Identification of Candidate Variants Associated with Milk Production, Health and Reproductive Traits for Holstein Cows in Southern China
Abstract
1. Introduction
2. Materials and Methods
2.1. Animals and Phenotypes
2.2. Deregressed Estimated Breeding Values
2.3. Genotyping and Quality Control
2.4. Estimation of Population Structure and Linkage Disequilibrium
2.5. GWAS Analysis
2.6. Functional Annotation for GWAS Signals
2.7. Haplotype Association Analysis
2.8. Fine-Mapping Analysis
3. Results
3.1. Descriptive Statistics
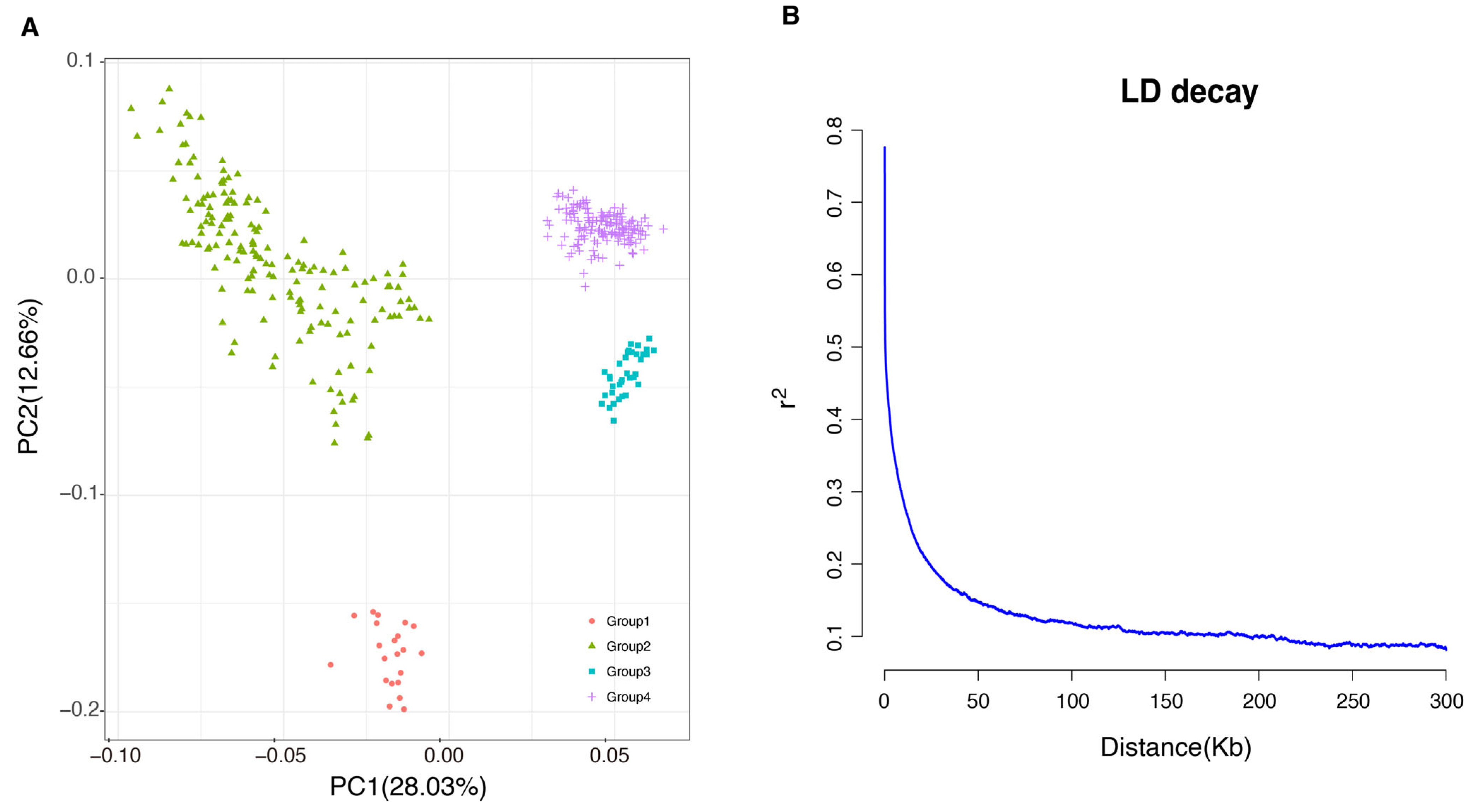
3.2. Population Stratification and LD Decay Analysis
3.3. GWAS for Milk Production Traits
3.4. GWAS for Health Traits
3.5. GWAS for Reproductive Traits
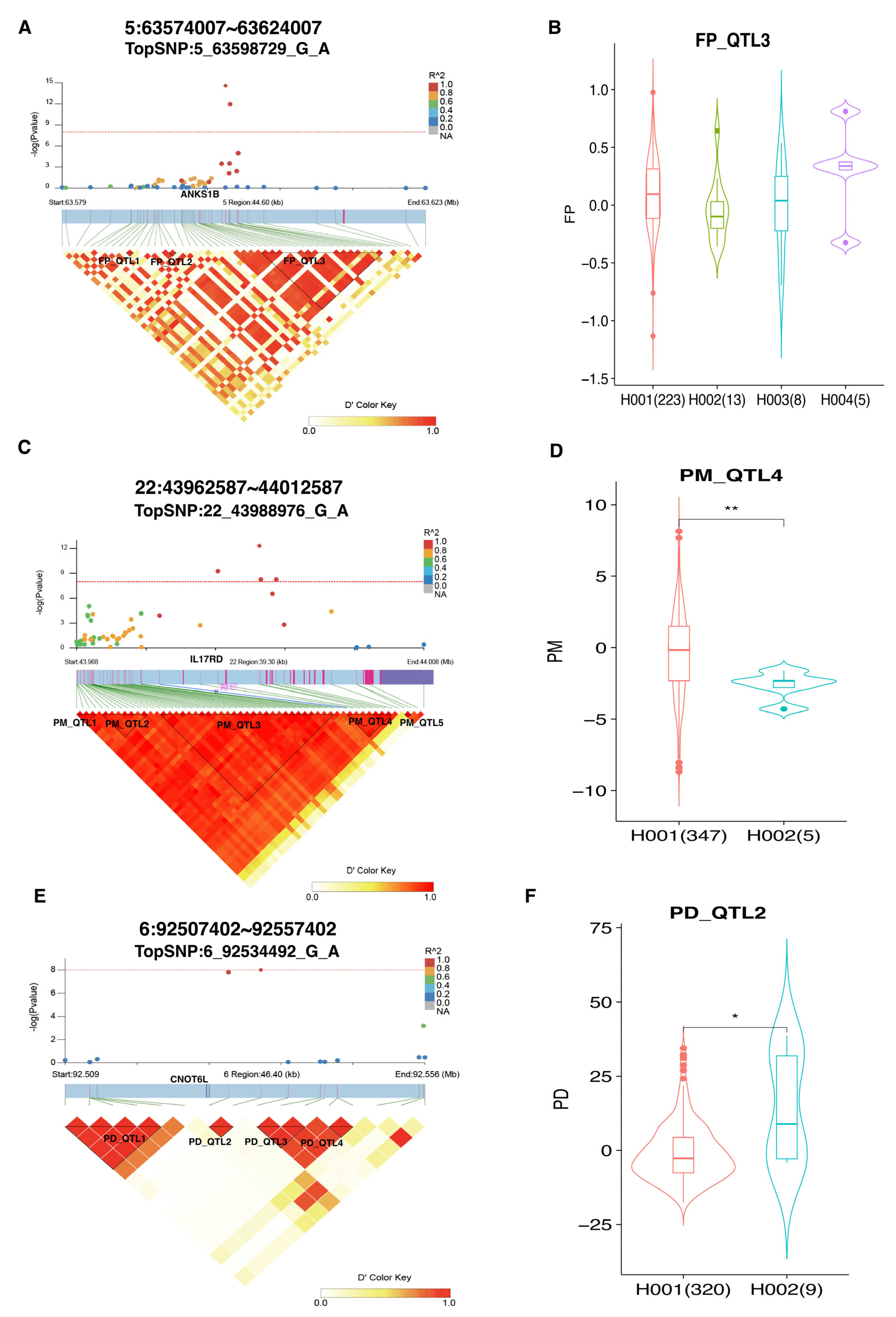
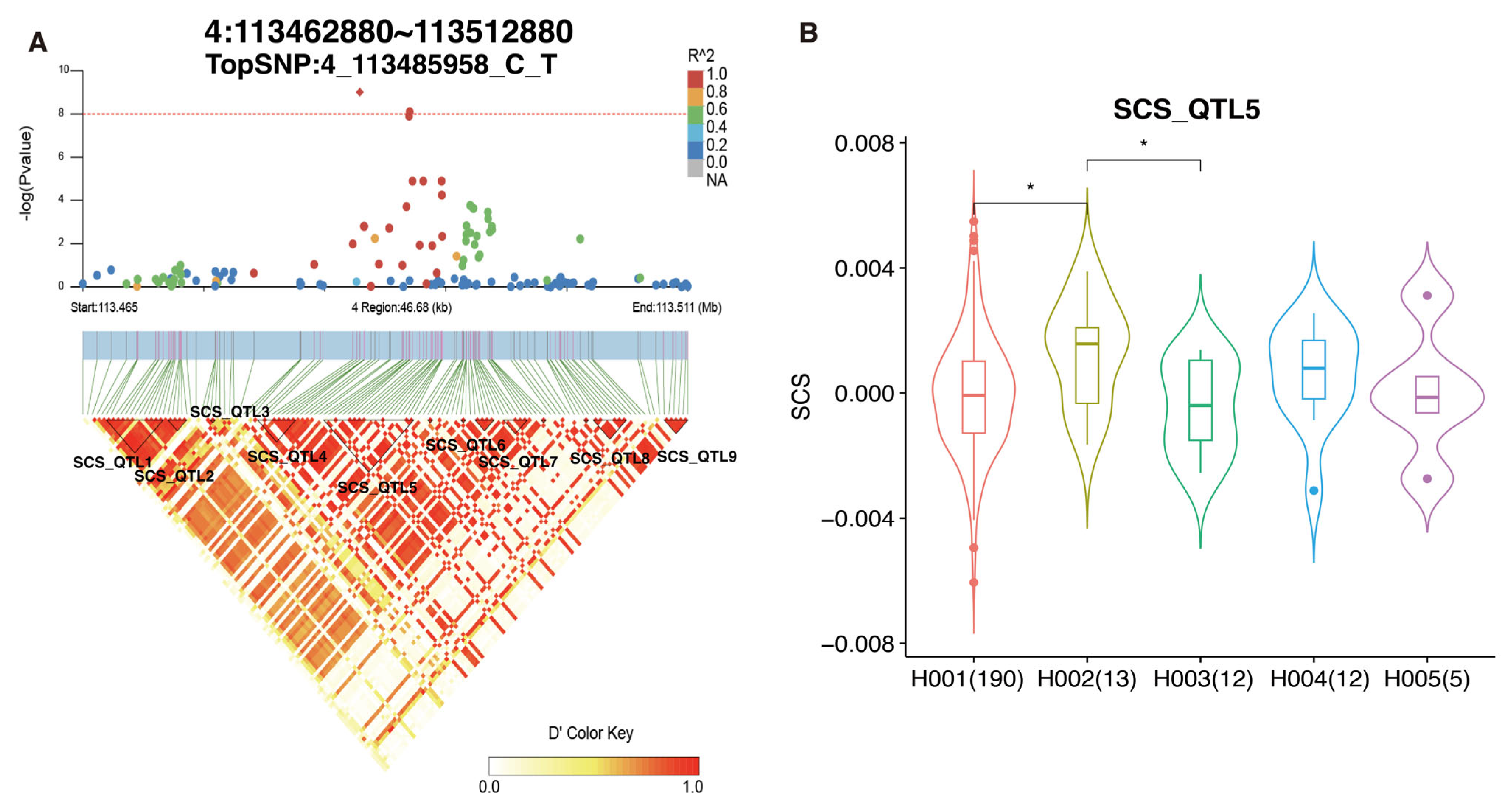
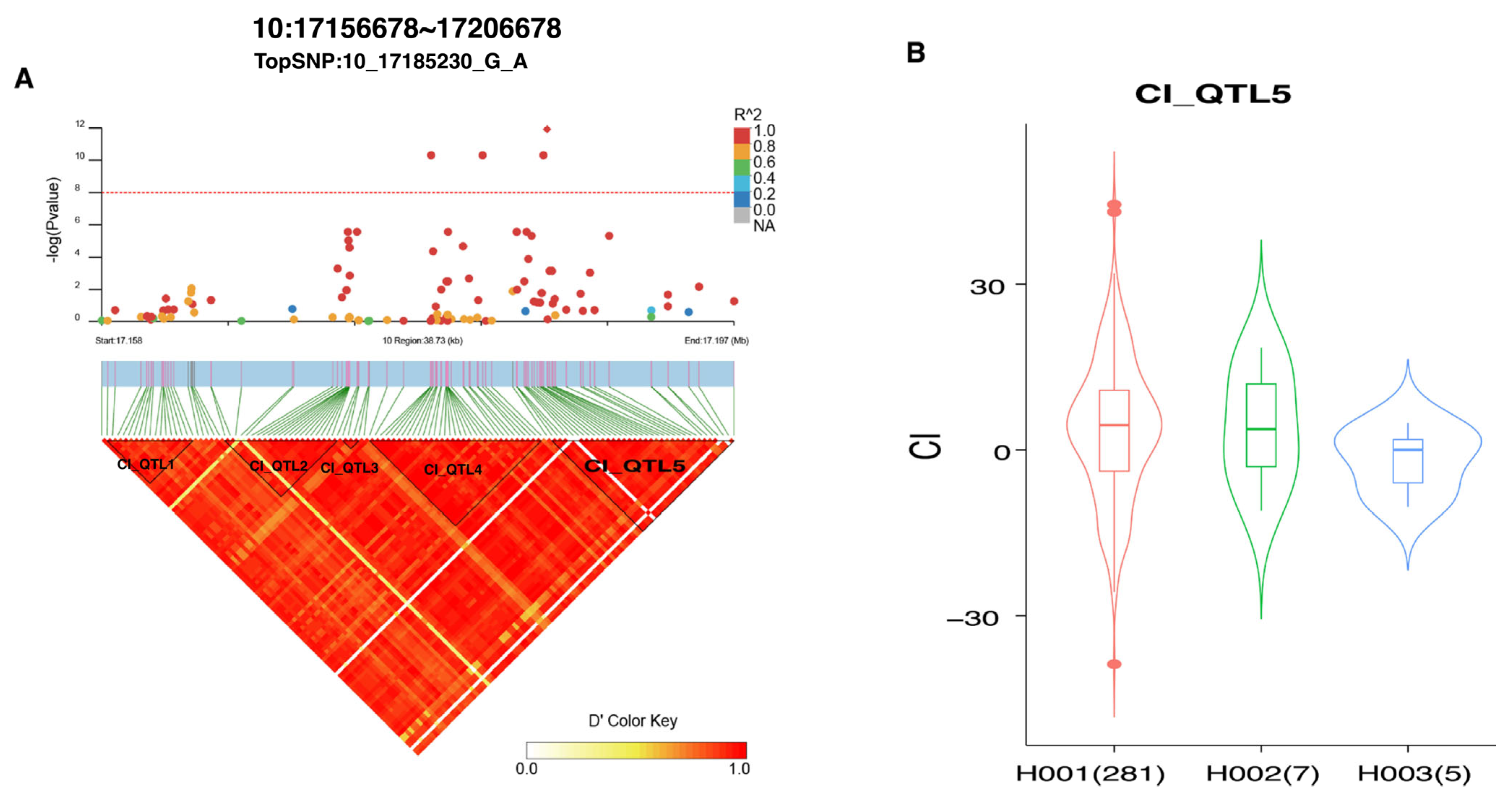
3.6. Fine-Mapping Analyses
4. Discussion
4.1. Milk Production Traits
4.2. Health Trait
4.3. Reproductive Trait
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| GWAS | Genome-wide association studies |
| QTL | quantitative trait loci |
| LD | linkage disequilibrium |
| PPC | posterior probabilities of causality |
| MY305 | 305-day milk yield |
| FP | fat percentage |
| PP | protein percentage |
| PM | peak milk yield |
| PD | days to peak milk yield |
| CI | calving interval |
| SCS | somatic cell score |
| EBV | estimated breeding value |
| dEBV | deregressed estimated breeding value |
| HYS | herd–year–season |
| CY | calving year |
| CS | calving season |
| GO | Gene Ontology |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
| GATK | Genome Analysis Toolkit |
| QC | quality control |
| MAF | minor allele frequency |
| HWE | Hardy–Weinberg equilibrium |
| BTA | Bos taurus chromosomes |
| PCA | principal component analysis |
References
- Li, S.; Ge, F.; Chen, L.; Liu, Y.; Chen, Y.; Ma, Y. Genome-Wide Association Analysis of Body Conformation Traits in Chinese Holstein Cattle. BMC Genom. 2024, 25, 1174. [Google Scholar] [CrossRef] [PubMed]
- Oliveira, C.P.; de Sousa, F.C.; da Silva, A.L.; Schultz, É.B.; Valderrama Londoño, R.I.; de Souza, P.A.R. Heat Stress in Dairy Cows: Impacts, Identification, and Mitigation Strategies-A Review. Animals 2025, 15, 249. [Google Scholar] [CrossRef] [PubMed]
- Summer, A.; Lora, I.; Formaggioni, P.; Gottardo, F. Impact of Heat Stress on Milk and Meat Production. Anim. Front. 2019, 9, 39–46. [Google Scholar] [CrossRef] [PubMed]
- Lemal, P.; Schroyen, M.; Gengler, N. Genetic Parameters and Relevance for Heat Stress Assessment in Dairy Cattle of 2 Udder Health Traits: Somatic Cell Score and Differential Somatic Cell Count. J. Dairy Sci. 2025, 108, 9930–9945. [Google Scholar] [CrossRef]
- Wolfenson, D.; Roth, Z. Impact of Heat Stress on Cow Reproduction and Fertility. Anim. Front. 2019, 9, 32–38. [Google Scholar] [CrossRef]
- Rejeb, M.; Sadraoui, R.; Najar, T.; M’rad, M.B. A Complex Interrelationship between Rectal Temperature and Dairy Cows’ Performance under Heat Stress Conditions. Open J. Anim. Sci. 2016, 6, 24–30. [Google Scholar] [CrossRef]
- Becker, C.A.; Collier, R.J.; Stone, A.E. Invited Review: Physiological and Behavioral Effects of Heat Stress in Dairy Cows. J. Dairy Sci. 2020, 103, 6751–6770. [Google Scholar] [CrossRef]
- Bernabucci, U.; Lacetera, N.; Baumgard, L.H.; Rhoads, R.P.; Ronchi, B.; Nardone, A. Metabolic and Hormonal Acclimation to Heat Stress in Domesticated Ruminants. Anim. Int. J. Anim. Biosci. 2010, 4, 1167–1183. [Google Scholar] [CrossRef]
- Schneider, H.; Segelke, D.; Tetens, J.; Thaller, G.; Bennewitz, J. A Genomic Assessment of the Correlation Between Milk Production Traits and Claw and Udder Health Traits in Holstein Dairy Cattle. J. Dairy Sci. 2023, 106, 1190–1205. [Google Scholar] [CrossRef]
- Teng, J.; Wang, D.; Zhao, C.; Zhang, X.; Chen, Z.; Liu, J.; Sun, D.; Tang, H.; Wang, W.; Li, J.; et al. Longitudinal Genome-Wide Association Studies of Milk Production Traits in Holstein Cattle Using Whole-Genome Sequence Data Imputed from Medium-Density Chip Data. J. Dairy Sci. 2023, 106, 2535–2550. [Google Scholar] [CrossRef]
- Križanac, A.-M.; Reimer, C.; Heise, J.; Liu, Z.; Pryce, J.E.; Bennewitz, J.; Thaller, G.; Falker-Gieske, C.; Tetens, J. Sequence-Based GWAS in 180,000 German Holstein Cattle Reveals New Candidate Variants for Milk Production Traits. Genet. Sel. Evol. GSE 2025, 57, 3. [Google Scholar] [CrossRef]
- Nguyen, T.T.T.; Bowman, P.J.; Haile-Mariam, M.; Nieuwhof, G.J.; Hayes, B.J.; Pryce, J.E. Short Communication: Implementation of a Breeding Value for Heat Tolerance in Australian Dairy Cattle. J. Dairy Sci. 2017, 100, 7362–7367. [Google Scholar] [CrossRef] [PubMed]
- Manzanilla-Pech, C.I.V.; L Vendahl, P.; Mansan Gordo, D.; Difford, G.F.; Pryce, J.E.; Schenkel, F.; Wegmann, S.; Miglior, F.; Chud, T.C.; Moate, P.J.; et al. Breeding for Reduced Methane Emission and Feed-Efficient Holstein Cows: An International Response. J. Dairy Sci. 2021, 104, 8983–9001. [Google Scholar] [CrossRef] [PubMed]
- Ribeiro, G.; Baldi, F.; Cesar, A.S.M.; Alexandre, P.A.; Peripolli, E.; Ferraz, J.B.S.; Fukumasu, H. Detection of Potential Functional Variants Based on Systems-Biology: The Case of Feed Efficiency in Beef Cattle. BMC Genom. 2022, 23, 774. [Google Scholar] [CrossRef] [PubMed]
- Civelek, M.; Lusis, A.J. Systems Genetics Approaches to Understand Complex Traits. Nat. Rev. Genet. 2014, 15, 34–48. [Google Scholar] [CrossRef]
- van den Berg, I.; Ho, P.N.; Nguyen, T.V.; Haile-Mariam, M.; MacLeod, I.M.; Beatson, P.R.; O’Connor, E.; Pryce, J.E. GWAS and Genomic Prediction of Milk Urea Nitrogen in Australian and New Zealand Dairy Cattle. Genet. Sel. Evol. GSE 2022, 54, 15. [Google Scholar] [CrossRef]
- Buss, C.E.; Afonso, J.; de Oliveira, P.S.N.; Petrini, J.; Tizioto, P.C.; Cesar, A.S.M.; Gustani-Buss, E.C.; Cardoso, T.F.; Rovadoski, G.A.; da Silva Diniz, W.J.; et al. Bivariate GWAS Reveals Pleiotropic Regions Among Feed Efficiency and Beef Quality-Related Traits in Nelore Cattle. Mamm. Genome 2023, 34, 90–103. [Google Scholar] [CrossRef]
- Tang, L.; Swedlund, B.; Dupont, S.; Harland, C.; Costa Monteiro Moreira, G.; Durkin, K.; Artesi, M.; Mullaart, E.; Sartelet, A.; Karim, L.; et al. GWAS Reveals Determinants of Mobilization Rate and Dynamics of an Active Endogenous Retrovirus of Cattle. Nat. Commun. 2024, 15, 2154. [Google Scholar] [CrossRef]
- Liu, J.J.; Liang, A.X.; Campanile, G.; Plastow, G.; Zhang, C.; Wang, Z.; Salzano, A.; Gasparrini, B.; Cassandro, M.; Yang, L.G. Genome-Wide Association Studies to Identify Quantitative Trait Loci Affecting Milk Production Traits in Water Buffalo. J. Dairy Sci. 2018, 101, 433–444. [Google Scholar] [CrossRef]
- El Nagar, A.G.; Salem, M.M.I.; Amin, A.M.S.; Khalil, M.H.; Ashour, A.F.; Hegazy, M.M.; Abdel-Shafy, H. A Single-Step Genome-Wide Association Study for Semen Traits of Egyptian Buffalo Bulls. Animals 2023, 13, 3758. [Google Scholar] [CrossRef]
- Deng, T.; Ma, X.; Duan, A.; Lu, X.; Abdel-Shafy, H. Genome-Wide Copy Number Variant Analysis Reveals Candidate Genes Associated with Milk Production Traits in Water Buffalo (Bubalus bubalis). J. Dairy Sci. 2024, 107, 7022–7037. [Google Scholar] [CrossRef]
- Hammons, V.; Ribeiro, L.; Munyard, K.; Sadeghi, R.; Miller, D.; Antczak, D.; Brooks, S.A. GWAS Identifies a Region Containing the SALL1 Gene in Variation of Pigmentation Intensity Within the Chestnut Coat Color of Horses. J. Hered. 2021, 112, 443–446. [Google Scholar] [CrossRef]
- Dias De Castro, L.L.; Oliveira Júnior, G.A.; Perez, B.C.; Carvalho, M.E.; De Souza Ramos, E.A.; Ferraz, J.B.S.; Molento, M.B. Genome-Wide Association Study in Thoroughbred Horses Naturally Infected with Cyathostomins. Anim. Biotechnol. 2023, 34, 2467–2479. [Google Scholar] [CrossRef] [PubMed]
- Pozharskiy, A.; Abdrakhmanova, A.; Beishova, I.; Shamshidin, A.; Nametov, A.; Ulyanova, T.; Bekova, G.; Kikebayev, N.; Kovalchuk, A.; Ulyanov, V.; et al. Genetic Structure and Genome-Wide Association Study of the Traditional Kazakh Horses. Anim. Int. J. Anim. Biosci. 2023, 17, 100926. [Google Scholar] [CrossRef] [PubMed]
- Jiang, J.; Cao, Y.; Shan, H.; Wu, J.; Song, X.; Jiang, Y. The GWAS Analysis of Body Size and Population Verification of Related SNPs in Hu Sheep. Front. Genet. 2021, 12, 642552. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Li, J.; Wang, H.; Zhang, R.; An, X.; Yuan, C.; Guo, T.; Yue, Y. Genomic Selection for Live Weight in the 14th Month in Alpine Merino Sheep Combining GWAS Information. Animals 2023, 13, 3516. [Google Scholar] [CrossRef]
- Liu, D.; Li, X.; Wang, L.; Pei, Q.; Zhao, J.; Sun, D.; Ren, Q.; Tian, D.; Han, B.; Jiang, H.; et al. Genome-Wide Association Studies of Body Size Traits in Tibetan Sheep. BMC Genom. 2024, 25, 739. [Google Scholar] [CrossRef]
- Mahmoudi, P.; Rashidi, A.; Nazari-Ghadikolaei, A.; Rostamzadeh, J.; Razmkabir, M.; Huson, H.J. Genome-Wide Association Study Reveals Novel Candidate Genes for Litter Size in Markhoz Goats. Front. Vet. Sci. 2022, 9, 1045589. [Google Scholar] [CrossRef]
- Selionova, M.; Aibazov, M.; Sermyagin, A.; Belous, A.; Deniskova, T.; Mamontova, T.; Zharkova, E.; Zinovieva, N. Genome-Wide Association and Pathway Analysis of Carcass and Meat Quality Traits in Karachai Young Goats. Animals 2023, 13, 3237. [Google Scholar] [CrossRef]
- Selionova, M.; Trukhachev, V.; Aibazov, M.; Sermyagin, A.; Belous, A.; Gladkikh, M.; Zinovieva, N. Genome-Wide Association Study of Milk Composition in Karachai Goats. Animals 2024, 14, 327. [Google Scholar] [CrossRef]
- Ruan, D.; Zhuang, Z.; Ding, R.; Qiu, Y.; Zhou, S.; Wu, J.; Xu, C.; Hong, L.; Huang, S.; Zheng, E.; et al. Weighted Single-Step GWAS Identified Candidate Genes Associated with Growth Traits in a Duroc Pig Population. Genes 2021, 12, 117. [Google Scholar] [CrossRef]
- Wang, H.; Wang, X.; Li, M.; Sun, H.; Chen, Q.; Yan, D.; Dong, X.; Pan, Y.; Lu, S. Genome-Wide Association Study of Growth Traits in a Four-Way Crossbred Pig Population. Genes 2022, 13, 1990. [Google Scholar] [CrossRef] [PubMed]
- Roth, K.; Pröll-Cornelissen, M.J.; Henne, H.; Appel, A.K.; Schellander, K.; Tholen, E.; Große-Brinkhaus, C. Multivariate Genome-Wide Associations for Immune Traits in Two Maternal Pig Lines. BMC Genom. 2023, 24, 492. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.; Dong, L.; Gan, X.; Gan, F.; Xu, W.; Lu, L. Genome-Wide Association Studies and Haplotype Sharing Analysis Targeting the Growth Traits in Yandang Partridge Chickens. Anim. Biotechnol. 2023, 34, 1943–1949. [Google Scholar] [CrossRef] [PubMed]
- Cai, K.; Liu, R.; Wei, L.; Wang, X.; Cui, H.; Luo, N.; Wen, J.; Chang, Y.; Zhao, G. Genome-Wide Association Analysis Identify Candidate Genes for Feed Efficiency and Growth Traits in Wenchang Chickens. BMC Genom. 2024, 25, 645. [Google Scholar] [CrossRef]
- Hormozdiari, F.; Kichaev, G.; Yang, W.-Y.; Pasaniuc, B.; Eskin, E. Identification of Causal Genes for Complex Traits. Bioinformatics 2015, 31, i206–i213. [Google Scholar] [CrossRef]
- Farh, K.K.-H.; Marson, A.; Zhu, J.; Kleinewietfeld, M.; Housley, W.J.; Beik, S.; Shoresh, N.; Whitton, H.; Ryan, R.J.H.; Shishkin, A.A.; et al. Genetic and Epigenetic Fine Mapping of Causal Autoimmune Disease Variants. Nature 2015, 518, 337–343. [Google Scholar] [CrossRef]
- Huang, H.; Fang, M.; Jostins, L.; Umićević Mirkov, M.; Boucher, G.; Anderson, C.A.; Andersen, V.; Cleynen, I.; Cortes, A.; Crins, F.; et al. Fine-Mapping Inflammatory Bowel Disease Loci to Single-Variant Resolution. Nature 2017, 547, 173–178. [Google Scholar] [CrossRef]
- Kichaev, G.; Yang, W.-Y.; Lindstrom, S.; Hormozdiari, F.; Eskin, E.; Price, A.L.; Kraft, P.; Pasaniuc, B. Integrating Functional Data to Prioritize Causal Variants in Statistical Fine-Mapping Studies. PLoS Genet. 2014, 10, e1004722. [Google Scholar] [CrossRef]
- Chen, W.; McDonnell, S.K.; Thibodeau, S.N.; Tillmans, L.S.; Schaid, D.J. Incorporating Functional Annotations for Fine-Mapping Causal Variants in a Bayesian Framework Using Summary Statistics. Genetics 2016, 204, 933–958. [Google Scholar] [CrossRef]
- Yang, Z.; Wang, C.; Liu, L.; Khan, A.; Lee, A.; Vardarajan, B.; Mayeux, R.; Kiryluk, K.; Ionita-Laza, I. CARMA is a New Bayesian Model for Fine-Mapping in Genome-Wide Association Meta-Analyses. Nat. Genet. 2023, 55, 1057–1065. [Google Scholar] [CrossRef] [PubMed]
- Benner, C.; Spencer, C.C.A.; Havulinna, A.S.; Salomaa, V.; Ripatti, S.; Pirinen, M. FINEMAP: Efficient Variable Selection Using Summary Data from Genome-Wide Association Studies. Bioinformatics 2016, 32, 1493–1501. [Google Scholar] [CrossRef]
- Jiang, J.; Cole, J.B.; Freebern, E.; Da, Y.; VanRaden, P.M.; Ma, L. Functional Annotation and Bayesian Fine-Mapping Reveals Candidate Genes for Important Agronomic Traits in Holstein Bulls. Commun. Biol. 2019, 2, 212. [Google Scholar] [CrossRef]
- Finucane, H.K.; Bulik-Sullivan, B.; Gusev, A.; Trynka, G.; Reshef, Y.; Loh, P.-R.; Anttila, V.; Xu, H.; Zang, C.; Farh, K.; et al. Partitioning Heritability by Functional Annotation Using Genome-Wide Association Summary Statistics. Nat. Genet. 2015, 47, 1228–1235. [Google Scholar] [CrossRef] [PubMed]
- Mei, Q.; Fu, C.; Li, J.; Zhao, S.; Xiang, T. blupADC: An R Package and Shiny Toolkit for Comprehensive Genetic Data Analysis in Animal and Plant Breeding. bioRxiv 2021. [Google Scholar] [CrossRef]
- Garrick, D.J.; Taylor, J.F.; Fernando, R.L. Deregressing Estimated Breeding Values and Weighting Information for Genomic Regression Analyses. Genet. Sel. Evol. GSE 2009, 41, 55. [Google Scholar] [CrossRef]
- McKenna, A.; Hanna, M.; Banks, E.; Sivachenko, A.; Cibulskis, K.; Kernytsky, A.; Garimella, K.; Altshuler, D.; Gabriel, S.; Daly, M.; et al. The Genome Analysis Toolkit: A MapReduce Framework for Analyzing Next-Generation DNA Sequencing Data. Genome Res. 2010, 20, 1297–1303. [Google Scholar] [CrossRef]
- Browning, B.L.; Tian, X.; Zhou, Y.; Browning, S.R. Fast Two-Stage Phasing of Large-Scale Sequence Data. Am. J. Hum. Genet. 2021, 108, 1880–1890. [Google Scholar] [CrossRef]
- Zhang, Z.; Wang, A.; Hu, H.; Wang, L.; Gong, M.; Yang, Q.; Liu, A.; Li, R.; Zhang, H.; Zhang, Q.; et al. The Efficient Phasing and Imputation Pipeline of low-Coverage Whole Genome Sequencing Data Using a High-Quality and Publicly Available Reference Panel in Cattle. Anim. Res. One Health 2023, 1, 4–16. [Google Scholar] [CrossRef]
- Purcell, S.; Neale, B.; Todd-Brown, K.; Thomas, L.; Ferreira, M.A.R.; Bender, D.; Maller, J.; Sklar, P.; de Bakker, P.I.W.; Daly, M.J.; et al. PLINK: A Tool Set for Whole-Genome Association and Population-Based Linkage Analyses. Am. J. Hum. Genet. 2007, 81, 559–575. [Google Scholar] [CrossRef]
- Zhang, C.; Dong, S.-S.; Xu, J.-Y.; He, W.-M.; Yang, T.-L. PopLDdecay: A Fast and Effective Tool for Linkage Disequilibrium Decay Analysis Based on Variant Call Format Files. Bioinformatics 2018, 35, 1786–1788. [Google Scholar] [CrossRef]
- Yin, L.; Zhang, H.; Tang, Z.; Xu, J.; Yin, D.; Zhang, Z.; Yuan, X.; Zhu, M.; Zhao, S.; Li, X.; et al. rMVP: A Memory-Efficient, Visualization-Enhanced, and Parallel-Accelerated Tool for Genome-Wide Association Study. Genom. Proteom. Bioinform. 2021, 19, 619–628. [Google Scholar] [CrossRef] [PubMed]
- Quinlan, A.R.; Hall, I.M. BEDTools: A Flexible Suite of Utilities for Comparing Genomic Features. Bioinformatics 2010, 26, 841–842. [Google Scholar] [CrossRef] [PubMed]
- Xie, C.; Mao, X.; Huang, J.; Ding, Y.; Wu, J.; Dong, S.; Kong, L.; Gao, G.; Li, C.Y.; Wei, L. KOBAS 2.0: A Web Server for Annotation and Identification of Enriched Pathways and Diseases. Nucleic Acids Res. 2011, 39, W316–W322. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.; Jia, G.; Diao, X. geneHapR: An R Package for Gene Haplotypic Statistics and Visualization. BMC Bioinform. 2023, 24, 199. [Google Scholar] [CrossRef]
- Mugambe, J.; Ahmed, R.H.; Thaller, G.; Schmidtmann, C. Impact of Inbreeding on Production, Fertility, and Health Traits in German Holstein Dairy Cattle Utilizing Various Inbreeding Estimators. J. Dairy Sci. 2024, 107, 4714–4725. [Google Scholar] [CrossRef]
- Zhou, J.; Liu, L.; Chen, C.J.; Zhang, M.; Lu, X.; Zhang, Z.; Huang, X.; Shi, Y. Genome-Wide Association Study of Milk and Reproductive Traits in Dual-Purpose Xinjiang Brown Cattle. BMC Genom. 2019, 20, 827. [Google Scholar] [CrossRef]
- Iung, L.H.S.; Petrini, J.; Ramírez-Díaz, J.; Salvian, M.; Rovadoscki, G.A.; Pilonetto, F.; Dauria, B.D.; Machado, P.F.; Coutinho, L.L.; Wiggans, G.R.; et al. Genome-Wide Association Study for Milk Production Traits in a Brazilian Holstein Population. J. Dairy Sci. 2019, 102, 5305–5314. [Google Scholar] [CrossRef]
- Jayawardana, J.M.D.R.; Lopez-Villalobos, N.; McNaughton, L.R.; Hickson, R.E. Genomic Regions Associated with Milk Composition and Fertility Traits in Spring-Calved Dairy Cows in New Zealand. Genes 2023, 14, 860. [Google Scholar] [CrossRef]
- Buaban, S.; Lengnudum, K.; Boonkum, W.; Phakdeedindan, P. Genome-Wide Association Study on Milk Production and Somatic Cell Score for Thai Dairy Cattle Using Weighted Single-Step Approach with Random Regression Test-Day Model. J. Dairy Sci. 2022, 105, 468–494. [Google Scholar] [CrossRef]
- Jayawardana, J.M.D.R.; Lopez-Villalobos, N.; McNaughton, L.R.; Hickson, R.E. Heritabilities and Genetic and Phenotypic Correlations for Milk Production and Fertility Traits of Spring-Calved Once-Daily or Twice-Daily Milking Cows in New Zealand. J. Dairy Sci. 2023, 106, 1910–1924. [Google Scholar] [CrossRef]
- Liu, D.; Xu, Z.; Zhao, W.; Wang, S.; Li, T.; Zhu, K.; Liu, G.; Zhao, X.; Wang, Q.; Pan, Y.; et al. Genetic Parameters and Genome-Wide Association for Milk Production Traits and Somatic Cell Score in Different Lactation Stages of Shanghai Holstein Population. Front. Genet. 2022, 13, 940650. [Google Scholar] [CrossRef]
- Charton, C.; Guinard-Flament, J.; Lefebvre, R.; Barbey, S.; Gallard, Y.; Boichard, D.; Larroque, H. Genetic Parameters of Milk Production Traits in Response to a Short Once-Daily Milking Period in crossbred Holstein × Normande Dairy Cows. J. Dairy Sci. 2018, 101, 2235–2247. [Google Scholar] [CrossRef]
- Fonseca, M.; Kurban, D.; Roy, J.-P.; Santschi, D.E.; Molgat, E.; Yang, D.A.; Dufour, S. Usefulness of Differential Somatic Cell Count for Udder Health Monitoring: Identifying Referential Values for Differential Somatic Cell Count in Healthy Quarters and Quarters with Subclinical Mastitis. J. Dairy Sci. 2025, 108, 3917–3928. [Google Scholar] [CrossRef] [PubMed]
- Olori, V.E.; Meuwissen, T.H.E.; Veerkamp, R.F. Calving Interval and Survival Breeding Values as Measure of Cow Fertility in a Pasture-Based Production System with Seasonal Calving. J. Dairy Sci. 2002, 85, 689–696. [Google Scholar] [CrossRef] [PubMed]
- Ali, I.; Muhammad Suhail, S.; Shafiq, M. Heritability Estimates and Genetic Correlations of Various Production and Reproductive Traits of Different Grades of Dairy Cattle Reared Under Subtropical Condition. Reprod. Domest. Anim. 2019, 54, 1026–1033. [Google Scholar] [CrossRef] [PubMed]
- Roy, I.; Rahman, M.; Karunakaran, M.; Gayari, I.; Baneh, H.; Mandal, A. Genetic Relationships Between Reproductive and Production Traits in Jersey Crossbred Cattle. Gene 2024, 894, 147982. [Google Scholar] [CrossRef]
- Buitenhuis, B.; Janss, L.L.G.; Poulsen, N.A.; Larsen, L.B.; Larsen, M.K.; Sørensen, P. Genome-Wide Association and Biological Pathway Analysis for Milk-Fat Composition in Danish Holstein and Danish Jersey Cattle. BMC Genom. 2014, 15, 1112. [Google Scholar] [CrossRef]
- Magee, D.A.; Sikora, K.M.; Berkowicz, E.W.; Berry, D.P.; Howard, D.J.; Mullen, M.P.; Evans, R.D.; Spillane, C.; MacHugh, D.E. DNA Sequence Polymorphisms in a Panel of Eight Candidate Bovine Imprinted Genes and Their Association with Performance Traits in Irish Holstein-Friesian Cattle. BMC Genet. 2010, 11, 93. [Google Scholar] [CrossRef]
- Cole, J.B.; Wiggans, G.R.; Ma, L.; Sonstegard, T.S.; Lawlor, T.J.; Crooker, B.A.; Van Tassell, C.P.; Yang, J.; Wang, S.; Matukumalli, L.K.; et al. Genome-Wide Association Analysis of Thirty One Production, Health, Reproduction and Body Conformation Traits in Contemporary U.S. Holstein Cows. BMC Genom. 2011, 12, 408. [Google Scholar] [CrossRef]
- Jiang, J.; Liu, L.; Gao, Y.; Shi, L.; Li, Y.; Liang, W.; Sun, D. Determination of Genetic Associations Between Indels in 11 Candidate Genes and Milk Composition Traits in Chinese Holstein Population. BMC Genet. 2019, 20, 48. [Google Scholar] [CrossRef]
- Hung, C.-C.C.; Luan, J.; Sims, M.; Keogh, J.M.; Hall, C.; Wareham, N.J.; O’Rahilly, S.; Farooqi, I.S. Studies of the SIM1 Gene in Relation to Human Obesity and Obesity-Related Traits. Int. J. Obes. 2007, 31, 429–434. [Google Scholar] [CrossRef]
- Revelo, H.A.; López-Alvarez, D.; Palacios, Y.A.; Vergara, O.D.; Yánez, M.B.; Ariza, M.F.; Molina, S.L.C.; Sanchez, Y.O.; Alvarez, L.Á. Genome-Wide Association Study Reveals Candidate Genes for Traits Related to Meat Quality in Colombian Creole Hair Sheep. Trop. Anim. Health Prod. 2023, 55, 357. [Google Scholar] [CrossRef]
- Pande, S.; Yang, X.; Friesel, R. Interleukin-17 receptor D (Sef) is a Multi-Functional Regulator of Cell Signaling. Cell Commun. Signal. 2021, 19, 6. [Google Scholar] [CrossRef] [PubMed]
- Usman, T.; Wang, Y.; Liu, C.; He, Y.; Wang, X.; Dong, Y.; Wu, H.; Liu, A.; Yu, Y. Novel SNPs in IL-17F and IL-17A Genes Associated with Somatic Cell Count in Chinese Holstein and Inner-Mongolia Sanhe Cattle. J. Anim. Sci. Biotechnol. 2017, 8, 5. [Google Scholar] [CrossRef] [PubMed]
- Ghulam Mohyuddin, S.; Liang, Y.; Ni, W.; Adam Idriss Arbab, A.; Zhang, H.; Li, M.; Yang, Z.; Karrow, N.A.; Mao, Y. Polymorphisms of the IL-17A Gene Influence Milk Production Traits and Somatic Cell Score in Chinese Holstein Cows. Bioengineering 2022, 9, 448. [Google Scholar] [CrossRef] [PubMed]
- Gaffen, S.L.; Moutsopoulos, N.M. Regulation of Host-Microbe Interactions at Oral Mucosal Barriers by Type 17 Immunity. Sci. Immunol. 2020, 5, eaau4594. [Google Scholar] [CrossRef]
- Porcherie, A.; Gilbert, F.B.; Germon, P.; Cunha, P.; Trotereau, A.; Rossignol, C.; Winter, N.; Berthon, P.; Rainard, P. IL-17A Is an Important Effector of the Immune Response of the Mammary Gland to Escherichia coli Infection. J. Immunol. 2016, 196, 803–812. [Google Scholar] [CrossRef]
- Mittal, S.; Aslam, A.; Doidge, R.; Medica, R.; Winkler, G.S. The Ccr4a (CNOT6) and Ccr4b (CNOT6L) Deadenylase Subunits of the Human Ccr4-Not Complex Contribute to the Prevention of Cell Death and Senescence. Mol. Biol. Cell 2011, 22, 748–758. [Google Scholar] [CrossRef]
- Pangmao, S.; Thomson, P.C.; Khatkar, M.S. Genetic Parameters of Milk and Lactation Curve Traits of Dairy Cattle from Research Farms in Thailand. Anim. Biosci. 2022, 35, 1499–1511. [Google Scholar] [CrossRef]
- Ring, S.C.; Purfield, D.C.; Good, M.; Breslin, P.; Ryan, E.; Blom, A.; Evans, R.D.; Doherty, M.L.; Bradley, D.G.; Berry, D.P. Variance Components for Bovine Tuberculosis Infection and Multi-Breed Genome-Wide Association Analysis Using Imputed Whole Genome Sequence Data. PLoS ONE 2019, 14, e0212067. [Google Scholar] [CrossRef] [PubMed]
- Dreher, C.; Wellmann, R.; Stratz, P.; Schmid, M.; Preuß, S.; Hamann, H.; Bennewitz, J. Genomic Analysis of Perinatal Sucking Reflex in German Brown Swiss Calves. J. Dairy Sci. 2019, 102, 6296–6305. [Google Scholar] [CrossRef]
- Ding, Y.; Feng, Y.; Huang, Z.; Zhang, Y.; Li, X.; Liu, R.; Li, H.; Wang, T.; Ding, Y.; Jia, Z.; et al. SOX15 Transcriptionally Increases the Function of AOC1 to Modulate Ferroptosis and Progression in Prostate Cancer. Cell Death Dis. 2022, 13, 673. [Google Scholar] [CrossRef] [PubMed]
- Zemanova, M.; Langova, L.; Novotná, I.; Dvorakova, P.; Vrtkova, I.; Havlicek, Z. Immune Mechanisms, Resistance Genes, and Their Roles in the Prevention of Mastitis in Dairy Cows. Arch. Anim. Breed. 2022, 65, 371–384. [Google Scholar] [CrossRef] [PubMed]
| Traits | Mean | SD | PV | AV | PEV | RV | h2 | SE (h2) | Reliability |
|---|---|---|---|---|---|---|---|---|---|
| Milk production traits | |||||||||
| MY305 (kg) | 9754.66 | 2733.97 | 4,393,615.80 | 1,522,274 | 3.00 × 10−5 | 2,871,341.80 | 0.35 | 0.16 | 0.49 |
| FP (%) | 4.31 | 0.55 | 0.28 | 0.15 | 0 | 0.14 | 0.52 | 0.19 | 0.51 |
| PP (%) | 3.33 | 0.24 | 0.06 | 0.02 | 0.004 | 0.03 | 0.39 | 0.16 | 0.42 |
| PM (kg) | 45.02 | 14.33 | 95.51 | 18.71 | 3.66 | 73.14 | 0.20 | 0.12 | 0.36 |
| PD (days) | 88.38 | 55.90 | 2629.12 | 324.73 | 0 | 2304.45 | 0.12 | 0.07 | 0.30 |
| Health trait | |||||||||
| SCS | 2.92 | 0.01 | 0.0001 | 1.00 × 10−5 | 0 | 9.00 × 10−5 | 0.14 | 0.09 | 0.26 |
| Reproductive trait | |||||||||
| CI (days) | 413.86 | 99.43 | 9937.97 | 736.11 | 0 | 9201.85 | 0.07 | 0.06 | 0.20 |
| Traits | SNP | CHR | Position (bp) | p-Value | FDR | Nearest Gene | Distance (Kb) |
|---|---|---|---|---|---|---|---|
| MY305 | 1_72919372_T_G | 1 | 72919372 | 1.54 × 10−10 | 3.97 × 10−5 | TMEM44 | within |
| MY305 | 5_12384753_C_G | 5 | 12384753 | 7.67 × 10−14 | 7.89 × 10−8 | TMTC2 | within |
| MY305 | 5_102787852_T_A | 5 | 102787852 | 3.32 × 10−8 | 4.08 × 10−3 | CD163L1 | within |
| MY305 | 7_136097_G_A | 7 | 136097 | 4.72 × 10−10 | 9.73 × 10−5 | LOC101907627 | 16.19 |
| MY305 | 7_3235359_G_C | 7 | 3235359 | 3.57 × 10−8 | 4.08 × 10−3 | LOC104969038 | within |
| MY305 | 13_25815175_G_A | 13 | 25815175 | 2.62 × 10−9 | 4.50 × 10−4 | GPR158 | within |
| MY305 | 17_63091807_C_T | 17 | 63091807 | 3.33 × 10−11 | 1.71 × 10−5 | SPPL3 | within |
| MY305 | 18_43016754_A_G | 18 | 43016754 | 1.32 × 10−10 | 3.97 × 10−5 | DPY19L3 | 23.5 |
| MY305 | 21_20072203_C_G | 21 | 20072203 | 3.17 × 10−8 | 4.08 × 10−3 | LOC132343335 | 2.99 |
| FP | 3_15686591_C_T | 3 | 15686591 | 3.71 × 10−11 | 9.97 × 10−6 | PMVK | within |
| FP | 5_9957392_A_C | 5 | 9957392 | 3.87 × 10−11 | 9.97 × 10−6 | OTOGL | 29.17 |
| FP | 5_63598729_G_A | 5 | 63598729 | 2.63 × 10−15 | 2.71 × 10−9 | ANKS1B | within |
| FP | 5_63599285_G_T | 5 | 63599285 | 1.11 × 10−12 | 5.71 × 10−7 | ANKS1B | within |
| FP | 9_49700710_C_T | 9 | 49700710 | 3.38 × 10−9 | 3.87 × 10−4 | SIM1 | 141.51 |
| FP | 18_36490909_C_T | 18 | 36490909 | 2.33 × 10−9 | 3.12 × 10−4 | NIP7 | within |
| FP | 21_33857755_C_G | 21 | 33857755 | 4.24 × 10−9 | 4.37 × 10−4 | LMAN1L | within |
| FP | 21_33914609_G_T | 21 | 33914609 | 1.30 × 10−8 | 1.22 × 10−3 | CYP1A2 | 8.50 |
| FP | 21_33936823_G_T | 21 | 33936823 | 2.33 × 10−9 | 3.12 × 10−4 | CYP1A2 | 6.75 |
| FP | 29_49287451_G_A | 29 | 49287451 | 9.09 × 10−10 | 1.87 × 10−4 | TSPAN32 | 5.21 |
| FP | 29_50418489_T_C | 29 | 50418489 | 2.43 × 10−9 | 3.12 × 10−4 | AP2A2 | within |
| PP | 1_72709215_G_A | 1 | 72709215 | 3.45 × 10−8 | 4.18 × 10−3 | LOC132346096 | 93.63 |
| PP | 6_55498889_T_C | 6 | 55498889 | 4.26 × 10−11 | 1.46 × 10−5 | ARAP2 | within |
| PP | 6_84194728_C_A | 6 | 84194728 | 4.14 × 10−11 | 1.46 × 10−5 | LOC100138004 | within |
| PP | 10_41071426_T_C | 10 | 41071426 | 2.38 × 10−10 | 6.12 × 10−4 | MDGA2 | 323.43 |
| PP | 16_36517381_G_A | 16 | 36517381 | 1.19 × 10−9 | 2.45 × 10−4 | DPT | 183.56 |
| PP | 17_71197319_G_A | 17 | 71197319 | 1.97 × 10−8 | 2.89 × 10−3 | ZNF70 | 14.25 |
| PP | 24_690958_G_A | 24 | 690958 | 1.59 × 10−9 | 2.73 × 10−4 | CTDP1 | within |
| PP | X_49143795_A_C | X | 49143795 | 3.65 × 10−8 | 4.18 × 10−3 | PCDH19 | 154.91 |
| PP | X_54208034_T_G | X | 54208034 | 6.80 × 10−12 | 7.01 × 10−6 | IL1RAPL2 | within |
| PM | 3_15509105_C_A | 3 | 15509105 | 1.54 × 10−8 | 5.27 × 10−4 | EFNA3 | 3.28 |
| PM | 3_15583935_G_T | 3 | 15583935 | 3.50 × 10−11 | 2.77 ×10−6 | ZBTB7B | within |
| PM | 3_15686591_C_T | 3 | 15686591 | 1.43 × 10−28 | 1.47 × 10−22 | PMVK | within |
| PM | 3_15826776_C_T | 3 | 15826776 | 4.93 × 10−24 | 2.54 × 10−18 | KCNN3 | within |
| PM | 3_25219743_G_C | 3 | 25219743 | 9.88 × 10−9 | 3.63 × 10−4 | SPAG17 | within |
| PM | 5_56801630_G_A | 5 | 56801630 | 1.73 × 10−12 | 1.98 × 10−7 | BAZ2A | within |
| PM | 6_57627037_C_T | 6 | 57627037 | 3.56 × 10−9 | 1.89 × 10−4 | TBC1D1 | 198.39 |
| PM | 6_113000819_G_A | 6 | 113000819 | 4.54 × 10−9 | 2.11 × 10−4 | LOC107132586 | within |
| PM | 6_113005937_G_A | 6 | 113005937 | 6.73 × 10−9 | 2.57 × 10−4 | LOC107132586 | within |
| PM | 6_113008264_C_T | 6 | 113008264 | 1.55 × 10−16 | 3.98 × 10−11 | LOC107132586 | within |
| PM | 6_113031755_A_G | 6 | 113031755 | 3.03 × 10−10 | 1.95 × 10−5 | LOC107132586 | 2.36 |
| PM | 6_113032815_G_A | 6 | 113032815 | 1.97 × 10−11 | 1.84 × 10−6 | LOC107132586 | 3.42 |
| PM | 6_113032851_G_A | 6 | 113032851 | 3.87 × 10−9 | 1.89 × 10−4 | LOC107132586 | 3.46 |
| PM | 6_113032867_C_A | 6 | 113032867 | 2.27 × 10−11 | 1.95 × 10−6 | LOC107132586 | 3.47 |
| PM | 6_113033403_C_T | 6 | 113033403 | 4.92 × 10−9 | 2.11 × 10−4 | LOC107132586 | 4.01 |
| PM | 6_113033404_T_C | 6 | 113033404 | 4.92 × 10−9 | 2.11 × 10−4 | LOC107132586 | 4.08 |
| PM | 11_87592639_G_A | 11 | 87592639 | 5.51 × 10−15 | 1.13 × 10−9 | GRHL1 | within |
| PM | 21_56854166_T_G | 21 | 56854166 | 3.87 × 10−9 | 1.89 × 10−4 | CPSF2 | 8.50 |
| PM | 21_56858110_T_A | 21 | 56858110 | 1.24 × 10−14 | 1.83 × 10−9 | CPSF2 | 12.50 |
| PM | 21_56860640_G_T | 21 | 56860640 | 7.60 × 10−11 | 5.59 × 10−6 | CPSF2 | 15.03 |
| PM | 21_56871715_G_A | 21 | 56871715 | 2.18 × 10−10 | 1.49 × 10−5 | CPSF2 | 26.10 |
| PM | 22_43984285_G_T | 22 | 43984285 | 5.60 × 10−10 | 3.39 × 10−5 | IL17RD | within |
| PM | 22_43988976_G_A | 22 | 43988976 | 4.92 × 10−13 | 6.34 × 10−8 | IL17RD | within |
| PM | 22_43989144_G_A | 22 | 43989144 | 5.39 × 10−9 | 2.14 × 10−4 | IL17RD | within |
| PM | 22_43990890_G_A | 22 | 43990890 | 5.39 × 10−9 | 2.14 × 10−4 | IL17RD | within |
| PM | 23_5521491_G_T | 23 | 5521491 | 2.23 × 10−8 | 7.19 × 10−4 | FAM83B | within |
| PM | 25_6321969_A_G | 25 | 6321969 | 1.07 × 10−8 | 3.79 × 10−4 | RBFOX1 | within |
| PM | 26_397054_G_A | 26 | 397054 | 2.12 × 10−8 | 7.06 × 10−4 | OR5D18 | 4.73 |
| PM | 26_556311_G_T | 26 | 556311 | 2.05 × 10−19 | 7.04 × 10−14 | UBE2D1 | 110.44 |
| PM | 26_638379_C_T | 26 | 638379 | 9.98 × 10−15 | 1.71 × 10−9 | UBE2D1 | 28.37 |
| PM | 26_803096_C_T | 26 | 803096 | 3.58 × 10−9 | 1.89 × 10−4 | IPMK | within |
| PM | 29_45659217_C_T | 29 | 45659217 | 2.11 × 10−12 | 2.18 × 10−7 | KMT5B | 6.47 |
| PD | 6_60140889_C_T | 6 | 60140889 | 4.38 × 10−8 | 0.004 | UCHL1 | 6.69 |
| PD | 6_60141280_T_C | 6 | 60141280 | 3.87 × 10−8 | 0.004 | UCHL1 | 6.30 |
| PD | 6_92530313_G_A | 6 | 92530313 | 1.56 × 10−8 | 0.004 | CNOT6L | 2.43 |
| PD | 6_92534492_G_A | 6 | 92534492 | 9.82 × 10−9 | 0.004 | CNOT6L | 6.61 |
| PD | 6_93430541_C_T | 6 | 93430541 | 3.34 × 10−8 | 0.004 | ANXA3 | 57.16 |
| PD | 6_93782255_G_A | 6 | 93782255 | 4.22 × 10−8 | 0.004 | PAQR3 | 104.48 |
| PD | 6_99689795_T_C | 6 | 99689795 | 3.26 × 10−8 | 0.004 | WDFY3 | within |
| PD | 6_99724549_T_C | 6 | 99724549 | 2.96 × 10−8 | 0.004 | WDFY3 | within |
| PD | 6_99766367_G_A | 6 | 99766367 | 2.49 × 10−8 | 0.004 | WDFY3 | within |
| PD | 6_99766578_T_G | 6 | 99766578 | 1.99 × 10−8 | 0.004 | WDFY3 | within |
| PD | 22_12760734_A_G | 22 | 12760734 | 3.41 × 10−8 | 0.004 | LOC101903734 | within |
| Traits | SNP | CHR | Position (bp) | p-Value | FDR | Nearest Gene | Distance (Kb) |
|---|---|---|---|---|---|---|---|
| SCS | 4_113485958_C_T | 4 | 113485958 | 9.85 × 10−10 | 2.54 × 10−4 | AOC1 | 22.70 |
| SCS | 4_113489752_G_A | 4 | 113489752 | 1.28 × 10−8 | 1.32 × 10−3 | AOC1 | 26.50 |
| SCS | 4_113489803_G_A | 4 | 113489803 | 8.06 × 10−9 | 9.22 × 10−4 | AOC1 | 26.55 |
| SCS | 6_94098782_G_T | 6 | 94098782 | 2.26 × 10−9 | 3.87 × 10−4 | GK2 | 16.99 |
| SCS | 8_15818198_G_T | 8 | 15818198 | 1.92 × 10−9 | 3.87 × 10−4 | LINGO2 | within |
| SCS | 9_56683722_C_T | 9 | 56683722 | 5.68 × 10−9 | 7.31 × 10−4 | EPHA7 | 67.57 |
| SCS | 15_19068193_A_G | 15 | 19068193 | 3.26 × 10−12 | 3.36 × 10−6 | C15H11orf87 | 261.28 |
| SCS | 17_16535314_G_A | 17 | 16535314 | 4.58 × 10−9 | 6.74 × 10−4 | ZNF330 | within |
| SCS | 21_69672126_G_A | 21 | 69672126 | 2.54 × 10−10 | 8.74 × 10−5 | PACS2 | within |
| SCS | 29_50103110_C_A | 29 | 50103110 | 2.88 × 10−11 | 1.49 × 10−5 | BRSK2 | 1.95 |
| Traits | SNP | CHR | Position (bp) | p-Value | FDR | Nearest Gene | Distance (Kb) |
|---|---|---|---|---|---|---|---|
| CI | 3_15494215_C_A | 3 | 15494215 | 3.64 × 10−8 | 3.75 ×10−3 | EFNA3 | 18.17 |
| CI | 7_1588817_A_C | 7 | 1588817 | 1.36 × 10−8 | 1.75 × 10−3 | MAML1 | within |
| CI | 10_17178126_G_A | 10 | 17178126 | 4.94 × 10−11 | 8.47 × 10−6 | LOC132346336 | 98.85 |
| CI | 10_17181283_C_T | 10 | 17181283 | 4.94 × 10−11 | 8.47 × 10−6 | LOC132346336 | 95.69 |
| CI | 10_17185002_G_A | 10 | 17185002 | 4.94 × 10−11 | 8.47 × 10−6 | LOC132346336 | 91.97 |
| CI | 10_17185230_G_A | 10 | 17185230 | 1.19 × 10−12 | 6.14 × 10−7 | LOC132346336 | 91.74 |
| CI | 13_16401282_T_C | 13 | 16401282 | 2.24 × 10−8 | 2.56 × 10−3 | SFMBT2 | within |
| CI | 14_9758134_C_T | 14 | 9758134 | 4.09 × 10−14 | 4.21 × 10−8 | ADCY8 | within |
| CI | 18_10045332_G_A | 18 | 10045332 | 2.89 × 10−9 | 4.25 × 10−4 | CDH13 | within |
| CI | X_77899540_G_A | X | 77899540 | 2.04 × 10−11 | 7.01 × 10−6 | PHKA1 | 21.95 |
| Traits | Nearest Gene | Distance (Kb) | Type | SNP | CHR | POS | GWAS p-Value | Casualty p-Value | PPC |
|---|---|---|---|---|---|---|---|---|---|
| PD | CNOT6L | 2.43 | Protein coding | 6_92530313_G_A | 6 | 92530313 | 1.56 × 10−8 | 1.33 × 10−8 | 0.885 |
| PD | CNOT6L | 6.61 | Protein coding | 6_92534492_G_A | 6 | 92534492 | 9.82 × 10−9 | 1.08 × 10−7 | 0.115 |
| SCS | KCNH2 | 36.167 | Protein coding | 4_113490019_G_A | 4 | 113490019 | 1.28 × 10−5 | 5.15 × 10−6 | 0.059 |
| SCS | KCNH2 | 35.336 | Protein coding | 4_113490850_A_G | 4 | 113490850 | 1.28 × 10−5 | 5.15 × 10−6 | 0.059 |
| SCS | KCNH2 | 33.921 | Protein coding | 4_113492265_T_C | 4 | 113492265 | 1.28 × 10−5 | 5.15 × 10−6 | 0.059 |
| SCS | SLC25A13 | within | Protein coding | 4_113485958_C_T | 4 | 113485958 | 9.85 × 10−10 | 1.25 × 10−5 | 0.023 |
| CI | LOC132346336 | 91.74 | lncRNA | a 10_17185230_G_A | 10 | 17185230 | 1.19 × 10−12 | 2.34 × 10−12 | 0.979 |
| CI | LOC132346336 | 67.86 | lncRNA | 10_17209112_T_G | 10 | 17209112 | 1.36 × 10−5 | 7.36 × 10−5 | 0.963 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Deng, T.; Cheng, L.; Liu, C.; Xiang, M.; Liu, Q.; Yu, B.; Chen, H. Identification of Candidate Variants Associated with Milk Production, Health and Reproductive Traits for Holstein Cows in Southern China. Agriculture 2025, 15, 2019. https://doi.org/10.3390/agriculture15192019
Deng T, Cheng L, Liu C, Xiang M, Liu Q, Yu B, Chen H. Identification of Candidate Variants Associated with Milk Production, Health and Reproductive Traits for Holstein Cows in Southern China. Agriculture. 2025; 15(19):2019. https://doi.org/10.3390/agriculture15192019
Chicago/Turabian StyleDeng, Tingxian, Lei Cheng, Chenhui Liu, Min Xiang, Qing Liu, Bo Yu, and Hongbo Chen. 2025. "Identification of Candidate Variants Associated with Milk Production, Health and Reproductive Traits for Holstein Cows in Southern China" Agriculture 15, no. 19: 2019. https://doi.org/10.3390/agriculture15192019
APA StyleDeng, T., Cheng, L., Liu, C., Xiang, M., Liu, Q., Yu, B., & Chen, H. (2025). Identification of Candidate Variants Associated with Milk Production, Health and Reproductive Traits for Holstein Cows in Southern China. Agriculture, 15(19), 2019. https://doi.org/10.3390/agriculture15192019







