DNA Methylation and mRNA Exon Sequence Variations in the Salt Stress Adaptation of Paspalum vaginatum
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Material and Treatment
2.2. Transcriptome Data Processing and Gene Expression Analysis
2.3. DNA Methylation Analysis
2.4. Identification of mRNA Exon and DNA Variant Sites
2.5. Screening of mRNA Exon Variant Genes and Enrichment Analysis
2.6. DNA Methylation and mRNA Exon Variants Association Analysis
3. Results
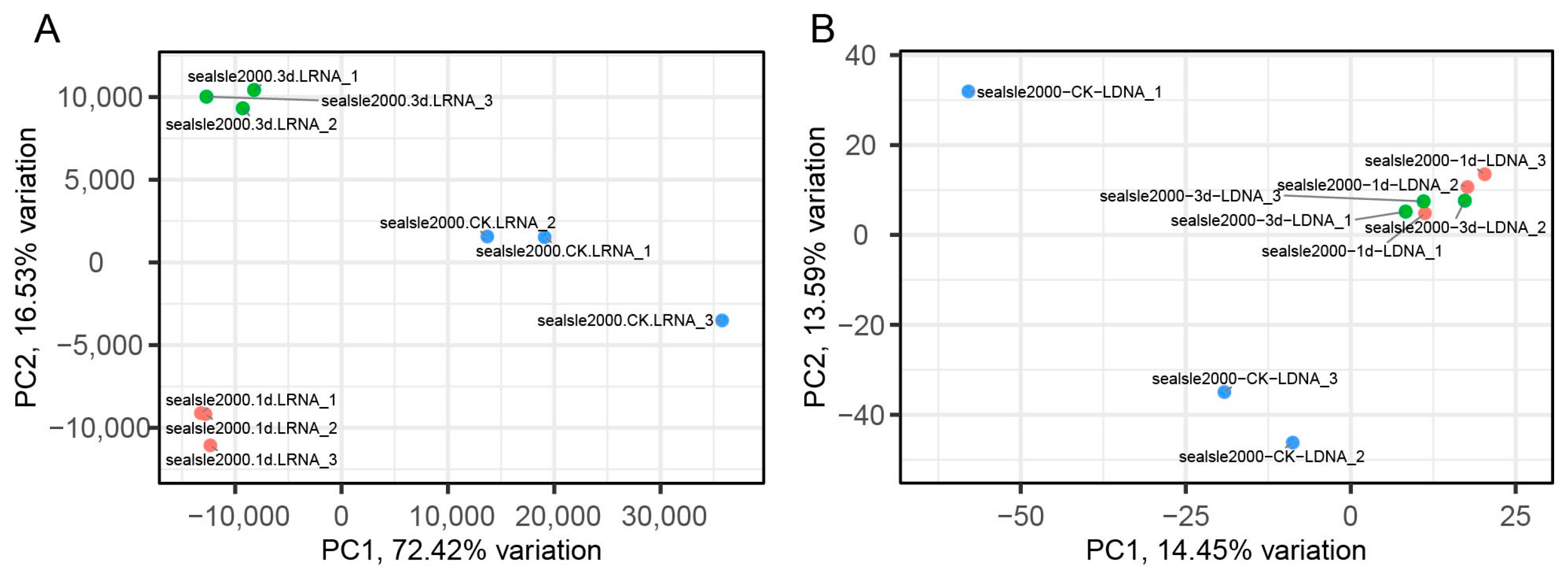
3.1. The Control Group and the Experimental Group Differ Under Salt Stress
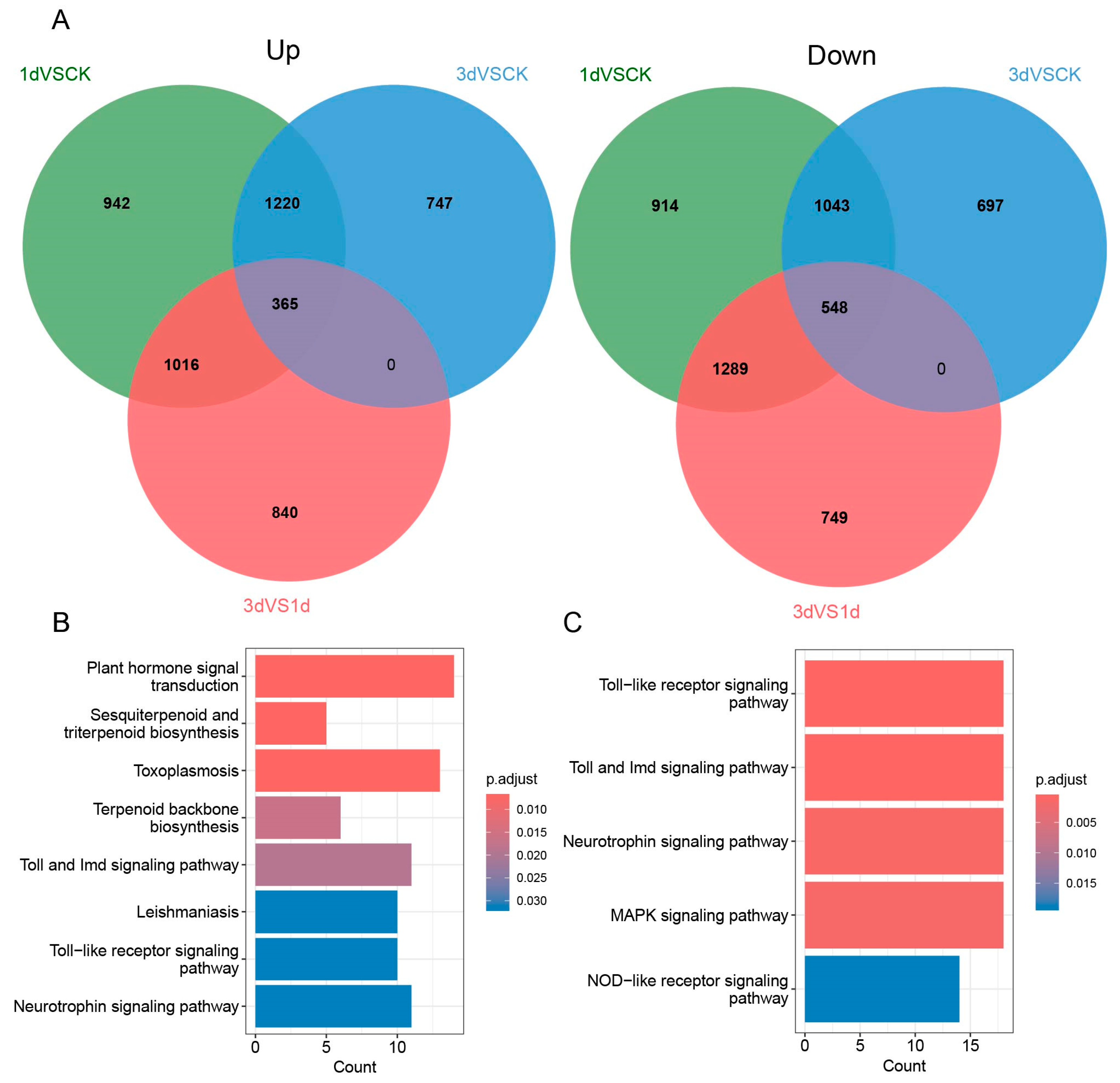
3.2. Analysis Results of Differentially Expressed Genes
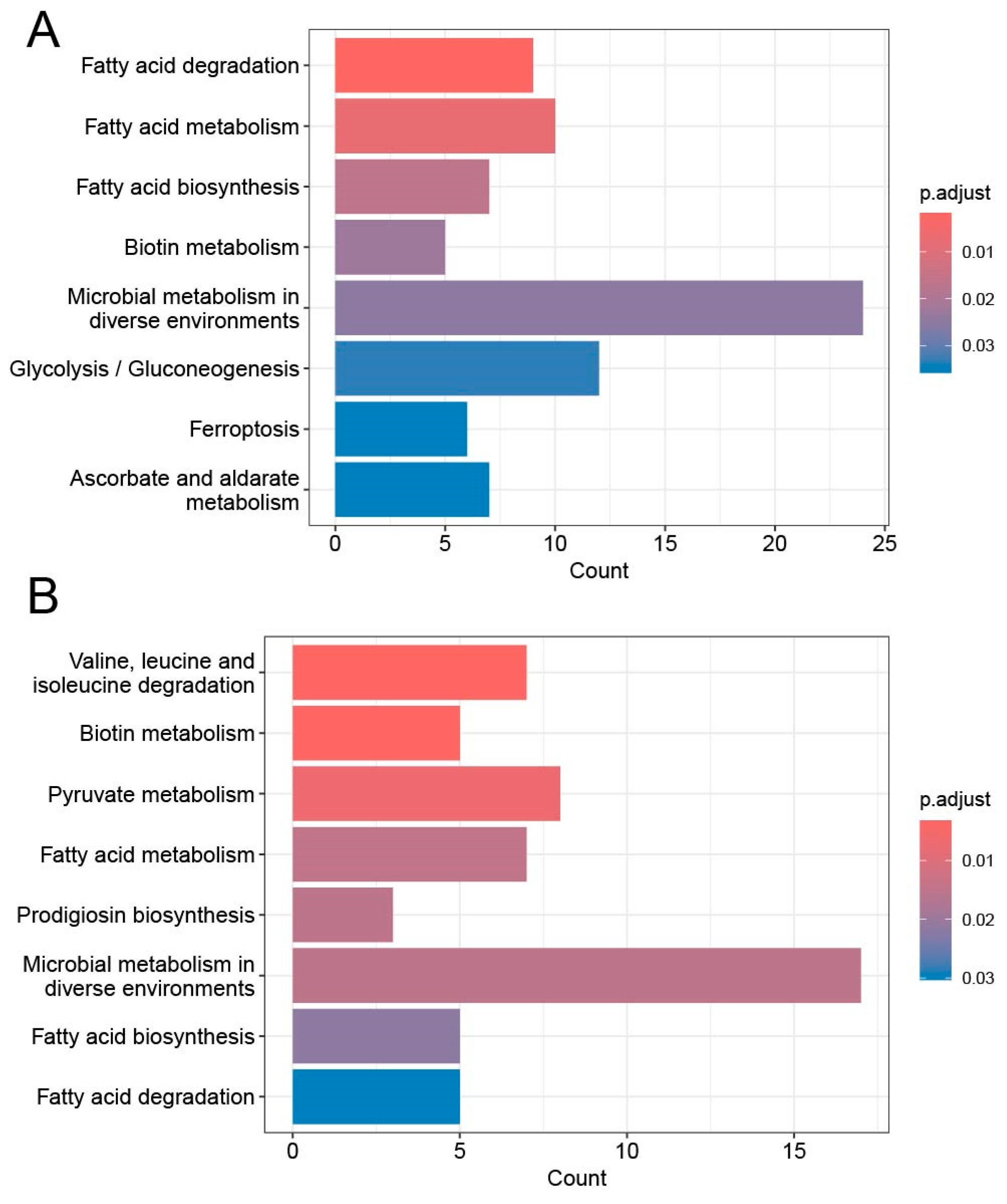
3.3. Genes with Increased Variation in mRNA Exon Levels Are Involved in Multiple Metabolic Pathways Associated with Salt Tolerance
3.4. Characteristics of DNA Methylation Distribution of P. vaginatum
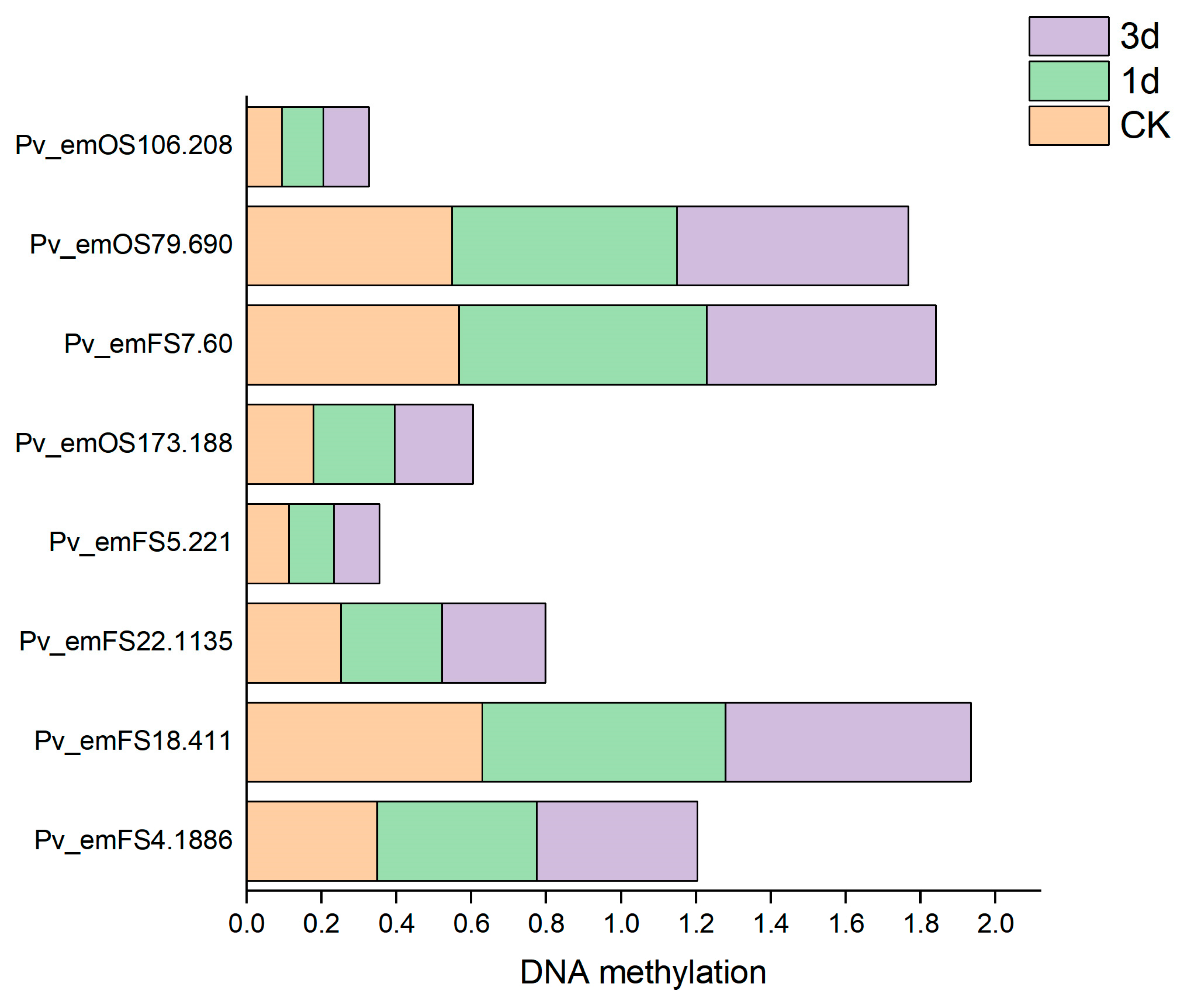
3.5. Association Analysis of DNA Methylation and mRNA Exon Variations
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Vanyushin, B.F.; Ashapkin, V.V. DNA methylation in higher plants: Past, present and future. Biochim. Biophys. Acta 2011, 1809, 360–368. [Google Scholar] [CrossRef] [PubMed]
- Sun, M.H.; Yang, Z.; Liu, L.; Duan, L. DNA methylation in plant responses and adaption to abiotic stresses. Int. J. Mol. Sci. 2022, 23, 6910. [Google Scholar] [CrossRef]
- Pan, L.Y.; Ma, J.J.; Li, J.M.; Yin, B.B.; Fu, C. Advances of salt stress-responsive transcription factors in plants. Chin. J. Biotechnol. 2022, 38, 50–65. [Google Scholar]
- Baek, D.; Jiang, J.F.; Chung, J.-S.; Wang, B.S.; Chen, J.P.; Xin, Z.G.; Shi, H.Z. Regulated AtHKT1 gene expression by a distal enhancer element and DNA methylation in the promoter plays an important role in salt tolerance. Plant Cell Physiol. 2011, 52, 149–161. [Google Scholar] [CrossRef]
- Kumar, S.; Beena, A.S.; Awana, M.; Singh, A. Salt-induced tissue-specific cytosine methylation downregulates expression of HKT genes in contrasting wheat (Triticum aestivum L.) genotypes. DNA Cell Biol. 2017, 36, 283–294. [Google Scholar] [CrossRef]
- Xu, R.; Wang, Y.H.; Zheng, H.; Lu, W.; Wu, C.; Huang, J.G.; Yan, K.; Yang, G.D.; Zheng, C.C. Salt-induced transcription factor MYB74 is regulated by the RNA-directed DNA methylation pathway in Arabidopsis. J. Exp. Bot. 2015, 66, 5997–6008. [Google Scholar] [CrossRef]
- Zhang, W.X.; Wang, N.; Yang, J.T.; Guo, H.; Liu, Z.H.; Zheng, X.J.; Li, S.; Xiang, F.N. The salt-induced transcription factor GmMYB84 confers salinity tolerance in soybean. Plant Sci. 2020, 291, 110326. [Google Scholar] [CrossRef] [PubMed]
- Peng, H.; Zhang, J. Plant genomic DNA methylation in response to stresses: Potential applications and challenges in plant breeding. Prog. Nat. Sci. 2009, 19, 1037–1045. [Google Scholar] [CrossRef]
- Zhang, H.M.; Lang, Z.B.; Zhu, J.K. Dynamics and function of DNA methylation in plants. Nat. Rev. Mol. Cell Biol. 2018, 19, 489–506. [Google Scholar] [CrossRef]
- Li, H.; Zhang, Y.F.; Li, X.G.; Wang, Z.H.; Lin, J.; Chang, Y.H. Identification of Salt-Tolerant Transcription Factors in the Roots of Pyrus betulaefolia by the Association Analysis of Genome-Wide DNA Methylation and Transcriptome. Sci. Agric. Sin. 2023, 56, 1377–1390. [Google Scholar]
- He, X.; Chen, Y.; Xia, Y.; Hong, X.; You, H.; Zhang, R.; Liang, Z.; Cui, Q.; Zhang, S.; Zhou, M. DNA methylation regulates biosynthesis of tanshinones and phenolic acids during growth of Salvia miltiorrhiza. Plant Physiol. 2024, 194, 2086–2100. [Google Scholar] [CrossRef] [PubMed]
- Gao, H.; Xiao, G.A.; Bao, Y.; Xia, Q.M.; Xie, K.D.; Wu, X.M.; Guo, W.W. Integrative DNA methylome and transcriptome analysis illuminate leaf phenotype alterations in tetraploid citrus. Hortic. Adv. 2024, 2, 28. [Google Scholar] [CrossRef]
- Sun, T.Y.; Xu, Y.Y.; Xiang, Y.; Ou, J.H.; Soderblom, E.J.; Diao, Y.R. Crosstalk between RNA m6A and DNA methylation regulates transposable element chromatin activation and cell fate in human pluripotent stem cells. Nat. Genet. 2023, 55, 1324–1335. [Google Scholar] [CrossRef] [PubMed]
- Deng, S.; Zhang, J.L.; Su, J.C.; Zuo, Z.X.; Zeng, L.X.; Liu, K.J.; Zheng, Y.F.; Huang, X.D.; Bai, R.H.; Zhuang, L.S. RNA m6A regulates transcription via DNA demethylation and chromatin accessibility. Nat. Genet. 2022, 54, 1427–1437. [Google Scholar] [CrossRef]
- Zhang, M.; Yang, L.L.; Jia, Y.H.; Wang, T.Y. Research Progress in the Roles of DNA and Histone Methylations in Epigenetic Regulation. Biotechnol. Bull. 2022, 38, 23–30. [Google Scholar]
- Chen, S.F.; Zhou, Y.Q.; Chen, Y.R.; Gu, J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef]
- Kim, D.; Paggi, J.M.; Park, C.; Bennett, C.; Salzberg, S.L. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 2019, 37, 907–915. [Google Scholar] [CrossRef]
- Ferragina, P.; Manzini, G. Opportunistic data structures with applications. In Proceedings of the 41st Annual Symposium on Foundations of Computer Science, Washington, DC, USA, 12–14 November 2000; pp. 390–398. [Google Scholar]
- Xu, Z.J.; Jiang, Z.Y.; Wang, H.F.; Xie, L. Performance Evaluation of Whole Genome Bisulfite Sequencing Alignment Software for Plant Data Analysis. Genom. Appl. Biol. 2024, 43, 986–996. [Google Scholar]
- Liao, Y.; Smyth, G.K.; Shi, W. featureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2014, 30, 923–930. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Lever, J.; Krzywinski, M.; Altman, N. Points of significance: Principal component analysis. Nat. Methods 2017, 14, 641–643. [Google Scholar] [CrossRef]
- Paul, D.S.; Guilhamon, P.; Karpathakis, A.; Butcher, L.M.; Thirlwell, C.; Feber, A.; Beck, S. Assessment of RainDrop BS-seq as a method for large-scale, targeted bisulfite sequencing. Epigenetics 2014, 9, 678–684. [Google Scholar] [CrossRef]
- Zhou, Q.W.; Lim, J.Q.; Sung, W.K.; Li, G.L. An integrated package for bisulfite DNA methylation data analysis with Indel-sensitive mapping. BMC Bioinform. 2019, 20, 47. [Google Scholar] [CrossRef]
- McKenna, A.; Hanna, M.; Banks, E.; Sivachenko, A.; Cibulskis, K.; Kernytsky, A.; Garimella, K.; Altshuler, D.; Gabriel, S.; Daly, M. The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010, 20, 1297–1303. [Google Scholar] [CrossRef]
- Danecek, P.; Auton, A.; Abecasis, G.; Albers, C.A.; Banks, E.; DePristo, M.A.; Handsaker, R.E.; Lunter, G.; Marth, G.T.; Sherry, S.T. The variant call format and VCFtools. Bioinformatics 2011, 27, 2156–2158. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Li, M.Y.; Hakonarson, H. ANNOVAR: Functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010, 38, e164. [Google Scholar] [CrossRef]
- Kanehisa, M.; Goto, S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef] [PubMed]
- Mastan, S.G.; Rathore, M.S.; Bhatt, V.D.; Yadav, P.; Chikara, J. Assessment of changes in DNA methylation by methylation-sensitive amplification polymorphism in Jatropha curcas L. subjected to salinity stress. Gene 2012, 508, 125–129. [Google Scholar] [CrossRef] [PubMed]
- Hsieh, T.F.; Ibarra, C.A.; Silva, P.; Zemach, A.; Eshed-Williams, L.; Fischer, R.L.; Zilberman, D. Genome-wide demethylation of Arabidopsis endosperm. Science 2009, 324, 1451–1454. [Google Scholar] [CrossRef]
- Zemach, A.; Kim, M.Y.; Silva, P.; Rodrigues, J.A.; Dotson, B.; Brooks, M.D.; Zilberman, D. Local DNA hypomethylation activates genes in rice endosperm. Proc. Natl. Acad. Sci. USA 2010, 107, 18729–18734. [Google Scholar] [CrossRef]
- Zhong, S.L.; Fei, Z.J.; Chen, Y.R.; Zheng, Y.; Huang, M.Y.; Vrebalov, J.; McQuinn, R.; Gapper, N.; Liu, B.; Xiang, J. Single-base resolution methylomes of tomato fruit development reveal epigenome modifications associated with ripening. Nat. Biotechnol. 2013, 31, 154–159. [Google Scholar] [CrossRef]
- Huang, H.; Liu, R.; Niu, Q.F.; Tang, K.; Zhang, B.; Zhang, H.; Chen, K.S.; Zhu, J.K.; Lang, Z.B. Global increase in DNA methylation during orange fruit development and ripening. Proc. Natl. Acad. Sci. USA 2019, 116, 1430–1436. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Gallego-Bartolomé, J.; Zhou, Y.; Zhong, Z.; Wang, M.; Wongpalee, S.P.; Gardiner, J.; Feng, S.; Kuo, P.H.; Jacobsen, S.E. Ectopic targeting of CG DNA methylation in Arabidopsis with the bacterial SssI methyltransferase. Nat. Commun. 2021, 12, 3130. [Google Scholar] [CrossRef]
- Zhou, L.L.; Tian, S.P.; Qin, G.Z. RNA methylomes reveal the m 6 A-mediated regulation of DNA demethylase gene SlDML2 in tomato fruit ripening. Genome Biol. 2019, 20, 1–23. [Google Scholar] [CrossRef]
- Sinha, P.; Singh, V.K.; Saxena, R.K.; Kale, S.M.; Li, Y.; Garg, V.; Meifang, T.; Khan, A.W.; Kim, K.D.; Chitikineni, A. Genome—Wide analysis of epigenetic and transcriptional changes associated with heterosis in pigeonpea. Plant Biotechnol. J. 2020, 18, 1697–1710. [Google Scholar] [CrossRef]
- Boulias, K.; Greer, E.L. Biological roles of adenine methylation in RNA. Nat. Rev. Genet. 2023, 24, 143–160. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Li, K.; Yang, Z.; Hou, Q.; Zhao, W.W.; Sun, Q. RNase H1C collaborates with ssDNA binding proteins WHY1/3 and recombinase RecA1 to fulfill the DNA damage repair in Arabidopsis chloroplasts. Nucleic Acids Res. 2021, 49, 6771–6787. [Google Scholar] [CrossRef] [PubMed]
- Qi, Y.; Ding, L.; Zhang, S.W.; Yao, S.Z.; Ong, J.N.; Li, Y.; Wu, H.; Du, P. A plant immune protein enables broad antitumor response by rescuing microRNA deficiency. Cell 2022, 185, 1888–1904.e24. [Google Scholar] [CrossRef]
- Kim, M.H.; Jeon, J.; Lee, S.; Lee, J.H.; Gao, L.; Lee, B.-H.; Park, J.M.; Kim, Y.J.; Kwak, J.M. Proteasome subunit RPT2a promotes PTGS through repressing RNA quality control in Arabidopsis. Nat. Plants 2019, 5, 1273–1282. [Google Scholar] [CrossRef]


| Samples | RNA Clean Data | DNA Clean Data | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Sum_seqs (M) | Sum_ Base (Gb) | Mapping Efficiency | Sum_seqs (M) | Sum_ BASE (Gb) | Unique Mapping Efficiency | mC/C | Methylation Site Sequence | |||
| mCG/CG | mCHG/CHG | mCHH/CHH | ||||||||
| CK-1 | 20.7 | 3.1 | 93.9% | 52.2 | 7.3 | 71.5% | 11.6% | 42.6% | 24.8% | 1.1% |
| CK-2 | 21.0 | 3.2 | 92.3% | 55.7 | 7.8 | 72.8% | 12.0% | 43.9% | 25.9% | 1.1% |
| CK-3 | 19.3 | 2.9 | 94.2% | 55.5 | 7.8 | 72.9% | 12.2% | 45.1% | 26.3% | 1.1% |
| 1D-1 | 25.3 | 3.8 | 93.2% | 60.7 | 8.5 | 71.6% | 12.5% | 45.6% | 27.0% | 1.1% |
| 1D-2 | 21.5 | 3.2 | 93.1% | 54.1 | 7.6 | 72.0% | 12.5% | 46.7% | 27.3% | 1.1% |
| 1D-3 | 20.6 | 3.1 | 92.8% | 53.8 | 7.5 | 72.9% | 12.8% | 47.5% | 28.2% | 1.1% |
| 3D-1 | 19.5 | 2.9 | 92.8% | 68.5 | 9.6 | 71.0% | 12.5% | 48.3% | 27.7% | 1.2% |
| 3D-2 | 21.4 | 3.2 | 93.4% | 55.0 | 7.7 | 72.3% | 12.7% | 48.2% | 28.0% | 1.2% |
| 3D-3 | 21.2 | 3.2 | 93.1% | 54.2 | 7.6 | 72.8% | 12.5% | 47.4% | 27.4% | 1.2% |
| Gene Name | Variant Type (0 d vs. 1 d vs. 3 d) | Position | Average DNA 5mC Methylation Level | FPKM Log2FoldChange (0 d vs. 1 d) | Functional Description | ||
|---|---|---|---|---|---|---|---|
| CK | 1 d | 3 d | |||||
| Pv_emFS18.411 | frameshift deletion 0/1:1/1:0/1 | exon10 | 0.6286 | 0.6502 | 0.6547 | −1.6183 | Ubiquitin-like modifier-activating enzyme ATG7 N-terminus (ATG7). |
| Pv_emFS22.1135 | frameshift deletion 0/0:0/1:0/1 | exon7 | 0.2511 | 0.2713 | 0.2756 | −1.1210 | Belongs to the peroxidase family (APX7). |
| Pv_emFS4.1886 | synonymous SNV 0/0:0/1:0/1 | exon5 | 0.3484 | 0.4265 | 0.4293 | −2.1420 | Belongs to the beta-ketoacyl-ACP synthases family (Ketoacyl-synt_C). |
| Pv_emFS5.221 | frameshift deletion 0/1:1/1:0/0 | exon3 | 0.1125 | 0.1209 | 0.1212 | 5.1240 | Belongs to the peroxidase family (peroxidase). |
| Pv_emOS173.188 | frameshift deletion 0/0:1/1:0/1 | exon4 | 0.1783 | 0.2174 | 0.2087 | −1.5996 | Encodes a plasma membrane-localized ABC transporter (ABC2_membrane). |
| nonframeshift deletion 0/0:0/1:0/1 | exon19 | ||||||
| Pv_emOS79.1286 | frameshift deletion 0/0:1/1:0/0 | exon11 | 0.5669 | 0.6612 | 0.6122 | −1.3358 | Encodes a member of the casein kinase 1 protein family that is localized to the cytoplasm and nucleus (Pkinase). |
| Pv_emOS79.690 | nonframeshift deletion 0/0:1/1:0/1 | exon5 | 0.5477 | 0.6010 | 0.6185 | −0.8127 | NULL |
| Pv_emOS106.208 | frameshift deletion 0/1:1/1:0/0 | exon2 | 0.0939 | 0.1114 | 0.1210 | 1.4275 | PC-Esterase |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wei, Y.; Zhu, Q.; Zheng, X.; Wang, Z.; Tang, M. DNA Methylation and mRNA Exon Sequence Variations in the Salt Stress Adaptation of Paspalum vaginatum. Agriculture 2025, 15, 1875. https://doi.org/10.3390/agriculture15171875
Wei Y, Zhu Q, Zheng X, Wang Z, Tang M. DNA Methylation and mRNA Exon Sequence Variations in the Salt Stress Adaptation of Paspalum vaginatum. Agriculture. 2025; 15(17):1875. https://doi.org/10.3390/agriculture15171875
Chicago/Turabian StyleWei, Youhao, Qing Zhu, Xinyi Zheng, Zhiyong Wang, and Minqiang Tang. 2025. "DNA Methylation and mRNA Exon Sequence Variations in the Salt Stress Adaptation of Paspalum vaginatum" Agriculture 15, no. 17: 1875. https://doi.org/10.3390/agriculture15171875
APA StyleWei, Y., Zhu, Q., Zheng, X., Wang, Z., & Tang, M. (2025). DNA Methylation and mRNA Exon Sequence Variations in the Salt Stress Adaptation of Paspalum vaginatum. Agriculture, 15(17), 1875. https://doi.org/10.3390/agriculture15171875






