The Genetic Basis of Wheat Spike Architecture
Abstract
1. Introduction
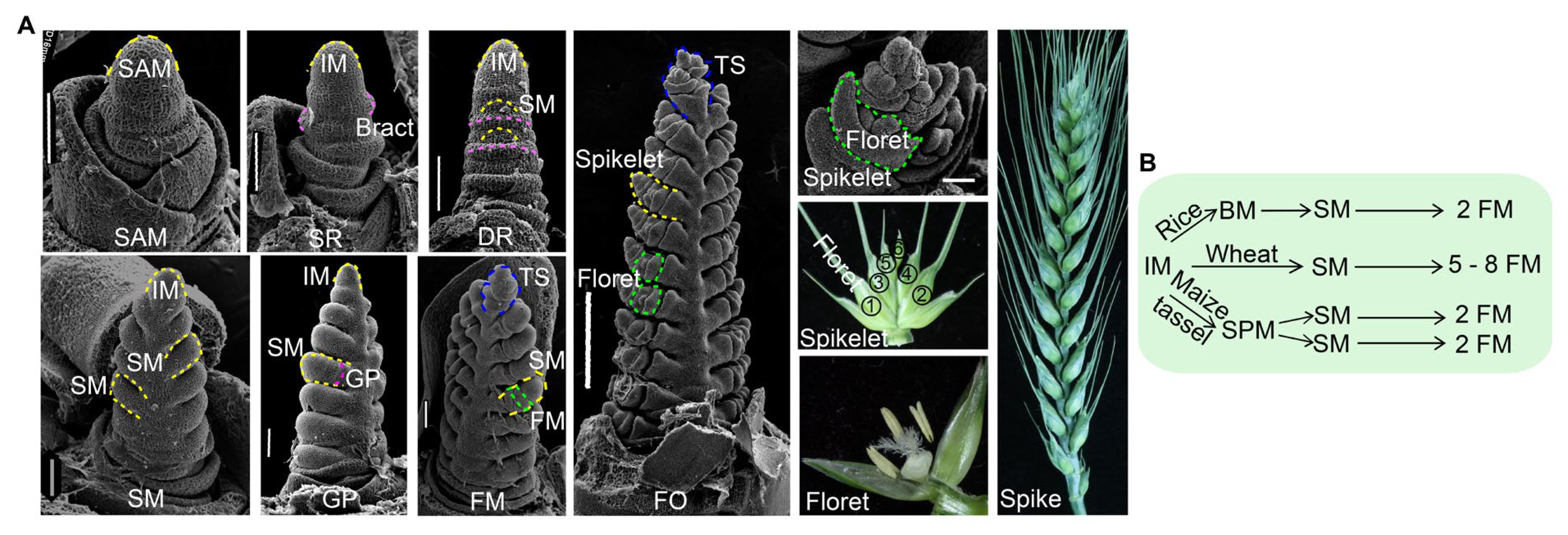
2. The Development of Wheat Inflorescence
3. The Major QTLs Controlling Wheat Spike Architecture
3.1. QTLs Controlling Spike Length and Spike Compactness
3.2. QTLs Controlling Spikelet Number per Spike
3.3. QTLs Controlling Grain Number per Spike
4. The Genetic Regulating Network in Shaping Spike Morphology
4.1. The Transition from the Shoot Apical Meristem to Inflorescence Meristem: The Photoperiod and Vernalization Pathway
4.2. The Regulator Network of Spikelet Development: The Normal Spikelet and Supernumerary Spikelets
4.3. The Genes Controlling Floret Development: The Floret Number and Floral Organ Identity
5. Perspective
5.1. Wheat Gene Cloning Is Becoming Easier
5.2. Improving Wheat Yield Through Spike Traits: Remaining Challenges
5.3. Future Directions in Wheat Spike Trait Research
5.4. Enhancing Wheat Yield: Via Genetics and Agronomy
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| SAM | Shoot Apical Meristem |
| IM | Inflorescence Meristem |
| SM | Spikelet Meristem |
| TS | Terminal Spikelets |
| SR | Single Ridge |
| DR | Double Ridge |
| SL | Spike Length |
| SC | Spikelet Compactness |
| SNS | Total Spikelet Number Per Spike |
| GNS | Grain Number Per Spike |
| SS | Supernumerary Spikelets |
References
- International Wheat Genome Sequencing Consortium (IWGSC). Shifting the limits in wheat research and breeding using a fully annotated reference genome. Science 2018, 361, eaar7191. [Google Scholar] [CrossRef] [PubMed]
- Jia, J.; Zhao, G.; Li, D.; Wang, K.; Kong, C.; Deng, P.; Yan, X.; Zhang, X.; Lu, Z.; Xu, S.; et al. Genome resources for the elite bread wheat cultivar Aikang 58 and mining of elite homeologous haplotypes for accelerating wheat improvement. Mol. Plant 2023, 16, 1893–1910. [Google Scholar] [CrossRef] [PubMed]
- Jiao, C.; Xie, X.; Hao, C.; Chen, L.; Xie, Y.; Garg, V.; Zhao, L.; Wang, Z.; Zhang, Y.; Li, T.; et al. Pan-genome bridges wheat structural variations with habitat and breeding. Nature 2024, 637, 384–393. [Google Scholar] [CrossRef]
- International Wheat Genome Sequencing Consortium (IWGSC). A chromosome-based draft sequence of the hexaploid bread wheat (Triticum aestivum) genome. Science 2014, 345, 1251788. [Google Scholar] [CrossRef]
- Shi, X.; Cui, F.; Han, X.; He, Y.; Zhao, L.; Zhang, N.; Zhang, H.; Zhu, H.; Liu, Z.; Ma, B.; et al. Comparative genomic and transcriptomic analyses uncover the molecular basis of high nitrogen-use efficiency in the wheat cultivar Kenong 9204. Mol. Plant 2022, 15, 1440–1456. [Google Scholar] [CrossRef]
- Sato, K.; Abe, F.; Mascher, M.; Haberer, G.; Gundlach, H.; Spannagl, M.; Shirasawa, K.; Isobe, S. Chromosome-scale genome assembly of the transformation-amenable common wheat cultivar ‘Fielder’. DNA Res. 2021, 28, dsab008. [Google Scholar] [CrossRef]
- Steuernagel, B.; Periyannan, S.K.; Hernández-Pinzón, I.; Witek, K.; Rouse, M.N.; Yu, G.; Hatta, A.; Ayliffe, M.; Bariana, H.; Jones, J.D.; et al. Rapid cloning of disease-resistance genes in plants using mutagenesis and sequence capture. Nat. Biotechnol. 2016, 34, 652–655. [Google Scholar] [CrossRef]
- Wang, D.; Li, Y.; Wang, H.; Xu, Y.; Yang, Y.; Zhou, Y.; Chen, Z.; Zhou, Y.; Gui, L.; Guo, Y.; et al. Boosting wheat functional genomics via an indexed EMS mutant library of KN9204. Plant Commun. 2023, 4, 100593. [Google Scholar] [CrossRef]
- Yuan, Y.; Lyu, B.; Qi, J.; Liu, X.; Wang, Y.; Delaplace, P.; Du, Y. A novel regulator of wheat tillering LT1 identified by using an upgraded BSA method, uni-BSA. Mol. Breed. 2024, 44, 47. [Google Scholar] [CrossRef]
- Zhang, H.; Wang, X.; Pan, Q.; Li, P.; Liu, Y.; Lu, X.; Zhong, W.; Li, M.; Han, L.; Li, J.; et al. QTG-Seq Accelerates QTL Fine Mapping through QTL Partitioning and Whole-Genome Sequencing of Bulked Segregant Samples. Mol. Plant 2019, 12, 426–437. [Google Scholar] [CrossRef]
- Liu, X.; Bie, X.M.; Lin, X.; Li, M.; Wang, H.; Zhang, X.; Yang, Y.; Zhang, C.; Zhang, X.S.; Xiao, J. Uncovering the transcriptional regulatory network involved in boosting wheat regeneration and transformation. Nat. Plants 2023, 9, 908–925. [Google Scholar] [CrossRef]
- Wang, K.; Riaz, B.; Ye, X. Wheat genome editing expedited by efficient transformation techniques: Progress and perspectives. Crop J. 2018, 6, 22–31. [Google Scholar] [CrossRef]
- Wang, K.; Shi, L.; Liang, X.; Zhao, P.; Wang, W.; Liu, J.; Chang, Y.; Hiei, Y.; Yanagihara, C.; Du, L.; et al. The gene TaWOX5 overcomes genotype dependency in wheat genetic transformation. Nat. Plants 2022, 8, 110–117. [Google Scholar] [CrossRef]
- Krasileva, K.V.; Vasquez-Gross, H.A.; Howell, T.; Bailey, P.; Paraiso, F.; Clissold, L.; Simmonds, J.; Ramirez-Gonzalez, R.H.; Wang, X.; Borrill, P.; et al. Uncovering hidden variation in polyploid wheat. Proc. Natl. Acad. Sci. USA 2017, 114, 913–921. [Google Scholar] [CrossRef]
- Song, L.; Liu, J.; Cao, B.; Liu, B.; Zhang, X.; Chen, Z.; Dong, C.; Liu, X.; Zhang, Z.; Wang, W.; et al. Reducing brassinosteroid signalling enhances grain yield in semi-dwarf wheat. Nature 2023, 617, 118–124. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Jia, H.; Li, T.; Wu, J.; Nagarajan, R.; Lei, L.; Powers, C.; Kan, C.C.; Hua, W.; Liu, Z.; et al. TaCol-B5 modifies spike architecture and enhances grain yield in wheat. Science 2022, 376, 180–183. [Google Scholar] [CrossRef] [PubMed]
- Cao, S.; Xu, D.; Hanif, M.; Xia, X.; He, Z. Genetic architecture underpinning yield component traits in wheat. Theor. Appl. Genet. 2020, 133, 1811–1823. [Google Scholar] [CrossRef] [PubMed]
- Luo, X.; Yang, Y.; Lin, X.; Xiao, J. Deciphering spike architecture formation towards yield improvement in wheat. J. Genet. Genom. 2023, 50, 835–845. [Google Scholar] [CrossRef]
- Shaheen, A.; Li, Z.; Yang, Y.; Xie, J.; Zhu, L.; Li, C.; Nie, F.; Wang, M.; Rasheed, A.; Li, H.; et al. Genetic regulation of wheat plant architecture and future prospects for its improvement. New Crops 2025, 2, 100048. [Google Scholar] [CrossRef]
- Xiao, J.; Liu, B.; Yao, Y.; Guo, Z.; Jia, H.; Kong, L.; Zhang, A.; Ma, W.; Ni, Z.; Xu, S.; et al. Wheat genomic study for genetic improvement of traits in China. Sci. China Life Sci. 2022, 65, 1718–1775. [Google Scholar] [CrossRef]
- Yao, Y.; Guo, W.; Gou, J.; Hu, Z.; Liu, J.; Ma, J.; Zong, Y.; Xin, M.; Chen, W.; Li, Q.; et al. Wheat2035: Integrating Pan-omics and Advanced Biotechnology for Future Wheat Design. Mol. Plant 2025, 18, 272–297. [Google Scholar] [CrossRef]
- Bommert, P.; Satoh-Nagasawa, N.; Jackson, D.; Hirano, H.Y. Genetics and evolution of inflorescence and flower development in grasses. Plant Cell Physiol. 2005, 46, 69–78. [Google Scholar] [CrossRef]
- Gauley, A.; Boden, S.A. Genetic pathways controlling inflorescence architecture and development in wheat and barley. J. Integr. Plant Biol. 2019, 61, 296–309. [Google Scholar] [CrossRef]
- Koppolu, R.; Schnurbusch, T. Developmental pathways for shaping spike inflorescence architecture in barley and wheat. J. Integr. Plant Biol. 2019, 61, 278–295. [Google Scholar] [CrossRef]
- Gao, X.Q.; Wang, N.; Wang, X.L.; Zhang, X.S. Architecture of Wheat Inflorescence: Insights from Rice. Trends Plant Sci. 2019, 24, 802–809. [Google Scholar] [CrossRef]
- Ikeda, K.; Sunohara, H.; Nagato, Y. Developmental Course of Inflorescence and Spikelet in Rice. Breed. Sci. 2004, 54, 147–156. [Google Scholar] [CrossRef]
- Vollbrecht, E.; Springer, P.S.; Goh, L.; Iv, E.S.B.; Martienssen, R. Architecture of floral branch systems in maize and related grasses. Nature 2005, 436, 1119–1126. [Google Scholar] [CrossRef]
- Guo, Z.; Zhao, Y.; Röder, M.S.; Reif, J.C.; Ganal, M.W.; Chen, D.; Schnurbusch, T. Manipulation and prediction of spike morphology traits for the improvement of grain yield in wheat. Sci. Rep. 2018, 8, 14435. [Google Scholar] [CrossRef] [PubMed]
- Hu, W.; Wu, H.; Lu, C.; Zheng, X.; Jia, J.; Xu, W. Genetic dissection of quantitative trait loci for spikelets compactness in two Yanzhan1-derived recombinant inbred line wheat populations. Plant Breed. 2022, 141, 719–732. [Google Scholar] [CrossRef]
- Li, T.; Deng, G.; Su, Y.; Yang, Z.; Tang, Y.; Wang, J.; Qiu, X.; Pu, X.; Li, J.; Liu, Z.; et al. Identification and validation of two major QTLs for spike compactness and length in bread wheat (Triticum aestivum L.) showing pleiotropic effects on yield-related traits. Theor. Appl. Genet. 2021, 134, 3625–3641. [Google Scholar] [CrossRef]
- You, J.; Liu, H.; Wang, S.; Luo, W.; Gou, L.; Tang, H.; Mu, Y.; Deng, M.; Jiang, Q.; Chen, G.; et al. Spike Density Quantitative Trait Loci Detection and Analysis in Tetraploid and Hexaploid Wheat Recombinant Inbred Line Populations. Front. Plant Sci. 2021, 12, 796397. [Google Scholar] [CrossRef]
- Chai, L.; Chen, Z.; Bian, R.; Zhai, H.; Cheng, X.; Peng, H.; Yao, Y.; Hu, Z.; Xin, M.; Guo, W.; et al. Dissection of two quantitative trait loci with pleiotropic effects on plant height and spike length linked in coupling phase on the short arm of chromosome 2D of common wheat (Triticum aestivum L.). Theor. Appl. Genet. 2019, 132, 1815–1831. [Google Scholar] [CrossRef]
- Heidari, B.; Sayed-Tabatabaei, B.E.; Saeidi, G.; Kearsey, M.; Suenaga, K. Mapping QTL for grain yield, yield components, and spike features in a doubled haploid population of bread wheat. Genome 2011, 54, 517–527. [Google Scholar] [CrossRef] [PubMed]
- Kumar, N.; Kulwal, P.L.; Balyan, H.S.; Gupta, P.K. QTL mapping for yield and yield contributing traits in two mapping populations of bread wheat. Mol. Breed. 2007, 19, 163–177. [Google Scholar] [CrossRef]
- Li, Y.; Gao, J.; Zhang, R.; Song, G.; Zhang, S.; Li, W.; Li, G. Identification of new QTL for yield-related traits in Chinese landrace and elite wheat varieties through a genome-wide linkage mapping. Euphytica 2020, 216. [Google Scholar] [CrossRef]
- Liu, Y.; Yu, R.; Shen, L.; Sun, M.; Peng, Y.; Zeng, Q.; Shen, K.; Yu, X.; Wu, H.; Ye, B.; et al. Genomic insights into the modifications of spike morphology traits during wheat breeding. Plant Cell Environ. 2024, 47, 5470–5482. [Google Scholar] [CrossRef]
- Sourdille, P.; Cadalen, T.; Guyomarc’h, H.; Snape, J.W.; Perretant, M.R.; Charmet, G.; Boeuf, C.; Bernard, S.; Bernard, M. An update of the Courtot x Chinese Spring intervarietal molecular marker linkage map for the QTL detection of agronomic traits in wheat. Theor. Appl. Genet. 2003, 106, 530–538. [Google Scholar] [CrossRef]
- Wu, X.; Cheng, R.; Xue, S.; Kong, Z.; Wan, H.; Li, G.; Huang, Y.; Jia, H.; Jia, J.; Zhang, L.; et al. Precise mapping of a quantitative trait locus interval for spike length and grain weight in bread wheat (Triticum aestivum L.). Mol. Breed. 2014, 33, 129–138. [Google Scholar] [CrossRef]
- Xu, X.; Li, X.; Zhang, D.; Zhao, J.; Jiang, X.; Sun, H.; Ru, Z. Identification and validation of QTLs for kernel number per spike and spike length in two founder genotypes of wheat. BMC Plant Biol. 2022, 22, 146. [Google Scholar] [CrossRef]
- Xiong, H.; Zhou, C.; Fu, M.; Guo, H.; Xie, Y.; Zhao, L.; Gu, J.; Zhao, S.; Ding, Y.; Li, Y.; et al. Cloning and functional characterization of Rht8, a “Green Revolution” replacement gene in wheat. Mol. Plant 2022, 15, 373–376. [Google Scholar] [CrossRef]
- Kong, X.; Wang, F.; Wang, Z.; Gao, X.; Geng, S.; Deng, Z.; Zhang, S.; Fu, M.; Cui, D.; Liu, S.; et al. Grain yield improvement by genome editing of TaARF12 that decoupled peduncle and rachis development trajectories via differential regulation of gibberellin signalling in wheat. Plant Biotechnol. J. 2023, 21, 1990–2001. [Google Scholar] [CrossRef]
- He, G.; Zhang, Y.; Liu, P.; Jing, Y.; Zhang, L.; Zhu, Y.; Kong, X.; Zhao, H.; Zhou, Y.; Sun, J. The transcription factor TaLAX1 interacts with Q to antagonistically regulate grain threshability and spike morphogenesis in bread wheat. New Phytol. 2021, 230, 988–1002. [Google Scholar] [CrossRef]
- Zhu, J.; Huang, F.; Zhai, H.; Zheng, Y.; Yu, J.; Chen, Z.; Fan, Y.; Zhao, H.; Sun, Q.; Liang, R.; et al. The Tetratricopeptide repeat protein TaTPR-B1 regulates spike compactness in bread wheat. Plant Physiol. 2024, 197, kiae546. [Google Scholar] [CrossRef]
- Yan, L.; Loukoianov, A.; Tranquilli, G.; Helguera, M.; Fahima, T.; Dubcovsky, J. Positional cloning of the wheat vernalization gene VRN1. Proc. Natl. Acad. Sci. USA 2003, 100, 6263–6268. [Google Scholar] [CrossRef]
- Yan, L.; Loukoianov, A.; Blechl, A.; Tranquilli, G.; Ramakrishna, W.; SanMiguel, P.; Bennetzen, J.L.; Echenique, V.; Dubcovsky, J. The wheat VRN2 gene is a flowering repressor down-regulated by vernalization. Science 2004, 303, 1640–1644. [Google Scholar] [CrossRef] [PubMed]
- Glenn, P.; Zhang, J.; Brown-Guedira, G.; DeWitt, N.; Cook, J.P.; Li, K.; Akhunov, E.; Dubcovsky, J. Identification and characterization of a natural polymorphism in FT-A2 associated with increased number of grains per spike in wheat. Theor. Appl. Genet. 2022, 135, 679–692. [Google Scholar] [CrossRef] [PubMed]
- Su, L.; Shan, J.X.; Gao, J.P.; Lin, H.X. OsHAL3, a Blue Light-Responsive Protein, Interacts with the Floral Regulator Hd1 to Activate Flowering in Rice. Mol. Plant 2016, 9, 233–244. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Wen, W.; Hanif, M.; Xia, X.; Wang, H.; Liu, S.; Liu, J.; Yang, L.; Gao, S.; He, Z. TaELF3-1DL, a homolog of ELF3, is associated with heading date in bread wheat. Mol. Breed. 2016, 36, 161. [Google Scholar] [CrossRef]
- Zikhali, M.; Wingen, L.U.; Leverington-Waite, M.; Specel, S.; Griffiths, S. The identification of new candidate genes triticum aestivum flowering locus T3-B1 (TAFT3-B1) and target of EAT1 (TATOE1-B1) controlling the short-day photoperiod response in bread wheat. Plant Cell Environ. 2017, 40, 2678–2690. [Google Scholar] [CrossRef]
- Corsi, B.; Obinu, L.; Zanella, C.M.; Cutrupi, S.; Day, R.; Geyer, M.; Lillemo, M.; Lin, M.; Mazza, L.; Percival-Alwyn, L.; et al. Identification of eight QTL controlling multiple yield components in a German multi-parental wheat population, including Rht24, WAPO-A1, WAPO-B1 and genetic loci on chromosomes 5A and 6A. Theor. Appl. Genet. 2021, 134, 1435–1454. [Google Scholar] [CrossRef]
- Dobrovolskaya, O.; Pont, C.; Sibout, R.; Martinek, P.; Badaeva, E.; Murat, F.; Chosson, A.; Watanabe, N.; Prat, E.; Gautier, N.; et al. Frizzy panicle drives supernumerary spikelets in bread wheat. Plant Physiol. 2015, 167, 189–199. [Google Scholar] [CrossRef]
- Du, D.; Zhang, D.; Yuan, J.; Feng, M.; Li, Z.; Wang, Z.; Zhang, Z.; Li, X.; Ke, W.; Li, R.; et al. FRIZZY PANICLE defines a regulatory hub for simultaneously controlling spikelet formation and awn elongation in bread wheat. New Phytol. 2021, 231, 814–833. [Google Scholar] [CrossRef]
- Kuzay, S.; Lin, H.; Li, C.; Chen, S.; Woods, D.P.; Zhang, J.; Lan, T.; von Korff, M.; Dubcovsky, J. WAPO-A1 is the causal gene of the 7AL QTL for spikelet number per spike in wheat. PLoS Genet. 2022, 18, e1009747. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Li, L.; Zhao, M.; Guo, L.; Guo, X.; Zhao, D.; Batool, A.; Dong, B.; Xu, H.; Cui, S.; et al. Wheat FRIZZY PANICLE activates VERNALIZATION1-A and HOMEOBOX4-A to regulate spike development in wheat. Plant Biotechnol. J. 2021, 19, 1141–1154. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Chen, J.; Yin, C.; Wang, Z.; Wu, H.; Shen, K.; Zhang, Z.; Kang, L.; Xu, S.; Bi, A.; et al. A high-resolution genotype–phenotype map identifies the TaSPL17 controlling grain number and size in wheat. Genome Biol. 2023, 24, 196. [Google Scholar] [CrossRef] [PubMed]
- Wittern, L.M.; Barrero, J.M.; Bovill, W.D.; Verbyla, K.L.; Hughes, T.; Swain, S.M.; Steed, G.; Webb, A.A.R.; Gardner, K.; Greenland, A.; et al. Overexpression of the WAPO-A1 gene increases the number of spikelets per spike in bread wheat. Sci. Rep. 2022, 12, 14229. [Google Scholar] [CrossRef]
- Chen, D.; Zhang, J.; Wang, J.; Yang, X.; Liu, W.; Gao, A.; Li, X.; Li, L. Inheritance and Availability of High Grain Number Per Spike in Two Wheat Germplasm Lines. J. Integr. Agric. 2012, 12, 1409–1416. [Google Scholar] [CrossRef]
- Fernandes, R.; Busanello, C.; Viana, V.; Venske, E.; de Oliveira, V.; Lopes, J.; da Maia, L.; Costa de Oliveira, A.; Pegoraro, C. Genetic variability and heritability of agronomic traits in a wheat collection used in southern Brazil. J. Crop Sci. Biotechnol. 2022, 25, 337–348. [Google Scholar] [CrossRef]
- Sakuma, S.; Golan, G.; Guo, Z.; Ogawa, T.; Tagiri, A.; Sugimoto, K.; Bernhardt, N.; Brassac, J.; Mascher, M.; Hensel, G.; et al. Unleashing floret fertility in wheat through the mutation of a homeobox gene. Proc. Natl. Acad. Sci. USA 2019, 116, 5182–5187. [Google Scholar] [CrossRef]
- Kong, X.; Wang, F.; Geng, S.; Guan, J.; Tao, S.; Jia, M.; Sun, G.; Wang, Z.; Wang, K.; Ye, X.; et al. The wheat AGL6-like MADS-box gene is a master regulator for floral organ identity and a target for spikelet meristem development manipulation. Plant Biotechnol. J. 2022, 20, 75–88. [Google Scholar] [CrossRef]
- Pearce, S.; Shaw, L.M.; Lin, H.; Cotter, J.D.; Li, C.; Dubcovsky, J. Night-break experiments shed light on the photoperiod1-mediated flowering. Plant Physiol. 2017, 174, 1139–1150. [Google Scholar] [CrossRef]
- Ream, T.S.; Woods, D.P.; Amasino, R.M. The molecular basis of vernalization in different plant groups. Cold Spring Harb. Symp. Quant. Biol. 2012, 77, 105–115. [Google Scholar] [CrossRef]
- Chouard, P. Vernalization and Its Relation to Dormancy. Annu. Rev. Plant Physiol. 1960, 11, 191–238. [Google Scholar] [CrossRef]
- Corbesier, L.; Vincent, C.; Jang, S.; Fornara, F.; Fan, Q.; Searle, I.; Giakountis, A.; Farrona, S.; Gissot, L.; Turnbull, C.; et al. FT protein movement contributes to long-distance signaling in floral induction of Arabidopsis. Science 2007, 316, 1030–1033. [Google Scholar] [CrossRef] [PubMed]
- Yan, L.; Fu, D.; Li, C.; Blechl, A.; Tranquilli, G.; Bonafede, M.; Sanchez, A.; Valarik, M.; Yasuda, S.; Dubcovsky, J. The wheat and barley vernalization gene VRN3 is an orthologue of FT. Proc. Natl. Acad. Sci. USA 2006, 103, 19581–19586. [Google Scholar] [CrossRef] [PubMed]
- Gauley, A.; Pasquariello, M.; Yoshikawa, G.V.; Alabdullah, A.K.; Hayta, S.; Smedley, M.A.; Dixon, L.E.; Boden, S.A. Photoperiod-1 regulates the wheat inflorescence transcriptome to influence spikelet architecture and flowering time. Curr. Biol. 2024, 34, 2330–2343. [Google Scholar] [CrossRef]
- Alvarez, M.A.; Li, C.; Lin, H.; Joe, A.; Padilla, M.; Woods, D.P.; Dubcovsky, J. EARLY FLOWERING 3 interactions with PHYTOCHROME B and PHOTOPERIOD1 are critical for the photoperiodic regulation of wheat heading time. PLoS Genet. 2023, 19, e1010655. [Google Scholar] [CrossRef]
- Chen, A.; Li, C.; Hu, W.; Lau, M.Y.; Lin, H.; Rockwell, N.C.; Martin, S.S.; Jernstedt, J.A.; Lagarias, J.C.; Dubcovsky, J. PHYTOCHROME C plays a major role in the acceleration of wheat flowering under long-day photoperiod. Proc. Natl. Acad. Sci. USA 2014, 111, 10037–10044. [Google Scholar] [CrossRef]
- Kippes, N.; VanGessel, C.; Hamilton, J.; Akpinar, A.; Budak, H.; Dubcovsky, J.; Pearce, S. Effect of phyB and phyC loss-of-function mutations on the wheat transcriptome under short and long day photoperiods. BMC Plant Biol. 2020, 20, 297. [Google Scholar] [CrossRef]
- Pearce, S.; Kippes, N.; Chen, A.; Debernardi, J.M.; Dubcovsky, J. RNA-seq studies using wheat PHYTOCHROME B and PHYTOCHROME C mutants reveal shared and specific functions in the regulation of flowering and shade-avoidance pathways. BMC Plant Biol. 2016, 16, 141. [Google Scholar] [CrossRef]
- Distelfeld, A.; Li, C.; Dubcovsky, J. Regulation of flowering in temperate cereals. Curr. Opin. Plant Biol. 2009, 12, 178–184. [Google Scholar] [CrossRef]
- Greenup, A.; Peacock, W.J.; Dennis, E.S.; Trevaskis, B. The molecular biology of seasonal flowering-responses in Arabidopsis and the cereals. Ann. Bot. 2009, 103, 1165–1172. [Google Scholar] [CrossRef]
- Xie, L.; Zhang, Y.; Wang, K.; Luo, X.; Xu, D.; Tian, X.; Li, L.; Ye, X.; Xia, X.; Li, W.; et al. TaVrt2, an SVP-like gene, cooperates with TaVrn1 to regulate vernalization-induced flowering in wheat. New Phytol. 2021, 23, 834–848. [Google Scholar] [CrossRef] [PubMed]
- Luo, X.; Liu, B.; Xie, L.; Wang, K.; Xu, D.; Tian, X.; Xie, L.; Li, L.; Ye, X.; He, Z.; et al. The TaSOC1-TaVRN1 module integrates photoperiod and vernalization signals to regulate wheat flowering. Plant Biotechnol. J. 2024, 22, 635–649. [Google Scholar] [CrossRef] [PubMed]
- Dixon, L.E.; Greenwood, J.R.; Bencivenga, S.; Zhang, P.; Cockram, J.; Mellers, G.; Ramm, K.; Cavanagh, C.; Swain, S.M.; Boden, S.A. TEOSINTE BRANCHED1 regulates inflorescence architecture and development in bread wheat (Triticum aestivum). Plant Cell 2018, 30, 563–581. [Google Scholar] [CrossRef] [PubMed]
- Shaw, L.M.; Lyu, B.; Turner, R.; Li, C.; Chen, F.; Han, X.; Fu, D.; Dubcovsky, J. FLOWERING LOCUS T2 regulates spike development and fertility in temperate cereals. J. Exp. Bot. 2019, 70, 193–204. [Google Scholar] [CrossRef]
- Li, C.; Lin, H.; Chen, A.; Lau, M.; Jernstedt, J.; Dubcovsky, J. Wheat VRN1, FUL2 and FUL3 play critical and redundant roles in spikelet development and spike determinacy. Development 2019, 146, dev175398. [Google Scholar] [CrossRef]
- Kobayashi, K.; Yasuno, N.; Sato, Y.; Yoda, M.; Yamazaki, R.; Kimizu, M.; Yoshida, H.; Nagamura, Y.; Kyozuka, J. Inflorescence meristem identity in rice is specified by overlapping functions of three AP1/FUL-Like MADS box genes and PAP2, a SEPALLATA MADS Box gene. Plant Cell 2012, 24, 1848–1859. [Google Scholar] [CrossRef]
- Wu, F.; Shi, X.; Lin, X.; Liu, Y.; Chong, K.; Theißen, G.; Meng, Z. The ABCs of flower development: Mutational analysis of AP1/FUL-like genes in rice provides evidence for a homeotic (A)-function in grasses. Plant J. 2017, 89, 310–324. [Google Scholar] [CrossRef]
- Li, K.; Debernardi, J.M.; Li, C.; Lin, H.; Zhang, C.; Jernstedt, J.; Korff, M.V.; Zhong, J.; Dubcovsky, J. Interactions between SQUAMOSA and SHORT VEGETATIVE PHASE MADS-box proteins regulate meristem transitions during wheat spike development. Plant Cell 2021, 33, 3621–3644. [Google Scholar] [CrossRef]
- Paraiso, F.; Lin, H.; Li, C.; Woods, D.P.; Lan, T.; Tumelty, C.; Debernardi, J.M.; Joe, A.; Dubcovsky, J. LEAFY and WAPO1 jointly regulate spikelet number per spike and floret development in wheat. Development 2024, 151, dev202803. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Burguener, G.F.; Paraiso, F.; Dubcovsky, J. Natural alleles of LEAFY and WAPO1 interact to regulate spikelet number per spike in wheat. Theor. Appl. Genet. 2024, 137, 257. [Google Scholar] [CrossRef] [PubMed]
- Lin, X.; Xu, Y.; Wang, D.; Yang, Y.; Zhang, X.; Bie, X.; Gui, L.; Chen, Z.; Ding, Y.; Mao, L.; et al. Systematic identification of wheat spike developmental regulators by integrated multi-omics, transcriptional network, GWAS, and genetic analyses. Mol. Plant 2024, 17, 438–459. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Cheng, X.; Liu, P.; Sun, J. miR156-targeted SBP-box transcription factors interact with DWARF53 to regulate teosinte branched1 and barren STALK1 expression in bread wheat. Plant Physiol. 2017, 174, 1931–1948. [Google Scholar] [CrossRef]
- Dixon, L.E.; Pasquariello, M.; Badgami, R.; Levin, K.A.; Poschet, G.; Ng, P.Q.; Orford, S.; Chayut, N.; Adamski, N.M.; Brinton, J.; et al. MicroRNA-resistant alleles of HOMEOBOX DOMAIN-2 modify inflorescence branching and increase grain protein content of wheat. Sci. Adv. 2022, 8, eabn5907. [Google Scholar] [CrossRef]
- Jiang, D.; Hua, L.; Zhang, C.; Li, H.; Wang, Z.; Li, J.; Wang, G.; Song, R.; Shen, T.; Li, H.; et al. Mutations in the miRNA165/166 binding site of the HB2 gene result in pleiotropic effects on morphological traits in wheat. Crop J. 2023, 11, 9–20. [Google Scholar] [CrossRef]
- Zhang, X.; Meng, W.; Liu, D.; Pan, D.; Yang, Y.; Chen, Z.; Ma, X.; Yin, W.; Niu, M.; Dong, N.; et al. Enhancing rice panicle branching and grain yield through tissue-specific brassinosteroid inhibition. Science 2024, 383, eadk8838. [Google Scholar] [CrossRef]
- Boden, S.A.; Cavanagh, C.; Cullis, B.R.; Ramm, K.; Greenwood, J.; Jean Finnegan, E.; Trevaskis, B.; Swain, S.M. Ppd-1 is a key regulator of inflorescence architecture and paired spikelet development in wheat. Nat. Plants 2015, 1, 14016. [Google Scholar] [CrossRef]
- Wang, Y.; Du, F.; Wang, J.; Wang, K.; Tian, C.; Qi, X.; Lu, F.; Liu, X.; Ye, X.; Jiao, Y. Improving bread wheat yield through modulating an unselected AP2/ERF gene. Nat. Plants 2022, 8, 930–939. [Google Scholar] [CrossRef]
- Clark, R.M.; Wagler, T.N.; Quijada, P.; Doebley, J. A distant upstream enhancer at the maize domestication gene tb1 has pleiotropic effects on plant and inflorescent architecture. Nat. Genet. 2006, 38, 594–597. [Google Scholar] [CrossRef]
- Studer, A.J.; Wang, H.; Doebley, J.F. Selection during maize domestication targeted a gene network controlling plant and inflorescence architecture. Genetics 2017, 207, 755–765. [Google Scholar] [CrossRef]
- Studer, A.; Zhao, Q.; Ross-Ibarra, J.; Doebley, J. Identification of a functional transposon insertion in the maize domestication gene tb1. Nat. Genet. 2011, 43, 1160–1163. [Google Scholar] [CrossRef]
- Studer, A.J.; Doebley, J.F. Evidence for a Natural Allelic Series at the Maize Domestication Locus teosinte branched1. Genetics 2012, 191, 951–958. [Google Scholar] [CrossRef][Green Version]
- Chuck, G.; Muszynski, M.; Kellogg, E.; Hake, S.; Schmidt, R.J. The control of spikelet meristem identity by the branched silkless1 gene in maize. Science 2002, 298, 1238–1241. [Google Scholar] [CrossRef] [PubMed]
- Komatsu, M.; Chujo, A.; Nagato, Y.; Shimamoto, K.; Kyozuka, J. Frizzy panicle is required to prevent the formation of axillary meristems and to establish floral meristem identity in rice spikelets. Development 2003, 130, 3841–3850. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Shi, F.; Wang, Y.; Yu, X.; Zhi, J.; Guan, Y.; Zhao, H.; Chang, J.; Chen, M.; Yang, G.; et al. TaSPL13 regulates inflorescence architecture and development in transgenic wheat (Triticum aestivum L.). Plant Sci. 2020, 296, 110516. [Google Scholar] [CrossRef] [PubMed]
- Gupta, A.; Hua, L.; Zhang, Z.; Yang, B.; Li, W. CRISPR-induced miRNA156-recognition element mutations in TaSPL13 improve multiple agronomic traits in wheat. Plant Biotechnol. J. 2023, 21, 536–548. [Google Scholar] [CrossRef]
- Feng, N.; Song, G.; Guan, J.; Chen, K.; Jia, M.; Huang, D.; Wu, J.; Zhang, L.; Kong, X.; Geng, S.; et al. Transcriptome profiling of wheat inflorescence development from spikelet initiation to floral patterning identified stage-specific regulatory Genes. Plant Physiol. 2017, 174, 1779–1794. [Google Scholar] [CrossRef]
- Su, Y.; Liu, J.; Liang, W.; Dou, Y.; Fu, R.; Li, W.; Feng, C.; Gao, C.; Zhang, D.; Kang, Z.; et al. Wheat AGAMOUS like 6 transcription factors function in stamen development by regulating the expression of Ta APETALA3. Development 2019, 146, dev177527. [Google Scholar] [CrossRef]
- Debernardi, J.M.; Greenwood, J.R.; Jean Finnegan, E.; Jernstedt, J.; Dubcovsky, J. APETALA 2-like genes AP2L2 and Q specify lemma identity and axillary floral meristem development in wheat. Plant J. 2020, 101, 171–187. [Google Scholar] [CrossRef]
- Zuo, J.; Li, J. Molecular dissection of complex agronomic traits of rice: A team effort by Chinese scientists in recent years. Natl. Sci. Rev. 2014, 1, 253–276. [Google Scholar] [CrossRef]
- Ai, G.; He, C.; Bi, S.; Zhou, Z.; Liu, A.; Hu, X.; Liu, Y.; Jin, L.; Zhou, J.; Zhang, H.; et al. Dissecting the molecular basis of spike traits by integrating gene regulatory networks and genetic variation in wheat. Plant Commun. 2024, 5, 100879. [Google Scholar] [CrossRef]
- Chen, Y.; Guo, Y.; Guan, P.; Wang, Y.; Wang, X.; Wang, Z.; Qin, Z.; Ma, S.; Xin, M.; Hu, Z.; et al. A wheat integrative regulatory network from large-scale complementary functional datasets enables trait-associated gene discovery for crop improvement. Mol. Plant 2023, 16, 393–414. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Abrouk, M.; Gourdoupis, S.; Koo, D.H.; Karafiátová, M.; Molnár, I.; Holušová, K.; Doležel, J.; Athiyannan, N.; Cavalet-Giorsa, E.; et al. An unusual tandem kinase fusion protein confers leaf rust resistance in wheat. Nat. Genet. 2023, 55, 914–920. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Liu, W.; Xu, J.; Huang, X.; Yang, L.; Xu, F. Decoding maize meristems maintenance and differentiation: Integrating single-cell and spatial omics. J. Genet. Genom. 2025, 52, 319–333. [Google Scholar] [CrossRef]
- Wang, Y.; Luo, Y.; Guo, X.; Li, Y.; Yan, J.; Shao, W.; Wei, W.; Wei, X.; Yang, T.; Chen, J.; et al. A spatial transcriptome map of the developing maize ear. Nat. Plants 2024, 10, 815–827. [Google Scholar] [CrossRef]
- Xu, X.; Crow, M.; Rice, B.R.; Li, F.; Harris, B.; Liu, L.; Demesa-Arevalo, E.; Lu, Z.; Wang, L.; Fox, N.; et al. Single-cell RNA sequencing of developing maize ears facilitates functional analysis and trait candidate gene discovery. Dev. Cell 2021, 56, 557–568. [Google Scholar] [CrossRef]
- Qin, X.; Zhang, F.; Liu, C.; Yu, H.; Cao, B.; Tian, S.; Liao, Y.; Siddique, K. Wheat yield improvements in China: Past trends and future directions. Field Crops Res. 2015, 177, 117–124. [Google Scholar] [CrossRef]
- Jian, C.; Pan, Y.; Liu, S.; Guo, M.; Huang, Y.; Cao, L.; Zhang, W.; Yan, L.; Zhang, X.; Hou, J.; et al. The TaGW2-TaSPL14 module regulates the trade-off between tiller number and grain weight in wheat. J. Integr. Plant Biol. 2024, 66, 1953–1965. [Google Scholar] [CrossRef]
- Xie, Q.; Sparkes, D.L. Dissecting the trade-off of grain number and size in wheat. Planta 2021, 254, 3. [Google Scholar] [CrossRef]
- Vicentin, L.; Canales, J.; Calderini, D.F. The trade-off between grain weight and grain number in wheat is explained by the overlapping of the key phases determining these major yield components. Front. Plant Sci. 2024, 15, 1380429. [Google Scholar] [CrossRef]
- Cell Editorial Team. Five decades of genetics and genomics. Cell 2024, 187, 1017–1018. [Google Scholar] [CrossRef]
- Wei, W.; Gao, C. Gene editing: From technologies to applications in research and beyond. Sci. China Life Sci. 2022, 65, 657–659. [Google Scholar] [CrossRef]
- Pacesa, M.; Pelea, O.; Jinek, M. Past, present, and future of CRISPR genome editing technologies. Cell 2024, 187, 1076–1100. [Google Scholar] [CrossRef] [PubMed]
- Bastos, L.M.; Carciochi, W.; Lollato, R.P.; Jaenisch, B.R.; Rezende, C.R.; Schwalbert, R.; Vara Prasad, P.V.; Zhang, G.; Fritz, A.K.; Foster, C.; et al. Winter Wheat Yield Response to Plant Density as a Function of Yield Environment and Tillering Potential: A Review and Field Studies. Front. Plant Sci. 2020, 11, 00054. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Wang, Q.; Liu, Y.; He, J.; Chen, W.; Xing, J.; Sun, M.; Gao, Z.; Wang, Z.; Zhang, M.; et al. Optimal water, nitrogen, and density management increased wheat yield by improving population uniformity. Agric. Water Manag. 2025, 310, 109362. [Google Scholar] [CrossRef]
- Yang, D.; Cai, T.; Luo, Y.; Wang, Z. Optimizing plant density and nitrogen application to manipulate tiller growth and increase grain yield and nitrogen-use efficiency in winter wheat. PeerJ 2019, 7, e6484. [Google Scholar] [CrossRef]
- Wyzińska, M.; Grabiński, J. The influence of autumn sowing date on the productivity of spring wheat (Triticum aestivum L.). Res. Rural. Dev. 2018, 2, 35–41. [Google Scholar] [CrossRef]
- Wyzińska, M.; Grabiński, J. The productivity of spring wheat (Triticum aestivum L.) on the autumn sowing date. Curr. Agron. 2020, 42, 51–56. [Google Scholar]
- Wen, P.; Wei, Q.; Zheng, L.; Rui, Z.; Niu, M.; Gao, C.; Guan, X.; Wang, T.; Xiong, S. Adaptability of wheat to future climate change: Effects of sowing date and sowing rate on wheat yield in three wheat production regions in the North China Plain. Sci. Total Environ. 2023, 901, 165906. [Google Scholar] [CrossRef]


| Traits | No. of QTLs | No. of Clustes | Chr Distribution |
|---|---|---|---|
| Spike length (SL) | 225 | 126 | 21 |
| Spike compactness (SC) | 99 | 49 | 17 (except chr1A/4D/6D/7D) |
| Grain number (GNS) | 525 | 347 | 21 |
| Spikelet number (SNS) | 309 | 221 | 21 |
| Chr | Spike Length Clusters | Interval (Mb) (Ref V1.0) | PVE (%) | Spike Compactness Clusters | Interval (Mb) (Ref V1.0) | PVE (%) | Genes |
|---|---|---|---|---|---|---|---|
| 1B | SL-cluster1B.4 | 634.2–634.3 | SC-cluster1B.2 | 638.14–656.9 | 6.04 | ||
| 2A | SL-cluster2A.5 | 747.6–753.1 | SC-cluster2A.2 | 754.17–755.9 | 4.42 | TaARF12-2A | |
| 2B | SL-cluster2B.9 | 739.12–776.5 | SC-cluster2B.2/2B.3 | 741.6–799.25 | 1.53–12.74 | TaARF12-2B | |
| 2D | SL-cluster2D.1 | 17.61–32.9 | 13–37.5 | SC-cluster2D.1 | 14.36–35.02 | 12.55–22.4 | Rht8/RNHL-D1 |
| 3A | SL-cluster3A.3 | 711.2 | SC-cluster3A.2 | 711.3–711.65 | 4.23 | ||
| 3B | SL-cluster3B.2 | 25.4–26.5 | SC-cluster3B.1 | 25.4–26.5 | |||
| 3B | SL-cluster3B.7 | 577.8–593.7 | SC-cluster3B.2 | 531.4–636.44 | 13.21 | TaLAX1-3B | |
| 3D | SL-cluster3D.1 | 1.11–4.49 | SC-cluster3D.1 | 2.2–2.4 | |||
| 3D | SL-cluster3D.2 | 30.37 | 10.53 | SC-cluster3D.2 | 30.37 | 11.12 | |
| 4A | SL-cluster4A.5 | 698.74–745.95 | SC-cluster4A.1 | 713.52–719.31 | 4.21 | ||
| 4B | SL-cluster4B.1 | 22.56–44.53 | 10.39–14.54 | SC-cluster4B.1 | 31.89–37.17 | 2.44–11.23 | Rht-B1b, ZnF-B |
| 5A | SL-cluster5A.1 | 13.85–43.43 | 4.87–10.32 | SC-cluster5A.1 | 19.24–37.98 | 10.42 | |
| 5A | SL-cluster5A.5 | 468.03–540.1 | 11.8–26.6 | SC-cluster5A.3 | 478.6–541.2 | 12.02–26.6 | |
| 5A | SL-cluster5A.7 | 668.41–710.42 | SC-cluster5A.5 | 690.42–702.19 | 3.56–6.78 | TaVRN2, WSOC1-5A | |
| 5B | SL-cluster5B.2 | 388.5–406.9 | 11 | SC-cluster5B.2 | 388.5–406.9 | ||
| 5B | SL-cluster5B.5 | 536.05–550.23 | SC-cluster5B.4 | 538.29–590.15 | 3.42–35.62 | ||
| 6A | SL-cluster6A.3 | 50.28–63.58 | 7.13–9.37 | SC-cluster6A.1 | 50.28–77.5 | 11.89 | |
| 6A | SL-cluster6A.5 | 548.08–554.04 | 11.4 | SC-cluster6A.3 | 533.24–616.97 | 11.85–12.98 | SVP1-6A, TaPIN1-6A |
| 6B | SL-cluster6B.5 | 654.19–702.3 | 19.1 | SC-cluster6B.2 | 701.61–703.3 | 7.19–13.04 | TaTPR-B1 |
| 7A | SL-cluster7A.2 | 68.9–85.6 | SC-cluster7A.2 | 82.38–84.78 | 1.66–5.94 | ||
| 7A | SL-cluster7A.4 | 643.09–683.39 | SC-cluster7A.5 | 678.47–682.29 | 23.14 | WAPO1-7A | |
| 7B | SL-cluster7B.5 | 686.16–711.11 | 7.11 | SC-cluster7B.1 | 680.85–687.74 | 12 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ji, Z.; Liu, X.; Yan, F.; Wu, S.; Du, Y. The Genetic Basis of Wheat Spike Architecture. Agriculture 2025, 15, 1575. https://doi.org/10.3390/agriculture15151575
Ji Z, Liu X, Yan F, Wu S, Du Y. The Genetic Basis of Wheat Spike Architecture. Agriculture. 2025; 15(15):1575. https://doi.org/10.3390/agriculture15151575
Chicago/Turabian StyleJi, Zhen, Xin Liu, Fei Yan, Shouqing Wu, and Yanfang Du. 2025. "The Genetic Basis of Wheat Spike Architecture" Agriculture 15, no. 15: 1575. https://doi.org/10.3390/agriculture15151575
APA StyleJi, Z., Liu, X., Yan, F., Wu, S., & Du, Y. (2025). The Genetic Basis of Wheat Spike Architecture. Agriculture, 15(15), 1575. https://doi.org/10.3390/agriculture15151575






