Cloning and Functional Validation of the Candidate Gene LuWRKY39 Conferring Resistance to Septoria linicola (Speg.) Garassini from Flax
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials, Inoculation, and Treatments
2.2. Total RNA Extraction and cDNA Synthesis
2.3. Cloning and Sequence Analysis of the LuWRKY39 Gene
2.4. Expression Analysis of LuWRKY39 Gene
2.5. Real-Time Quantitative PCR (qRT-PCR) Analysis
2.6. Virus-Induced Gene Silencing (VIGS)
2.7. Statistical Analysis
3. Results
3.1. Cloning and Sequence Analysis of LuWRKY39
3.2. Subcellular Localization of the LuWRKY39 Protein
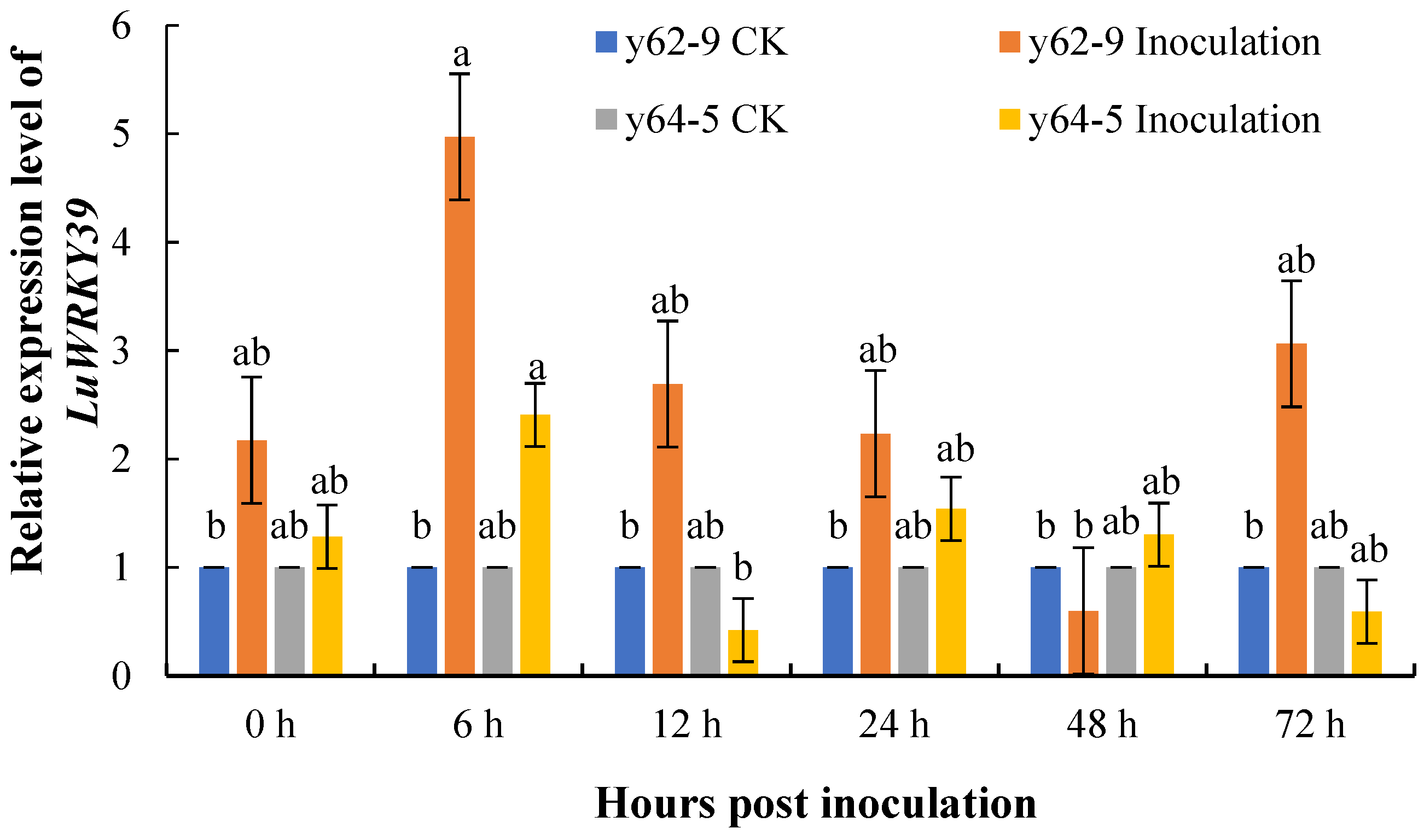
3.3. Expression Level Analysis of LuWRKY39 Induced by S. linicola
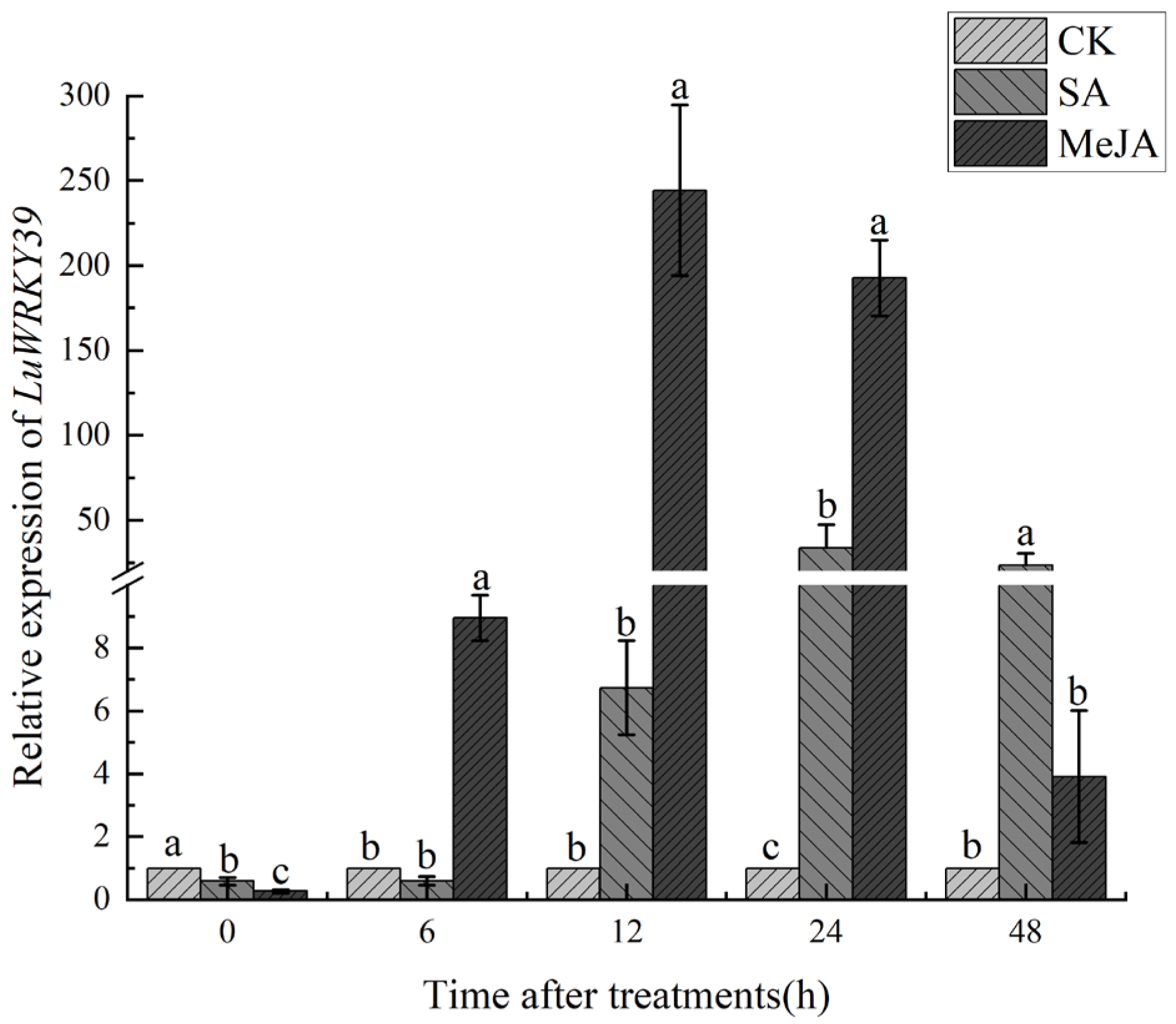
3.4. Expression Level Analysis of LuWRKY39 Induced by Exogenous Hormones
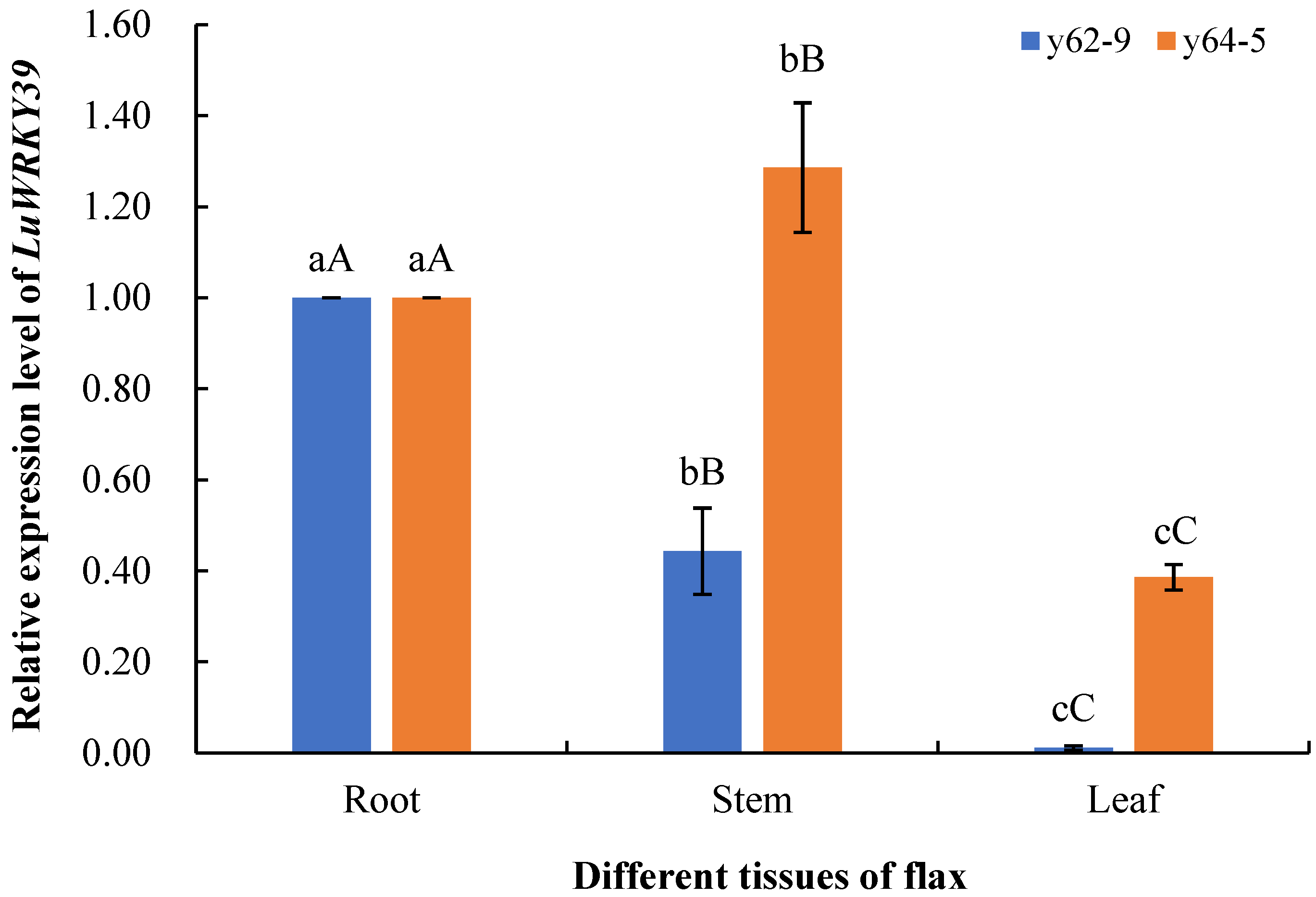
3.5. Expression Level Analysis of LuWRKY39 in Different Tissues
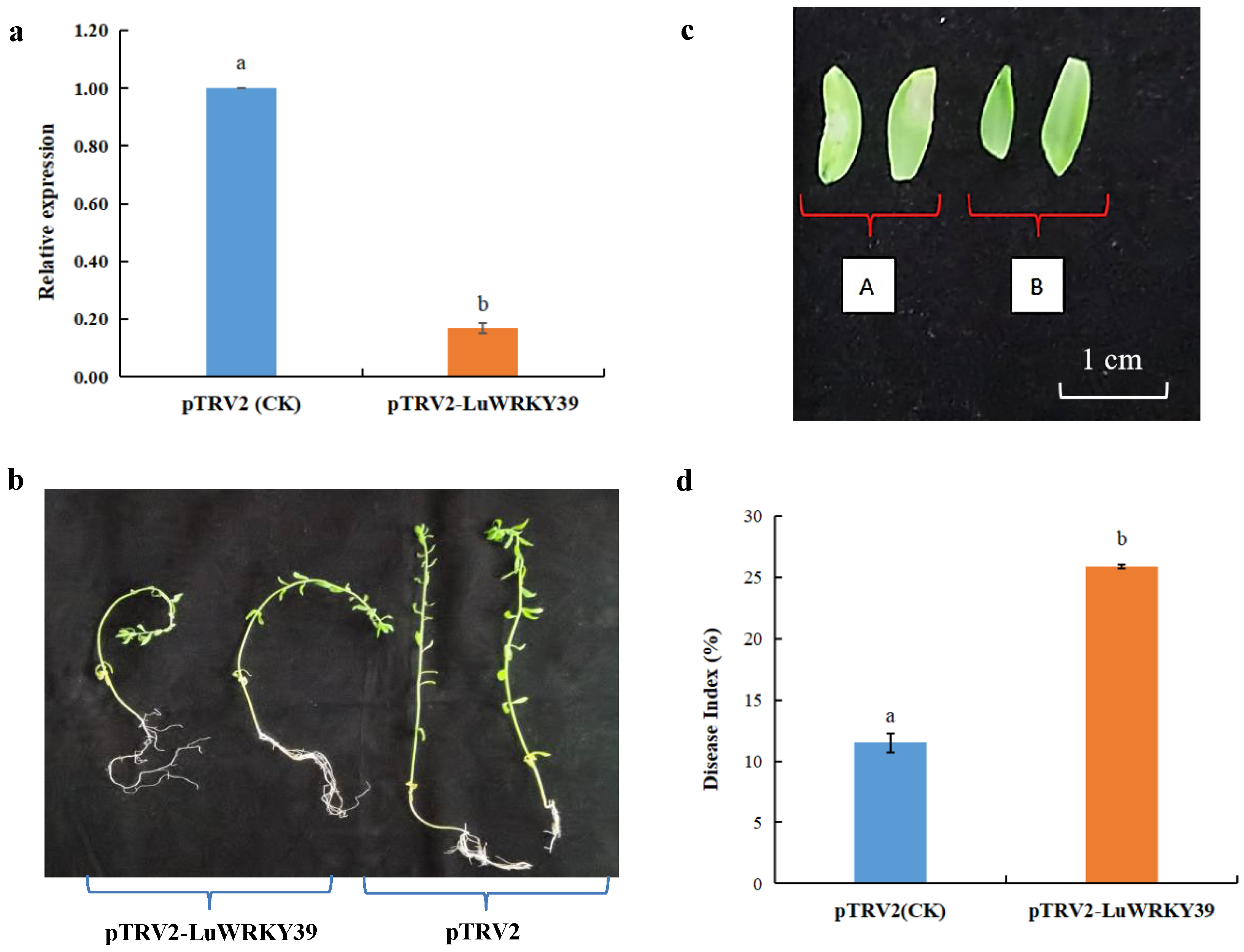
3.6. LuWRKY39-Silenced Plants Exhibited Greater Sensitivity to S. linicola
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Chen, S.; Li, Z.G.; Wu, G.W.; Yuan, H.M.; Huang, W.G.; Liu, Y.; Yao, Y.B.; Wu, G.W. Screening pasmo-resistant germplasm resources from flax varieties (lines). Crops 2019, 35, 63–67. [Google Scholar] [CrossRef]
- Pan, H.; Wu, G.W.; Song, X.X.; Huang, W.G.; Chen, H.; Zhao, D.S.; Jiang, W.D.; Kang, Q.H.; Liu, Y.; Guan, F.Z. Progress in physiological race of Fusarium oxysporum Schl.f.sp.lini. Plant Fiber Sci. China 2011, 33, 100–104. [Google Scholar]
- Yang, X. The study on occurrence characteristics and preventing technique of flax pasmo disease. Plant Fibers Prod. 2004, 26, 170–172. [Google Scholar] [CrossRef]
- Zhang, Q.; Jiang, G.; Yang, X.; Qu, Z.H.; Li, Y.; Lu, Y.; Duan, H. Analysis of genetic relationship among 17 flax varieties by using SSR markers. Chin. Agric. Sci. Bull. 2014, 30, 211–216. [Google Scholar] [CrossRef]
- Xie, D.W.; Lu, Y.; Zhao, D.B.; Yang, X.; Su, J.G.; Sun, J. Genome-wide analysis of NBS resistance genes in flax. Plant Fiber Sci. China 2015, 37, 113–119, 125. [Google Scholar] [CrossRef]
- Jaber, R.; Planchon, A.; Mathieu-Rivet, E.; Kiefer-Meyer, M.; Zahid, A.; Plasson, C. Identification of two compounds able to improve flax resistance towards Fusarium oxysporum infection. J. Plant Sci. 2020, 301, 110690. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Yuan, H.M.; Yang, X.; Chen, L.; Chen, J.; Liu, Y.; Wu, L.L.; Hu, Y.Y.; Huang, W.G.; Yao, Y.B.; et al. Identification and analysis of flax resistance genes to Septoria linicola (Speg.) Garassini. J. Nat. Fibers 2023, 20, 2163331. [Google Scholar] [CrossRef]
- Long, S.H.; Li, X.; Chen, X.B.; Qiao, R.Q.; Deng, X.; Qiu, C.S.; Guo, Y.; Hao, D.; Wang, Y. RACE clone and RNAi transformation of flax 4CL gene. Acta Bot. Boreal.-Occident. Sin. 2014, 34, 2405–2411. [Google Scholar] [CrossRef]
- Chen, X.J. Clone of the LuBR11 and LuBES1 Gene’s Core Fragements and Establishment of Regeneration System of Flax (Linum usitatissinmum L.). Master’s Thesis, Xinjiang University, Xinjiang, China, 2013. [Google Scholar]
- Wang, Y.Y. Cloning and Functional Analysis of Flax Transcription Factor ABI3 in Flax (Linum usitatissinmum L.). Master’s Dissertation, Huazhong Agricultural University, Wuhan, China, 2016. [Google Scholar]
- Jiang, H.X. Clone and Functional Analysis of BRs Related Genes in Flax (Linum usitassimum L.). Master’s Thesis, Xinjiang University, Xinjiang, China, 2017. [Google Scholar]
- Rushton, P.J.; Somssich, I.E.; Ringler, P.; Shen, Q.J. WRKY transcription factors. Trends Plant Sci. 2010, 15, 247–258. [Google Scholar] [CrossRef] [PubMed]
- Jiang, J.J.; Ma, S.H.; Ye, N.H.; Jiang, M.; Cao, J.S.; Zhang, J.H. WRKY transcription factors in plant responses to stresses. J. Integr. Plant Biol. 2017, 59, 86–101. [Google Scholar] [CrossRef] [PubMed]
- Lai, Z.B.; Vinod, K.; Zheng, Z.Y.; Fan, B.F.; Chen, Z.X. Roles of Arabidopsis WRKY3 and WRKY4 transcription factors in plant responses to pathogens. BMC Plant Biol. 2008, 8, 68. [Google Scholar] [CrossRef] [PubMed]
- Yin, L.H.; Yang, Y.; Ke, X.W.; Cui, S.P.; Zhang, J.P.; Zuo, Y.H. Cloning of VaWRKY33 gene from adzuki bean and expression analysis in response to adzuki bean rust infection. Mol. Plant Breed. 2020, 18, 7301–7308. [Google Scholar] [CrossRef]
- Tao, Z.; Liu, H.B.; Qiu, D.Y.; Zhou, Y.; Wang, S. A pair of allelic WRKY genes play opposite roles in rice-bacteria interactions. Plant Physiol. 2009, 151, 936–948. [Google Scholar] [CrossRef] [PubMed]
- Tang, Y.; Kuang, J.-F.; Wang, F.-Y.; Chen, L.; Hong, K.-Q.; Xiao, Y.-Y.; Xie, H.; Lu, W.-J.; Chen, J.-Y. Molecular characterization of PR and WRKY genes during SA- and MeJA-induced resistance against Colletotrichum musae in banana fruit. Postharvest Biol. Technol. 2013, 79, 62–68. [Google Scholar] [CrossRef]
- Wang, X.L.; Yan, Y.; Li, Y.Z.; Chu, X.Q.; Wu, C.A.; Guo, X.Q. GhWRKY40, a multiple stress-responsive cotton WRKY gene, plays an important role in the wounding response and enhances susceptibility to Ralstonia solanacearum infection in transgenic Nicotiana benthamiana. PLoS ONE 2014, 9, e93577. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Li, J.J.; Guo, X.F.; Ma, Y. PlWRKY13: A transcription factor involved in abiotic and biotic stress responses in Paeonia lactiflora. Int. J. Mol. Sci. 2019, 20, 5953. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Li, J.J.; Guo, J.; Qiao, Q.; Ma, Y. The WRKY transcription factor PlWRKY65 enhances the resistance of Paeonia lactiflora (herbaceous peony) to Alternaria tenuissima. Hortic. Res. 2020, 7, 57. [Google Scholar] [CrossRef] [PubMed]
- Burch-Smith, T.M.; Anderson, J.C.; Martin, G.B.; Dinesh-Kumar, S.P. Applications and advantages of virus-induced gene silencing for gene function studies in plants. Plant J. 2004, 39, 734–746. [Google Scholar] [CrossRef] [PubMed]
- Holzberg, S.; Brosio, P.; Gross, C.; Pogue, G.P. Barley stripe mosaic virus-induced gene silencing in a monocot plant. Plant J. 2002, 30, 315–327. [Google Scholar] [CrossRef] [PubMed]
- Atamian, H.S.; Eulgem, T.; Kaloshian, I. SlWRKY70 is required for Mi-1-mediated resistance to aphids and nematodes in tomato. Planta 2012, 235, 299–309. [Google Scholar] [CrossRef] [PubMed]
- Guo, W.L. Response to Chilling Stress in Pepper as Well as Characterization and Functional Analysis of Chilling-Related Genes. Ph.D. Dissertation, Northwest A&F University, Xianyang, China, 2013. [Google Scholar]
- Wang, J.E. Expression Analysis and Functional Identification of Carga1 and Capod Genes Induced by Phytophthora Capsici in Pepper. Ph.D. Dissertation, Northwest A&F Univeresity, Xianyang, China, 2013. [Google Scholar]
- Zhou, Y. Progresses in study of virus-based vectors of fruit trees. Sci. Agric. Sin. 2014, 47, 1119–1127. [Google Scholar] [CrossRef]
- Huang, B.Y.; Ji, W.Q.; Guo, A.G.; Sadequr, R.; Li, Z.Y. Post-transcriptional gene silencing (PTGS) and its application to crop genetic improvement. China Biotechnol. 2005, 25, 1–5. [Google Scholar] [CrossRef]
- Aihaiti, A.; Yao, Z.P.; Chen, Q.J.; Huang, Q.X.; Han, Y.H.; Rebiguli, R.; Qu, Y.Y. Cloning and functional analyses of fusarium wilt resistanse gene GbMPK16 in Gossypium barbadense L. Genom. Appl. Biol. 2019, 38, 5082–5090. [Google Scholar]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Yu, D.Q.; Chen, C.H.; Chen, Z.X. Evidence for an important role of WRKY DNA binding proteins in the regulation of NPR1 gene expression. Plant Cell 2001, 13, 1527–1540. [Google Scholar] [CrossRef] [PubMed]
- Shen, Q.H.; Saijo, Y.; Mauch, S.; Biskup, C.; Bieri, S.; Keller, B.; Seki, H.; Ulker, B.; Somssich, I.E.; Schulze-Lefert, P. Nuclear activity of MLA immune receptors links isolate-specific and basal disease-resistance responses. Science 2007, 315, 1098–1103. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Q.Q.; Qiu, H.R.; He, X.W.; Wang, X.P.; Liu, X.X.; Li, B.H.; Wu, S.J.; Chen, X.S. MdWRKY40 mediated improvement of the immune resistance of apple and Arabidopsis thaliana to Botryosphaeria dothidea. Sci. Agric. Sin. 2018, 51, 4052–4064. [Google Scholar] [CrossRef]
- Ulker, B.; Somssich, I.E. WRKY transcription factors: From DNA binding towards biological function. Curr. Opin. Plant Biol. 2004, 7, 491–498. [Google Scholar] [CrossRef] [PubMed]
- Hsu, F.C.; Chou, M.Y.; Chou, S.J.; Li, Y.R.; Shih, M.C. Submergence confers immunity mediated by the WRKY22 transcription factor in Arabidopsis. Plant Cell 2013, 25, 2699–2713. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Zhong, R.S.; Palva, E.T. WRKY70 and its homolog WRKY54 negatively modulate the cell wall-associated defenses to necrotrophic pathogens in Arabidopsis. PLoS ONE 2017, 12, e0183731. [Google Scholar] [CrossRef] [PubMed]
- Hara, K.; Yagi, M.; Kusano, T.; Sano, H. Rapid systemic accumulation of transcripts encoding a tobacco WRKY transcription factor on wounding. Mol. Gen. Genet. 2000, 263, 30–37. [Google Scholar] [CrossRef] [PubMed]
- Kim, C.Y.; Lee, S.H.; Park, H.C.; Bae, C.G.; Cheong, Y.H.; Choi, Y.J.; Han, C.-D.; Lee, S.Y.; Lim, C.O.; Cho, M.J. Identification of rice blast fungal elicitor-responsive genes by differential display analysis. Mol. Plant–Microbe Interact. 2000, 13, 470–474. [Google Scholar] [CrossRef] [PubMed]
- Cormack, R.S.; Eulgem, T.; Rushton, P.J.; Petra Köchner Hahlbrock, K.; Somssich, I.E. Leucine zipper-containing WRKY proteins widen the spectrum of immediate early elicitor-induced WRKY transcription factors in parsley. Biochim. Biophys. Sci. Acta 2002, 1576, 92–100. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.H.; Wang, J.W.; Wang, S.; Wang, J.Y.; Chen, X.Y. Characterization of GaWRKYl, a cotton transcription factorthat regulates the sesquiterpene synthase gene (+)-delta-cadinene synthase-A. Plant Physiol. 2004, 135, 507–515. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.Y.; Su, Y.X.; Zuo, T.; Chang, J.P.; Guo, L.M.; He, W. The differential expression analysis of the SA and JA signal transduction related genes in poplar varieties susceptible and resistant to canker. Chin. Agric. Sci. Bull. 2015, 31, 156–163. [Google Scholar] [CrossRef]
- Sun, H.; Wang, L.; Zhang, B.; Ma, J.; Christian, H.; Cao, G.; Sun, G.; Wu, J.; Wu, J. Scopoletin is a phytoalexin against Alternaria alternata in wild tobacco dependent on jasmonate signalling. J. Exp. Bot. 2014, 15, 4305–4315. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Wu, J.S. Scopolin, a glycoside form of the phytoalexin scopoletin, is likely involved in the resistance of Nicotiana attenuata against Alternaria alternata. J. Plant Pathol. 2016, 98, 641–644. [Google Scholar] [CrossRef]
- Sun, H.; Song, N.; Ma, L.; Li, J.; Ma, L.; Wu, J. Ethylene signalling is essential for the resistance of Nicotiana attenuata against Alternaria alternata and phytoalexin scopoletin biosynthesis. Plant Pathol. 2017, 66, 277–284. [Google Scholar] [CrossRef]
- Jiang, S.S.; Yao, J.; Ma, K.W.; Zhou, H.B.; Song, J.K.; He, S.Y.; Ma, W.B. Bacterial effector activates jasmonate signaling by directly targeting JAZ transcriptional repressors. PLoS Pathog. 2013, 9, e1003715. [Google Scholar] [CrossRef] [PubMed]
- Gimenez-Lbanez, S.; Boter, M.; Fernández-Barbero, G.; Chini, A.; Rathjen, J.; Solano, R. The bacterial effector HopX1 targets JAZ transcriptional repressors to activate jasmonate signaling and promote infection in Arabidopsis. PLoS Biol. 2014, 12, e1001792. [Google Scholar] [CrossRef]
- Yang, D.C.; Zhang, Y.X.; Zheng, G.S. Gene cloning and expression analysis of pathogenesis-related Protein 1 of Paeonia suffruticosa. Acta Hortic. Sin. 2013, 40, 1583–1590. [Google Scholar] [CrossRef]
- Li, X.H.; Yang, R.; Chen, H.M. The Arabidopsis thaliana mediator subunit MED8 regulates plant immunity to Botrytis Cinerea through interacting with the basic helix-loop-helix (bHLH) transcription factor FAMA. PLoS ONE 2018, 13, e193458. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.J.; Qian, Y.J.; Li, Z.H.; Zhou, X.P. Virus-induced gene silencing and its application in plant functional genomics. Sci. China (Life Sci.) 2012, 55, 99–108. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.L.; Zhao, J. Progress in study of plant resistance gene function using virus induced gene silence system. Chin. Potato J. 2013, 27, 181–186. [Google Scholar] [CrossRef]
- Zou, S.; Wang, H.; Li, Y.; Kong, Z.; Tang, D. The NB-LRR gene Pm60 confers powdery mildew resistance in wheat. New Phytol. 2017, 218, 14964. [Google Scholar] [CrossRef] [PubMed]
- Hou, W.Q. Identification and BSMV-VIGS Analysis of Candidate Genes Resistant to Fusarium Head Blight. Ph.D. Dissertation, Shandong Agricultural University, Tai’an, China, 2014. [Google Scholar] [CrossRef]
- Jin, Y.; Zhang, T.; Guo, H.S. Application of host induced gene silencing in crop protection against fungal diseases. Chin. J. Biotechnol. 2017, 33, 161–169. [Google Scholar] [CrossRef]
- Ma, M. Development of an Effective System of Virus Induced Gene Silencing by Barley Stripe Mosaic Virus (BSMV) in Wheat Spikes. Master’s Dissertation, Northwest A&F University, Xianyang, China, 2010. [Google Scholar]
- Tsaballa, A.; Pasentsis, K.; Darzentas, N.; Tsaftaris, A.S. Multiple evidence for the role of an Ovate-like gene in determining fruit shape in pepper. BMC Plant Biol. 2011, 11, 46. [Google Scholar] [CrossRef] [PubMed]
- González, M.; Salazar, E.; Castillo, J.; Morales, P.; Mura-Jornet, I.; Maldonado, J.; Silva, H.; Carrasco, B. Genetictructure based on EST–SSR: A putative tool for fruit color selection in Japanese plum (Prunus salicina L.) breeding programs. Mol. Breed. 2016, 36, 1–15. [Google Scholar] [CrossRef]
- Vadim, G.L.; Natalya, M.S.; Oleg, P.M.; Tatyana, N.L.; Konstantin, V.K.; Konstantin, A.S. Transferability and polymorphism of SSR markers located in flavonoid pathway genes in Fragaria and Rubus Species. Genes 2019, 11, 11. [Google Scholar] [CrossRef] [PubMed]
- Liu, N.; Cheng, F. Association mapping for yield traits in Paeonia rockii based on SSR markers within transcription factors of comparative transcriptome. BMC Plant Biol. 2020, 20, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Biswas, M.K.; Nath, U.K.; Howlader, J.; Bagchi, M.; Natarajan, S.; Kayum, M.A.; Kim, H.-T.; Park, H.-T.; Kang, J.G.; Nou, I.-S. Exploration and exploitation of novel SSR markers for candidate transcription factor genes in Lilium Species. Genes 2018, 9, 97. [Google Scholar] [CrossRef] [PubMed]



| Primer Name | Nucleotide Sequence (5′–3′) | Purpose |
|---|---|---|
| LuGAPDH-F | AGGTTCTTCCCGCTCTCAAT | Actin primers |
| LuGAPDH-R | CCTCCTTGATAGCAGCCTTG | |
| LuWRKY39F | GCCATGGAGGCCAGTGAATTCATGGAAGGAGTTGAGGAAGCAAG | Full-length DNA and cDNA amplification |
| LuWRKY39R | CAGCTCGAGCTCGATGGATCCTTATGTAAGTGCAGACATTGAAGAAG | |
| LuWRKY39qF | GGGCTCATCTGTCACCCAAT | Specific primer for qRT-PCR |
| LuWRKY39qR | GAATCTGCCACGACCAACTAAG | |
| LuWRKY39(E)F | GTGAGTAAGGTTACCGAATTCCAGATTGGTTTGTATGGTATTCAT | Vector construction for VIGS |
| LuWRKY39(B)R | CGTGAGCTCGGTACCGGATCCCTGCTGCACTTGTAATATCC | |
| LuWRKY39(I)F | GTGTCAACAAAGATGGACATTGTT | Colony PCR identification |
| LuWRKY39(I)R | TAAAACTTCAGACACGGATCTACTT |
| Gene Name | LuWRKY39 | |
| Gene ID | Lus10021999.g (PAC:23160245) | |
| Amino acids | 315 | |
| Protein molecular weight/kDa | 34.97 | |
| Isoelectric point (pI) | 9.58 | |
| Instability Index | 52.41 | |
| Aliphatic Index | 64.98 | |
| GRAVY | −0.537 | |
| Transmembrane structure | - | |
| Signal | peptide | - |
| Subcellular | localization | Nucleus |
| Secondary structure | α-helix (%) | 24.13% |
| β-angle (%) | 6.03% | |
| Random coil (%) | 57.14% | |
| Extended chain (%) | 12.70% | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, S.; Yuan, H.; Wu, G.; Yang, X.; Liu, D.; Chen, L.; Chen, J.; Liu, Y.; Yin, W.; Li, C.; et al. Cloning and Functional Validation of the Candidate Gene LuWRKY39 Conferring Resistance to Septoria linicola (Speg.) Garassini from Flax. Agriculture 2025, 15, 1561. https://doi.org/10.3390/agriculture15141561
Chen S, Yuan H, Wu G, Yang X, Liu D, Chen L, Chen J, Liu Y, Yin W, Li C, et al. Cloning and Functional Validation of the Candidate Gene LuWRKY39 Conferring Resistance to Septoria linicola (Speg.) Garassini from Flax. Agriculture. 2025; 15(14):1561. https://doi.org/10.3390/agriculture15141561
Chicago/Turabian StyleChen, Si, Hongmei Yuan, Guangwen Wu, Xue Yang, Dandan Liu, Le Chen, Jing Chen, Yan Liu, Weiping Yin, Cen Li, and et al. 2025. "Cloning and Functional Validation of the Candidate Gene LuWRKY39 Conferring Resistance to Septoria linicola (Speg.) Garassini from Flax" Agriculture 15, no. 14: 1561. https://doi.org/10.3390/agriculture15141561
APA StyleChen, S., Yuan, H., Wu, G., Yang, X., Liu, D., Chen, L., Chen, J., Liu, Y., Yin, W., Li, C., Wu, L., Ma, J., Bian, D., & Zhang, L. (2025). Cloning and Functional Validation of the Candidate Gene LuWRKY39 Conferring Resistance to Septoria linicola (Speg.) Garassini from Flax. Agriculture, 15(14), 1561. https://doi.org/10.3390/agriculture15141561






