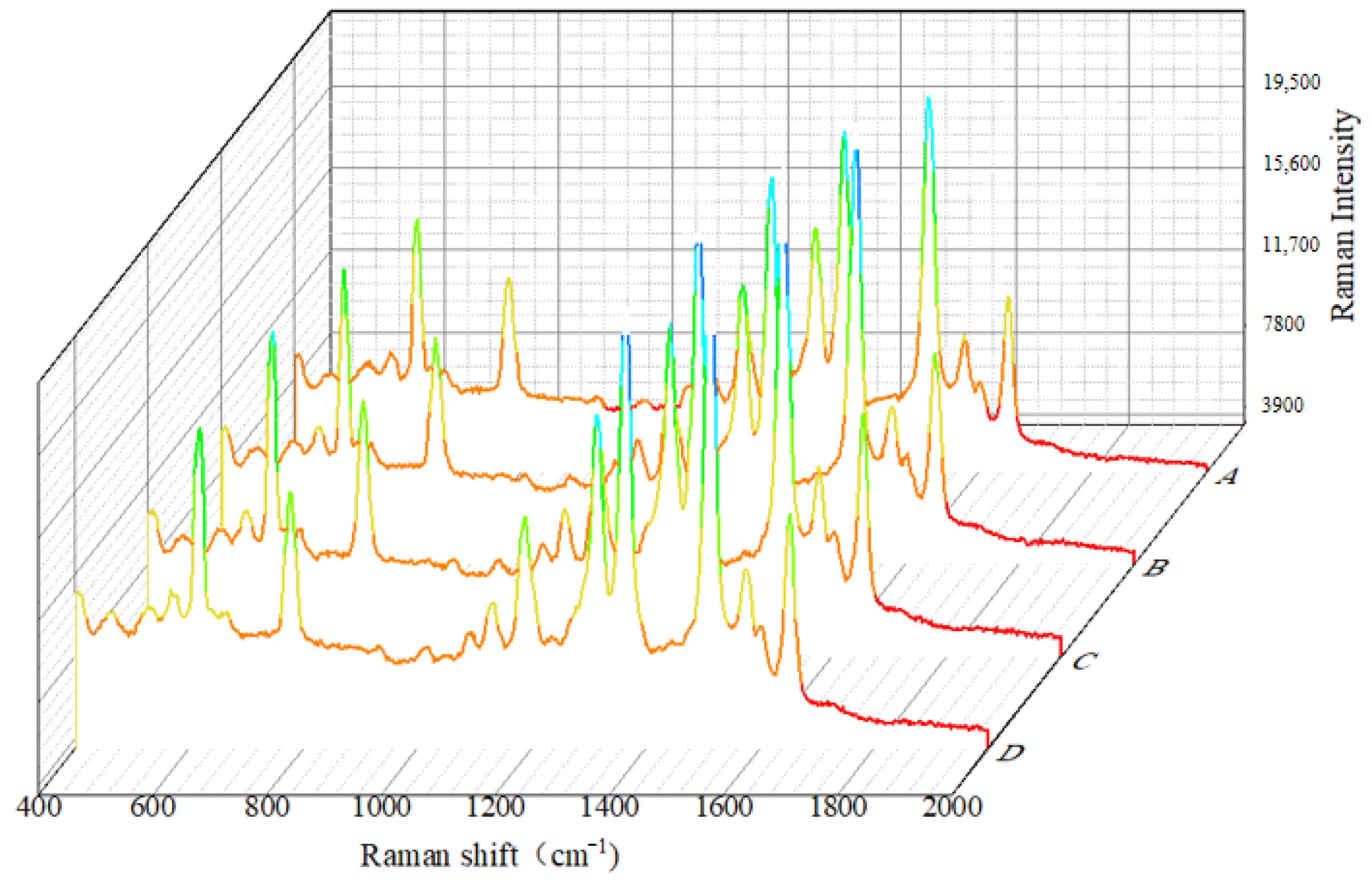
3.2. SERS Spectral Analysis Results of Crop Disease Spores
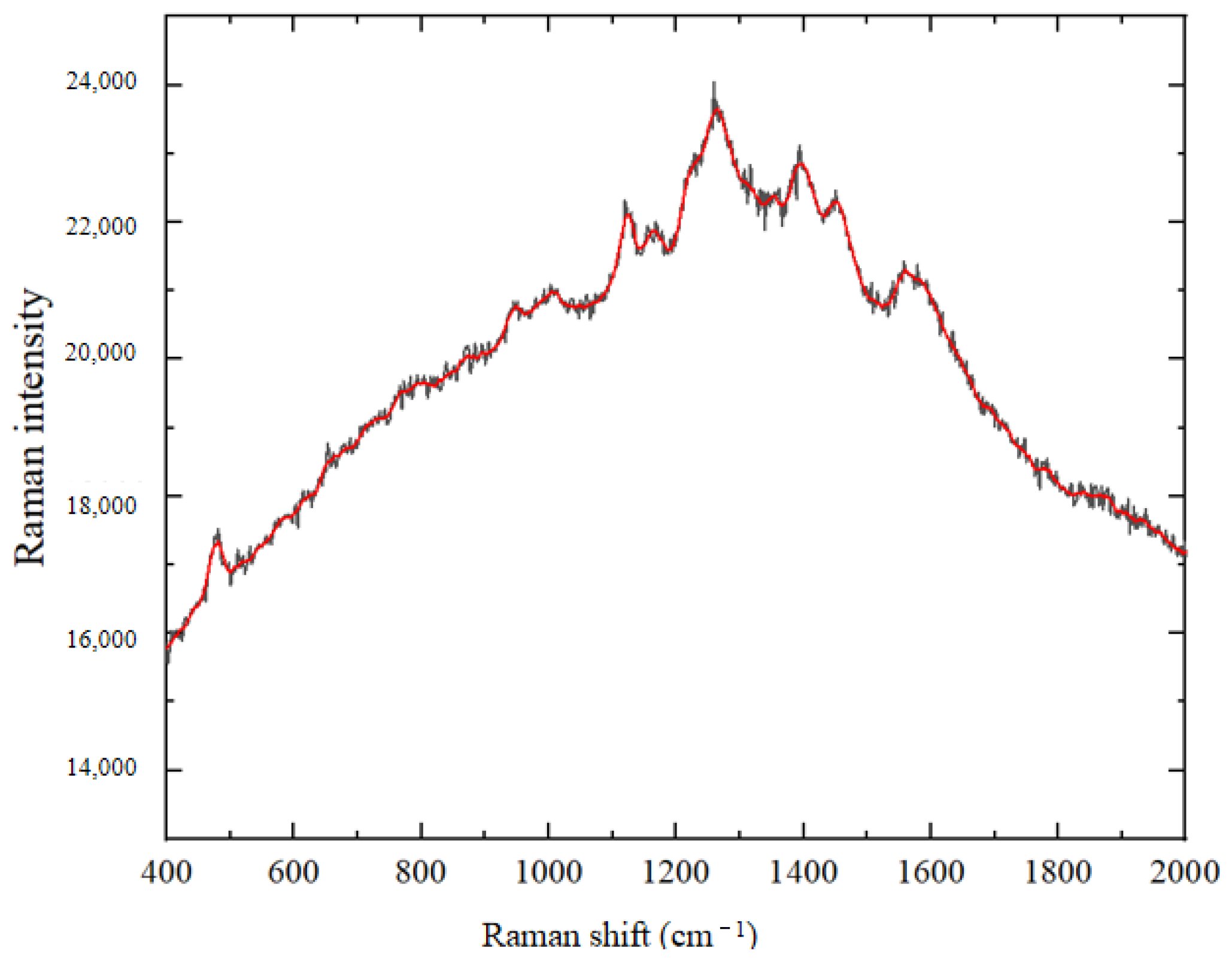
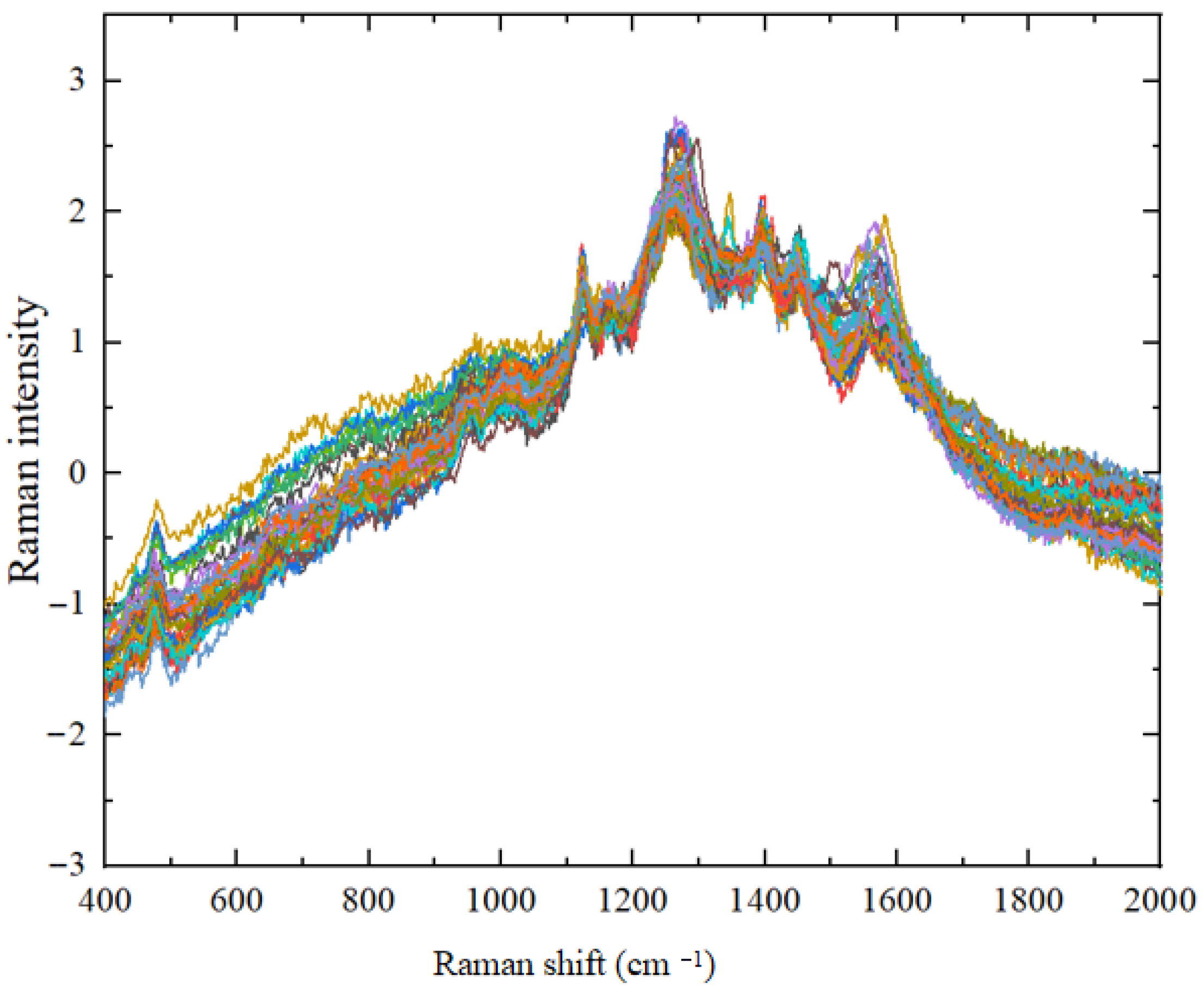
3.2.1. Spectral Smoothing
3.2.2. Spectral Standardization
3.2.3. Baseline Correction of Spectral Data for Crop Disease Spores
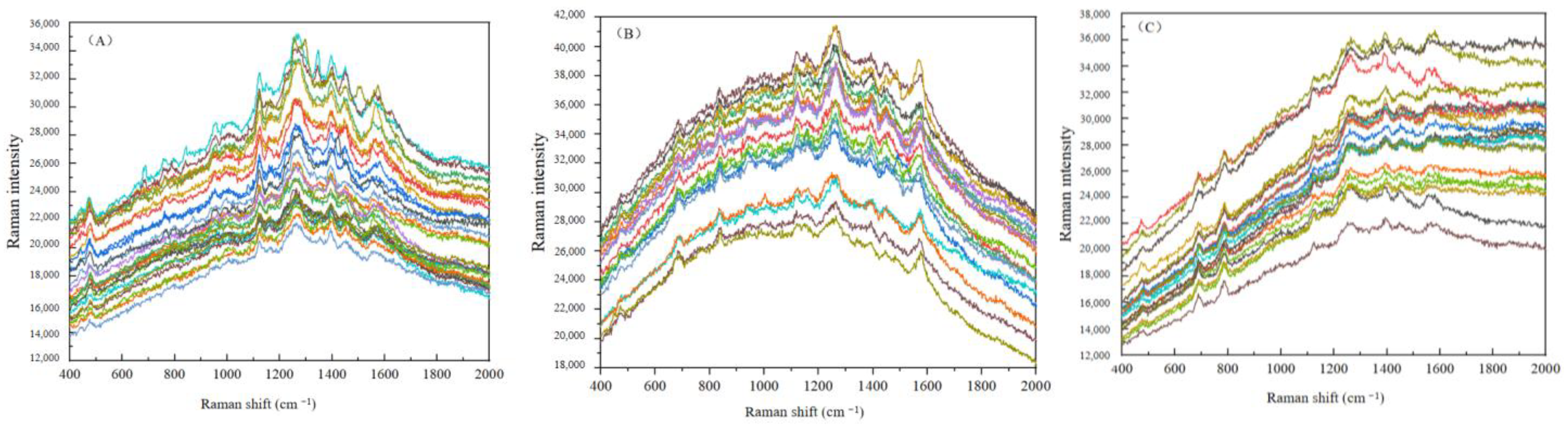
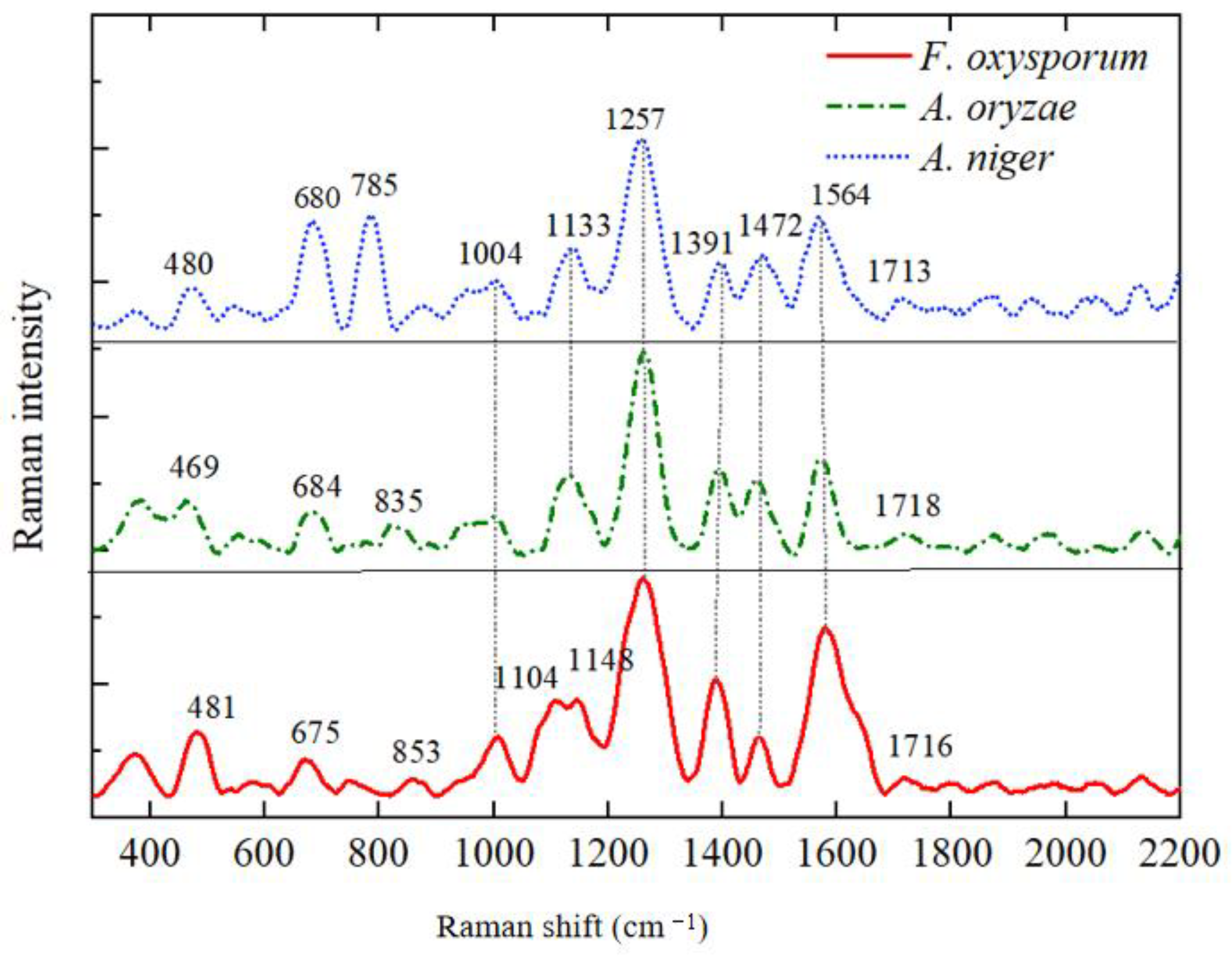
3.3. Analysis of SERS Fingerprint Information for Crop Diseases
3.4. Classification of SERS Spectra
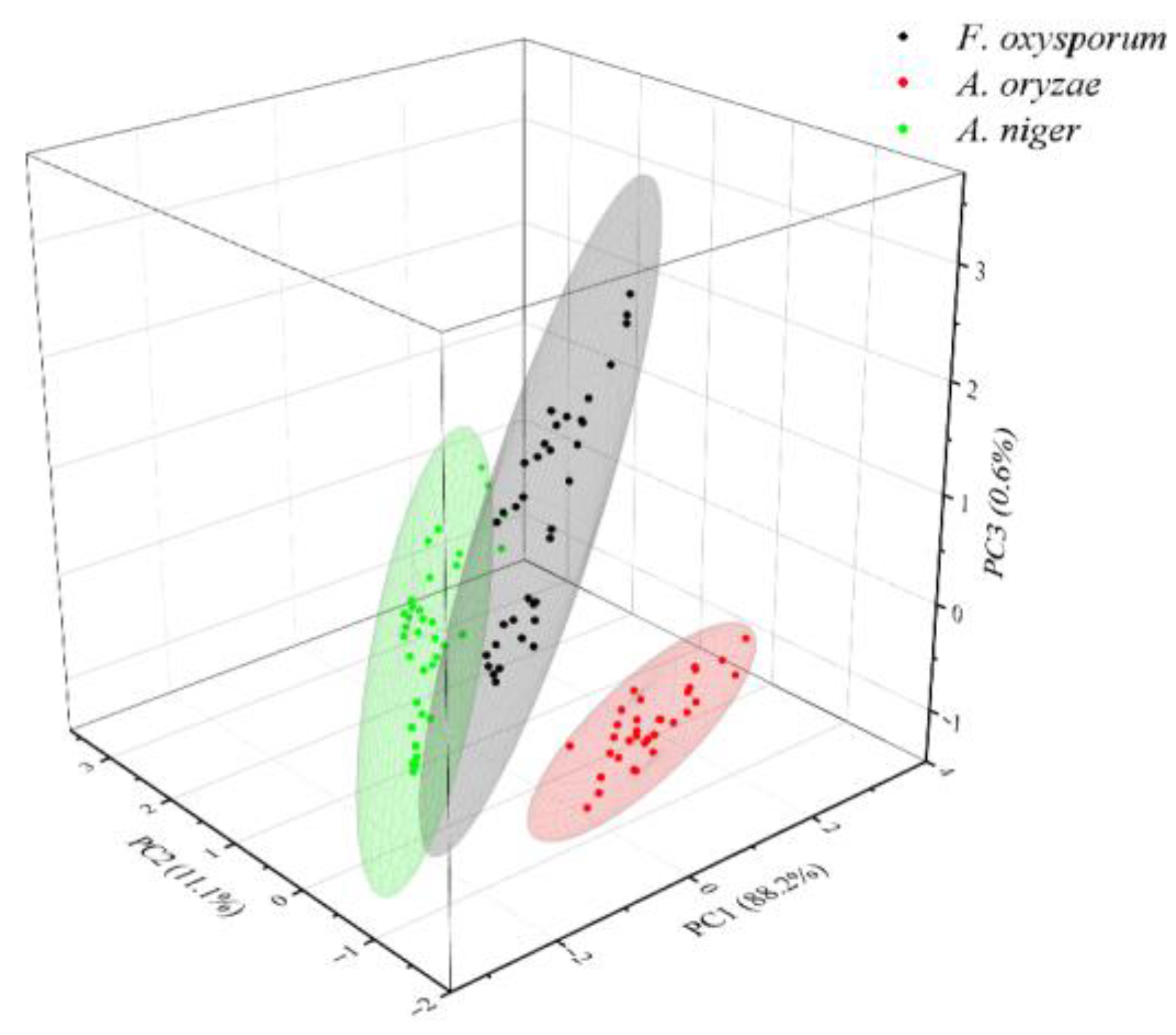
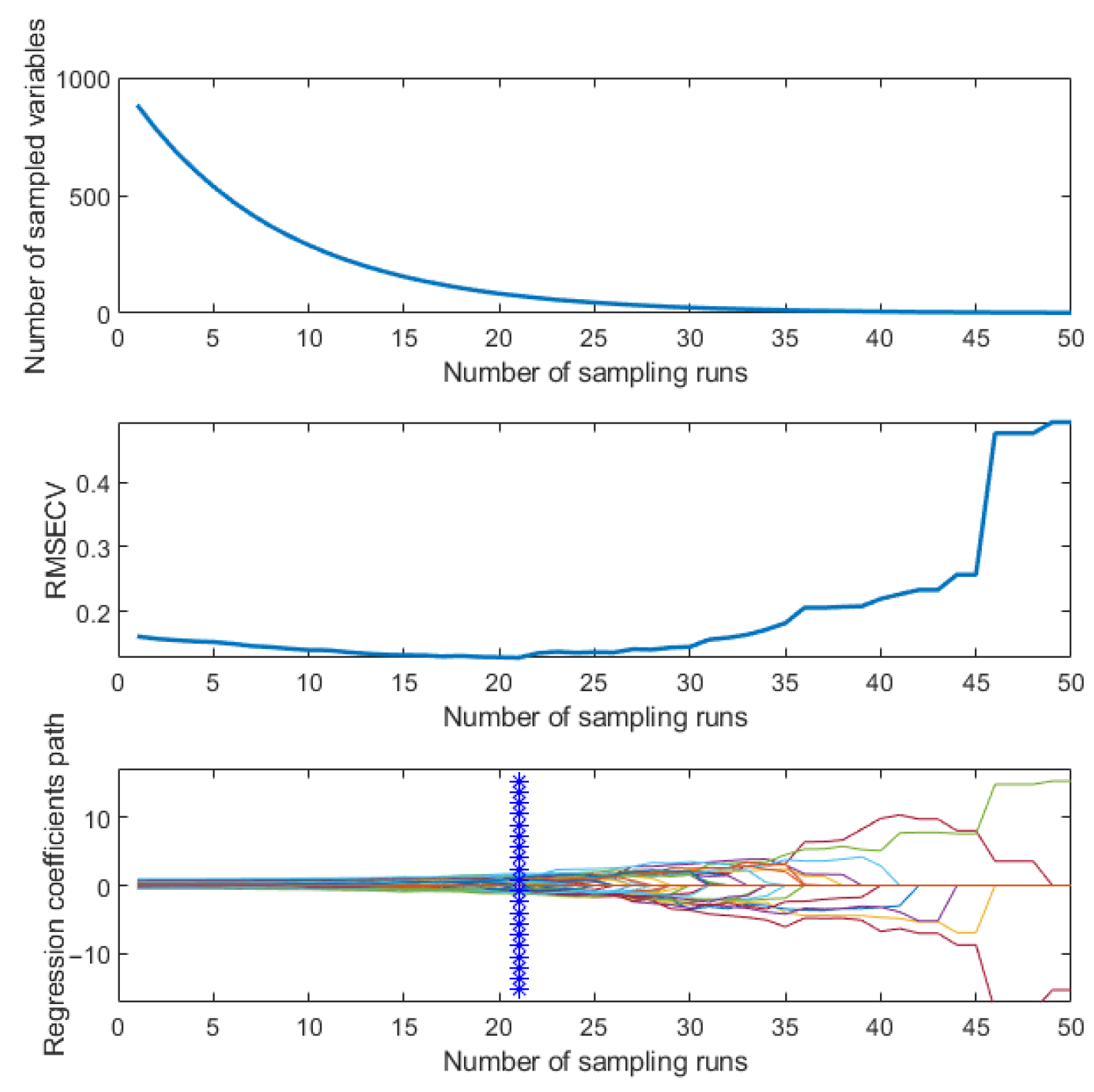
3.4.1. Dimensionality Reduction of SERS Spectra
3.4.2. SERS Spectrum Classification Model
4. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Savary, S.; Willocquet, L.; Pethybridge, S.J.; Esker, P.; McRoberts, N.; Nelson, A. The global burden of pathogens and pests on major food crops. Nat. Ecol. Evol. 2019, 3, 430–439. [Google Scholar] [CrossRef]
- Ngolong Ngea, G.L.; Qian, X.; Yang, Q.; Dhanasekaran, S.; Ianiri, G.; Ballester, A.-R.; Zhang, X.; Castoria, R.; Zhang, H. Securing fruit production: Opportunities from the elucidation of the molecular mechanisms of postharvest fungal infections. Compr. Rev. Food Sci. Food Saf. 2021, 20, 2508–2533. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Liu, D.; Yao, J. Aerosolization of fungal spores in indoor environments. Sci. Total Environ. 2022, 820, 153003. [Google Scholar] [CrossRef] [PubMed]
- Walsh, N.M.; Botts, M.R.; McDermott, A.J.; Ortiz, S.C.; Wüthrich, M.; Klein, B.; Hull, C.M. Infectious particle identity determines dissemination and disease outcome for the inhaled human fungal pathogen Cryptococcus. PLoS Pathog. 2019, 15, e1007777. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.; Lee, J. Beneficial bacteria and fungi in hydroponic systems: Types and characteristics of hydroponic food production methods. Sci. Hortic. 2015, 195, 206–215. [Google Scholar] [CrossRef]
- Ward, E. Plant pathogen diagnostics: Immunological and nucleic acid-based approaches. Ann. Appl. Biol. 2004, 145, 1–16. [Google Scholar] [CrossRef]
- Yang, N.; Ji, Y.; Wang, A.; Tang, J.; Liu, S.; Zhang, X.; Xu, L.; He, Y. An integrated nucleic acid detection method based on a microfluidic chip for collection and culture of rice false smut spores. Lab Chip 2022, 22, 4894–4904. [Google Scholar] [CrossRef]
- Lei, Y.; Yao, Z.; He, D. Automatic detection and counting of urediniospores of Puccinia striiformis f. sp. tritici using spore traps and image processing. Sci. Rep. 2018, 8, 13647. [Google Scholar] [CrossRef]
- Zhang, Y.; Li, J.; Tang, F.; Zhang, H.; Cui, Z.; Zhou, H. An Automatic Detector for Fungal Spores in Microscopic Images Based on Deep Learning. Appl. Eng. Agric. 2021, 37, 85–94. [Google Scholar] [CrossRef]
- Imbusch, F.; Liebe, S.; Erven, T.; Varrelmann, M. Dynamics of cercospora leaf spot disease determined by aerial spore dispersal in artificially inoculated sugar beet fields. Plant Pathol. 2021, 70, 853–861. [Google Scholar] [CrossRef]
- Munir, M.; Wang, H.; Dufault, N.S.; Anco, D.J. Early Detection of Airborne Inoculum of Nothopassalora personata in Spore Trap Samples from Peanut Fields Using Quantitative PCR. Plants 2020, 9, 1327. [Google Scholar] [CrossRef]
- Huayhongthong, S.; Khuntayaporn, P.; Thirapanmethee, K.; Wanapaisan, P.; Chomnawang, M.T. Raman spectroscopic analysis of food-borne microorganisms. LWT 2019, 114, 108419. [Google Scholar] [CrossRef]
- Breuch, R.; Klein, D.; Siefke, E.; Hebel, M.; Herbert, U.; Wickleder, C.; Kaul, P. Differentiation of meat-related microorganisms using paper-based surface-enhanced Raman spectroscopy combined with multivariate statistical analysis. Talanta 2020, 219, 121315. [Google Scholar] [CrossRef]
- Feng, J.; de la Fuente-Núñez, C.; Trimble, M.J.; Xu, J.; Hancock, R.E.; Lu, X. An in situ Raman spectroscopy-based microfluidic “lab-on-a-chip” platform for non-destructive and continuous characterization of Pseudomonas aeruginosa biofilms. Chem. Commun. 2015, 51, 8966–8969. [Google Scholar] [CrossRef]
- Gao, Y.; Pan, X.; Xu, S.; Liu, Z.; Wang, J.; Yu, K.; Wang, C.; Yuan, H.; Wu, S. Fluorescence-enhanced microfluidic sensor for highly sensitive in-situ detection of copper ions in lubricating oil. Mater. Des. 2020, 191, 108693. [Google Scholar] [CrossRef]
- Chen, T.; Sun, J.; Ma, T.; Li, T.; Liu, C.; Zhu, X.; Xue, N. Design and Analysis of Particulate Matter Air-Microfluidic Grading Chip Based on MEMS. Micromachines 2019, 10, 497. [Google Scholar] [CrossRef]
- Wang, P.; Yuan, S.; Yang, N.; Wang, A.; Fordjour, A.; Chen, S. The Collection Method for Crop Fungal Spores Based on an Efficient Microfluidic Device. Aerosol Air Qual. Res. 2020, 20, 72–79. [Google Scholar] [CrossRef]
- Yang, N.; Chen, C.; Li, T.; Li, Z.; Zou, L.; Zhang, R.; Mao, H. Portable Rice Disease Spores Capture and Detection Method Using Diffraction Fingerprints on Microfluidic Chip. Micromachines 2019, 10, 289. [Google Scholar] [CrossRef] [PubMed]
- Beeram, R.; Vepa, K.R.; Soma, V.R. Recent Trends in SERS-Based Plasmonic Sensors for Disease Diagnostics, Biomolecules Detection, and Machine Learning Techniques. Biosensors 2023, 13, 328. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Guo, B.; Wang, Y.; Hu, L.; Yang, N.; Mao, H. A Detection Method for Crop Fungal Spores Based on Microfluidic Separation Enrichment and AC Impedance Characteristics. J. Fungi 2022, 8, 1168. [Google Scholar] [CrossRef] [PubMed]
- Tian, F.; Bonnier, F.; Casey, A.; Shanahan, A.E.; Byrne, H.J. Surface enhanced Raman scattering with gold nanoparticles: Effect of particle shape. Anal. Methods 2014, 6, 9116–9123. [Google Scholar] [CrossRef]
- Zhou, X.; Liu, Q.; Shi, X.; Xu, C.; Li, B. Effect of aspect ratio on the chirality of gold nanorods prepared through conventional seed-mediated growth method. Anal. Chim. Acta 2021, 1152, 338277. [Google Scholar] [CrossRef] [PubMed]
- Fu, G.; Sun, D.-W.; Pu, H.; Wei, Q. Fabrication of gold nanorods for SERS detection of thiabendazole in apple. Talanta 2019, 195, 841–849. [Google Scholar] [CrossRef] [PubMed]
- Jiang, R.; Chen, H.; Shao, L.; Li, Q.; Wang, J. Unraveling the evolution and nature of the plasmons in (Au core)-(Ag shell) nanorods. Adv. Mater. 2012, 24, OP200–OP207. [Google Scholar] [CrossRef]
- Guo, Z.; Wang, M.; Barimah, A.O.; Chen, Q.; Li, H.; Shi, J.; El-Seedi, H.R.; Zou, X. Label-free surface enhanced Raman scattering spectroscopy for discrimination and detection of dominant apple spoilage fungus. Int. J. Food Microbiol. 2021, 338, 108990. [Google Scholar] [CrossRef]
- Guha, S.; Roy, S.; Banerjee, A. Fluorescent Au@Ag core-shell nanoparticles with controlled shell thickness and Hg(II) sensing. Langmuir 2011, 27, 13198–13205. [Google Scholar] [CrossRef]
- Guo, S.; Bocklitz, T.; Popp, J. Optimization of Raman-spectrum baseline correction in biological application. Analyst 2016, 141, 2396–2404. [Google Scholar] [CrossRef]
- Xu, Y.; Du, P.; Senger, R.; Robertson, J.; Pirkle, J.L. ISREA: An Efficient Peak-Preserving Baseline Correction Algorithm for Raman Spectra. Appl. Spectrosc. 2021, 75, 34–45. [Google Scholar] [CrossRef]
- Pinkus, A. Approximation theory of the MLP model in neural networks. Acta Numer. 1999, 8, 143–195. [Google Scholar] [CrossRef]
- Zheng, Y.; Zhong, X.; Li, Z.; Xia, Y. Successive, Seed-Mediated Growth for the Synthesis of Single-Crystal Gold Nanospheres with Uniform Diameters Controlled in the Range of 5–150 nm. Part. Part. Syst. Charact. 2014, 31, 266–273. [Google Scholar] [CrossRef]
- Okuno, Y.; Nishioka, K.; Kiya, A.; Nakashima, N.; Ishibashi, A.; Niidome, Y. Uniform and controllable preparation of Au–Ag core–shell nanorods using anisotropic silver shell formation on gold nanorods. Nanoscale 2010, 2, 1489–1493. [Google Scholar] [CrossRef]
- Garcia-Rico, E.; Alvarez-Puebla, R.A.; Guerrini, L. Direct surface-enhanced Raman scattering (SERS) spectroscopy of nucleic acids: From fundamental studies to real-life applications. Chem. Soc. Rev. 2018, 47, 4909–4923. [Google Scholar] [CrossRef]
- Lemma, T.; Wang, J.; Arstila, K.; Hytönen, V.P.; Toppari, J.J. Identifying yeasts using surface enhanced Raman spectroscopy. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2019, 218, 299–307. [Google Scholar] [CrossRef]
- Gelder, J.D.; Gussem, K.D.; Vandenabeele, P.; Moens, L. Reference database of Raman spectra of biological molecules. J. Raman Spectrosc. 2007, 38, 1133–1147. [Google Scholar] [CrossRef]
- Lin, Y.J.; Lin, H.K.; Lin, Y.H. Construction of Raman spectroscopic fingerprints for the detection of Fusarium wilt of banana in Taiwan. PLoS ONE 2020, 15, e0230330. [Google Scholar] [CrossRef]
- Alsamad, F.; Gobinet, C.; Vuiblet, V.; Jaisson, S.; Piot, O. Towards normalization selection of Raman data in the context of protein glycation: Application of validity indices to PCA processed spectra. Analyst 2020, 145, 2945–2957. [Google Scholar] [CrossRef]
- Strycker, B.D.; Han, Z.; Commer, B.; Shaw, B.D.; Sokolov, A.V.; Scully, M.O. CARS spectroscopy of Aspergillus nidulans spores. Sci. Rep. 2019, 9, 1789. [Google Scholar] [CrossRef] [PubMed]
- Xing, Z.; Du, C.; Shen, Y.; Ma, F.; Zhou, J. A method combining FTIR-ATR and Raman spectroscopy to determine soil organic matter: Improvement of prediction accuracy using competitive adaptive reweighted sampling (CARS). Comput. Electron. Agric. 2021, 191, 106549. [Google Scholar] [CrossRef]
- Chauhan, V.K.; Dahiya, K.; Sharma, A. Problem formulations and solvers in linear SVM: A review. Artif. Intell. Rev. 2019, 52, 803–855. [Google Scholar] [CrossRef]
- Karlik, B.; Olgaç, A.V. Performance Analysis of Various Activation Functions in Generalized MLP Architectures of Neural Networks. Int. J. Artif. Intell. Expert Syst. 2011, 1, 111–122. [Google Scholar]








| Reagent Name | CAS |
|---|---|
| Hexadecyl trimethyl ammonium Bromide (CTAB) | 57-09-0 |
| Hydrogen tetrachloroaurate (III) trihydrate | 16961-25-4 |
| Sodium borohydride | 16940-66-2 |
| Ascorbic acid | 50-81-7 |
| Silver nitrate | 7761-88-8 |
| Hydrochloric acid | 7647-01-0 |
| Sulfuric acid | 7664-93-9 |
| Cetyltrimethylammonium chloride (CTAC) | 112-02-7 |
| Group | /μL | CTAB/mL |
|---|---|---|
| 1 | 150 | 20 |
| 2 | 250 | 20 |
| 3 | 350 | 20 |
| Element | Line Type | k Factor | k Factor Type | Absorption Correction | Wt% | Wt% Sigma |
|---|---|---|---|---|---|---|
| C | K series | 2.760 | Theoretical | 1.00 | 30.30 | 0.26 |
| O | K series | 2.013 | Theoretical | 1.00 | 2.76 | 0.08 |
| Si | K series | 1.000 | Theoretical | 1.00 | 3.03 | 0.07 |
| Cr | K series | 1.117 | Theoretical | 1.00 | 0.24 | 0.03 |
| Fe | K series | 1.150 | Theoretical | 1.00 | 0.15 | 0.03 |
| Co | K series | 1.194 | Theoretical | 1.00 | 0.21 | 0.03 |
| Cu | K series | 1.261 | Theoretical | 1.00 | 18.46 | 0.17 |
| Ag | K series | 11.157 | Theoretical | 1.00 | 9.89 | 0.42 |
| Os | L series | 2.261 | Theoretical | 1.00 | 1.12 | 0.17 |
| Au | L series | 2.325 | Theoretical | 1.00 | 33.83 | 0.30 |
| Raman Shift | Fusarium oxysporum | Rice False Smut | Aspergillus niger | Spectral Assignment | References |
|---|---|---|---|---|---|
| 469–481 | 481 | 469 | 480 | Galactomannan, Chitin | [25] |
| 675–684 | 675 | 684 | 680 | Guanine, Thymine (Hydrogen-Bonded Ring) | [25,33] |
| 785 | - | - | 785 | L-Histidine | [34] |
| 835 | 853 | 835 | - | O-P-O Rotation in RNA | [33] |
| 1004–1008 | 1008 | 1004 | 1004 | Phenylalanine | [33] |
| 1104–1117 | 1104 | - | 1117 | Galactomannan | [25] |
| 1133–1148 | 1148 | 1133 | 1133 | C-O Ring Aromatic Amino Acids in Proteins | [12] |
| 1257–1260 | 1260 | 1260 | 1257 | Amide III (Random), Thymine | [33,35] |
| 1386–1391 | 1386 | 1391 | 1391 | D-Galactosamine | [34] |
| 1457–1472 | 1464 | 1457 | 1472 | L-Histidine | [34] |
| 1564–1580 | 1580 | 1574 | 1564 | Adenine, Guanine (Ring Stretching Vibration) | [12,35] |
| 1713–1718 | 1716 | 1718 | 1713 | L-Arginine in Proteins | [34] |
| Number | Combination | TP | TN | FP | FN | Training Set Accuracy (%) | Test Set Accuracy (%) |
|---|---|---|---|---|---|---|---|
| 1 | SVM | 45 | 17 | 5 | 5 | 86.65 | 86.32 |
| 2 | MPL | 46 | 17 | 5 | 4 | 88.46 | 87.61 |
| 3 | PCA-SVM | 49 | 20 | 2 | 1 | 97.25 | 95.24 |
| 4 | PCA-MPL | 49 | 21 | 1 | 1 | 97.34 | 96.55 |
| 5 | SCARS-SVM | 49 | 20 | 2 | 1 | 97.37 | 95.83 |
| 6 | SCARS-MLP | 50 | 21 | 1 | 0 | 98.9 | 97.92 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, Y.; Guo, J.; Bian, F.; Li, Z.; Guo, C.; Zheng, J.; Zhang, X. Crop Disease Spore Detection Method Based on Au@Ag NRS. Agriculture 2025, 15, 2076. https://doi.org/10.3390/agriculture15192076
Zhang Y, Guo J, Bian F, Li Z, Guo C, Zheng J, Zhang X. Crop Disease Spore Detection Method Based on Au@Ag NRS. Agriculture. 2025; 15(19):2076. https://doi.org/10.3390/agriculture15192076
Chicago/Turabian StyleZhang, Yixue, Jili Guo, Fei Bian, Zhaowei Li, Chuandong Guo, Jialiang Zheng, and Xiaodong Zhang. 2025. "Crop Disease Spore Detection Method Based on Au@Ag NRS" Agriculture 15, no. 19: 2076. https://doi.org/10.3390/agriculture15192076
APA StyleZhang, Y., Guo, J., Bian, F., Li, Z., Guo, C., Zheng, J., & Zhang, X. (2025). Crop Disease Spore Detection Method Based on Au@Ag NRS. Agriculture, 15(19), 2076. https://doi.org/10.3390/agriculture15192076





