Combined Genomic and Transcriptomic Screening of Candidate Genes for Asymmetric Oviduct Development in Hens
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection
2.2. Transcriptome Sequencing
2.3. Whole-Genome Resequencing
2.4. Joint Analysis of RNA-Seq and WGRS Data
2.5. RNA Extraction and Quantitative PCR (qPCR) Validation
2.6. DNA Extraction and Sanger Sequencing Validation
2.7. Protein Extraction and Western Blotting
2.8. Statistical Data
3. Results
3.1. Phenotypic Changes in the Left and Right Oviducts in Chick Embryos of Different Ages and the Phenotypic Morphology of Hens with Unilateral and Bilateral Oviducts
3.2. Analysis of Oviduct RNA-Seq Sequencing Data
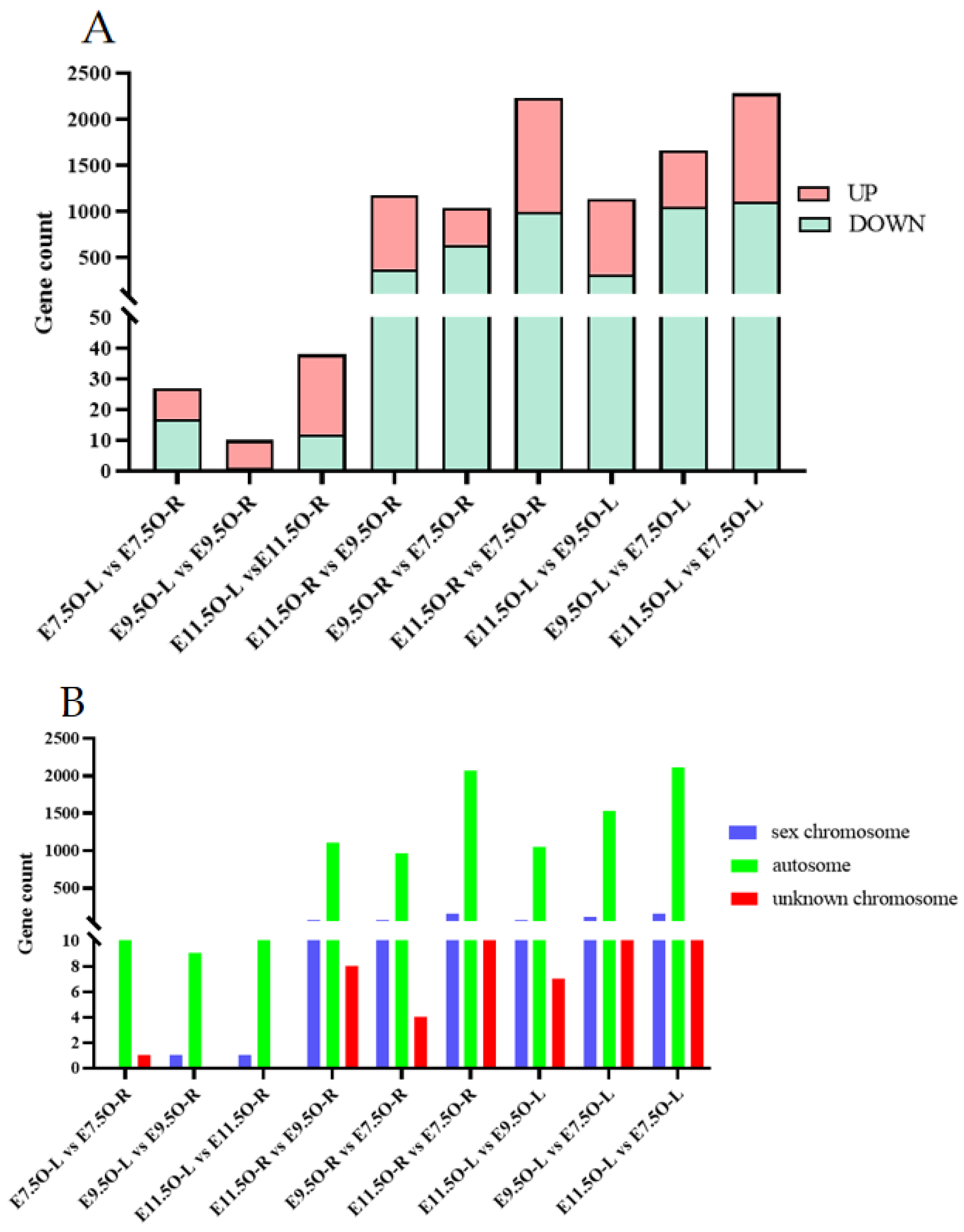
3.3. Analysis of Whole-Genome Resequencing Data
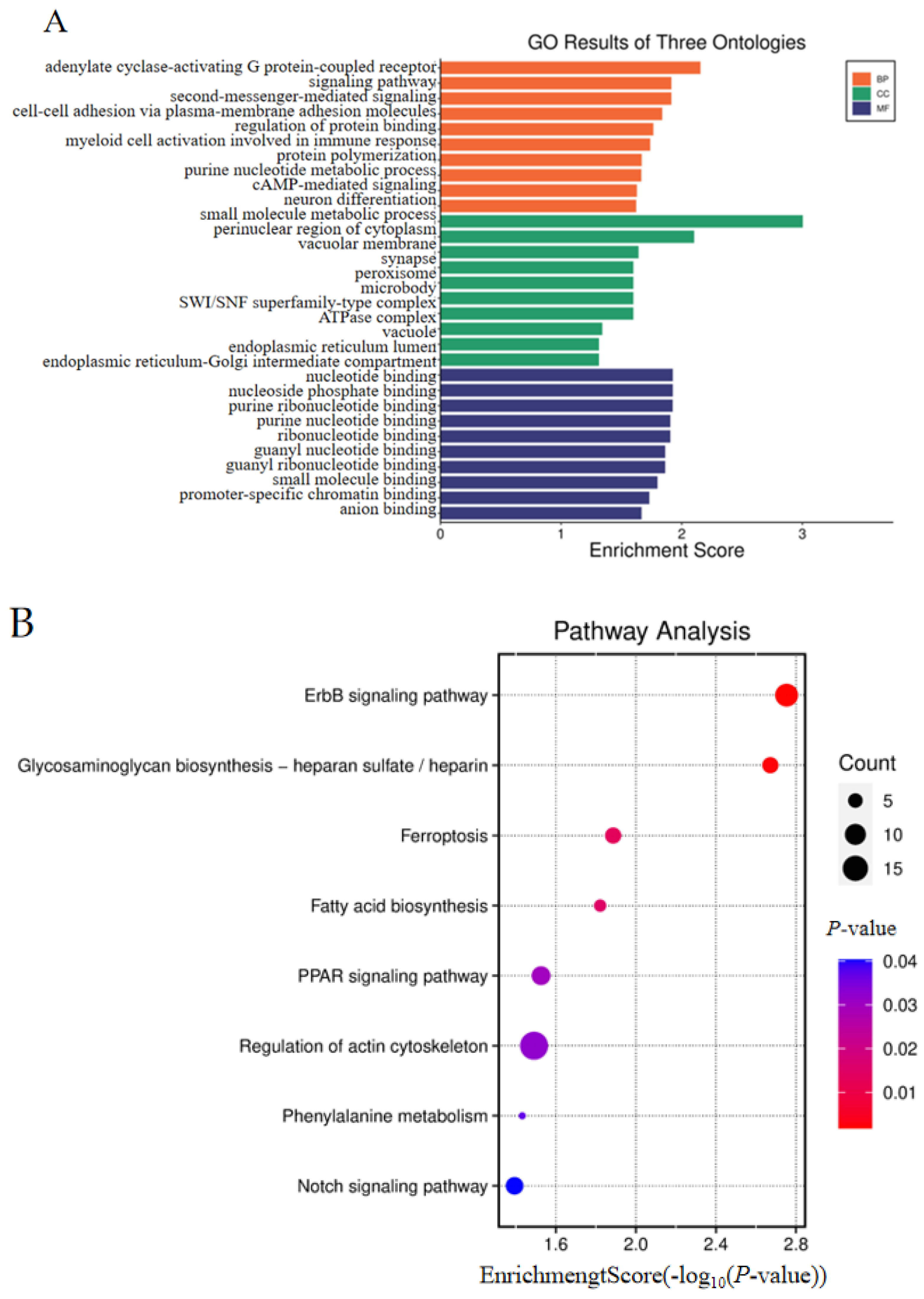
3.4. Combined Analysis of Oviduct RNA-Seq Data and Gonad RNA-Seq Data
3.5. Combined Analysis of WGRS with Oviduct RNA-Seq and Gonad RNA-Seq Data
3.6. RT-qPCR, Western Blot, and SNP Validation of Candidate Genes
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| WD | Wolffian ducts |
| MD | Müllerian ducts |
| WGRS | Whole-genome resequencing |
| DMGs | Differentially mutated genes |
| DEGs | Differentially expressed genes |
| E7.5O-R | E7.5 right-side oviducts |
| E7.5O-L | E7.5 left-side oviducts |
| E9.5O-R | E9.5 right-side oviducts |
| E9.5O-L | E9.5 left-side oviducts |
| E11.5O-R | E11.5 right-side oviducts |
| E11.5O-L | E11.5 left-side oviducts |
| D | Bilateral group |
| S | Unilateral group |
| FPKM | Fragments Per Kilo bases per Million fragments |
| GO | Gene Ontology |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
| NCBI | National Center for Biotechnology Information |
| SNP | Single-nucleotide polymorphism |
| InDel | Insertion–deletion |
Appendix A
Appendix A.1
| Gene Name | Gene Sequence | Gene ID |
|---|---|---|
| β-actin | TATGTGCAAGGCCGGTTTC | NM_205518.2 |
| TGTCTTTCTGGCCCATACCAA | ||
| RGCC | GCTGATCTGGACAGGACACT | XM_417029.8 |
| GGCAGCAGTTCTCCATGACT | ||
| ADAM19 | ATCACGACTCATCTGGCTGC | NM_001195122.2 |
| TTGGAAAGGCACATCCCTCC | ||
| RERG | GATGACACGGTGAAGCCGAA | XM_046904998.1 |
| TATGTAGACTCTGACTTGCCCAC | ||
| PDGFRB | TCTCTGCAGCTCCTCATCCT | XM_046900454.1 |
| ACGAGGGAGAAAGTGCTGTG | ||
| ADAMTS5 | CGCCACCACTCTCAAGAACT | XM_425541.8 |
| AACGTCTGCCATTCCCAGAG | ||
| ZMAT3 | TGCTAAAGGGGCTGATTCCAG | XM_423190.8 |
| TGTCACCACTTAGAGCAGCCT | ||
| PAK3 | TCTACACGCGCTCCGTAATC | XM_015278599.4 |
| TCCCACACTCACAATGCTCC | ||
| SLC6A11 | GGAATACTGCACGGTGGG | XM_040646635.2 |
| AATGCTGCCGCTTGAGAC | ||
| GPM6A | AGGACACCCCTCCATCTGTT | NM_001012579.2 |
| GGGGGAAATTTGGTCCGAGT | ||
| CREB3L3 | AGATGCCCAGCTCCCTACTA | XM_015300011.4 |
| ATCTTCTTCAGCACCCGCTC | ||
| PRKCB | TCTTCCGCTACATCGACTGG | XM_015294481.3 |
| GGTGGATGGCGAGTGAAAAAC | ||
| KCTD21 | GCGCTCACATCTTCTCCACT | XM_004938933.5 |
| GGTCAAATGGTTGGGGTCCT | ||
| PAX3 | AGCAGCCCAGTCTGATGAAG | NM_001397759.1 |
| AAACCCTCCTGGGATCAGGT | ||
| BMP7 | AGGTCCTCAGCAAACCTCTAA | XM_417496.8 |
| CAAATTCCCGTGGTTGCGTT | ||
| PITX2 | GGAGTGTGTCCAGAGCATGA | XM_416467.8 |
| GGCCCTGAATCCATCCATGT | ||
| SMC1B | GGAGTGTGTCCAGAGCATGA | XM_416467.8 |
| GGCCCTGAATCCATCCATGT | ||
| ETV4 | CAGATGAGGCCCTTTGCTGA | XM_015299450.4 |
| ACAGCCTGGCTACCTCTTCT |
Appendix A.2
| Gene Name | Gene Sequence | Gene ID |
|---|---|---|
| PITX2 | CTTGGGGGATGGAGTAGCAC | NC_052535.1 |
| TTACACTCCCCCTTGAGCCT | ||
| ASZ1 | CTACAGAGCTGGATGACGGC | NC_052532.1 |
| TAGGGGGATAGGGCTGAACG | ||
| SLC6A11-1 1 | GAAGAGAAGTCCCCTGGTGT | NC_052543.1 |
| AGTAAACAGAGGGCAGGGTAG | ||
| SLC6A11-2 2 | TATGCGAGGGCTTCTACCCT | |
| AGCCTTGCTGTATGGTAGGC |
Appendix A.3
| Group 1 | Co-Pathways 2 | RNA-seq | WGRS 3 |
|---|---|---|---|
| E4.5G-L vs. E4.5G-R | Ferroptosis | CYBB, MAP1LC3C | ACSL1, SLC39A8, FTH1, STEAP3, ACSL3, ACSL6 |
| E5.5G-L vs. E5.5G-R | Phenylalanine metabolism | TAT, HPD | PAH, MAOB, MAOA |
| E6.5G-L vs. E6.5G-R | / | / | / |
Appendix B
Appendix B.1
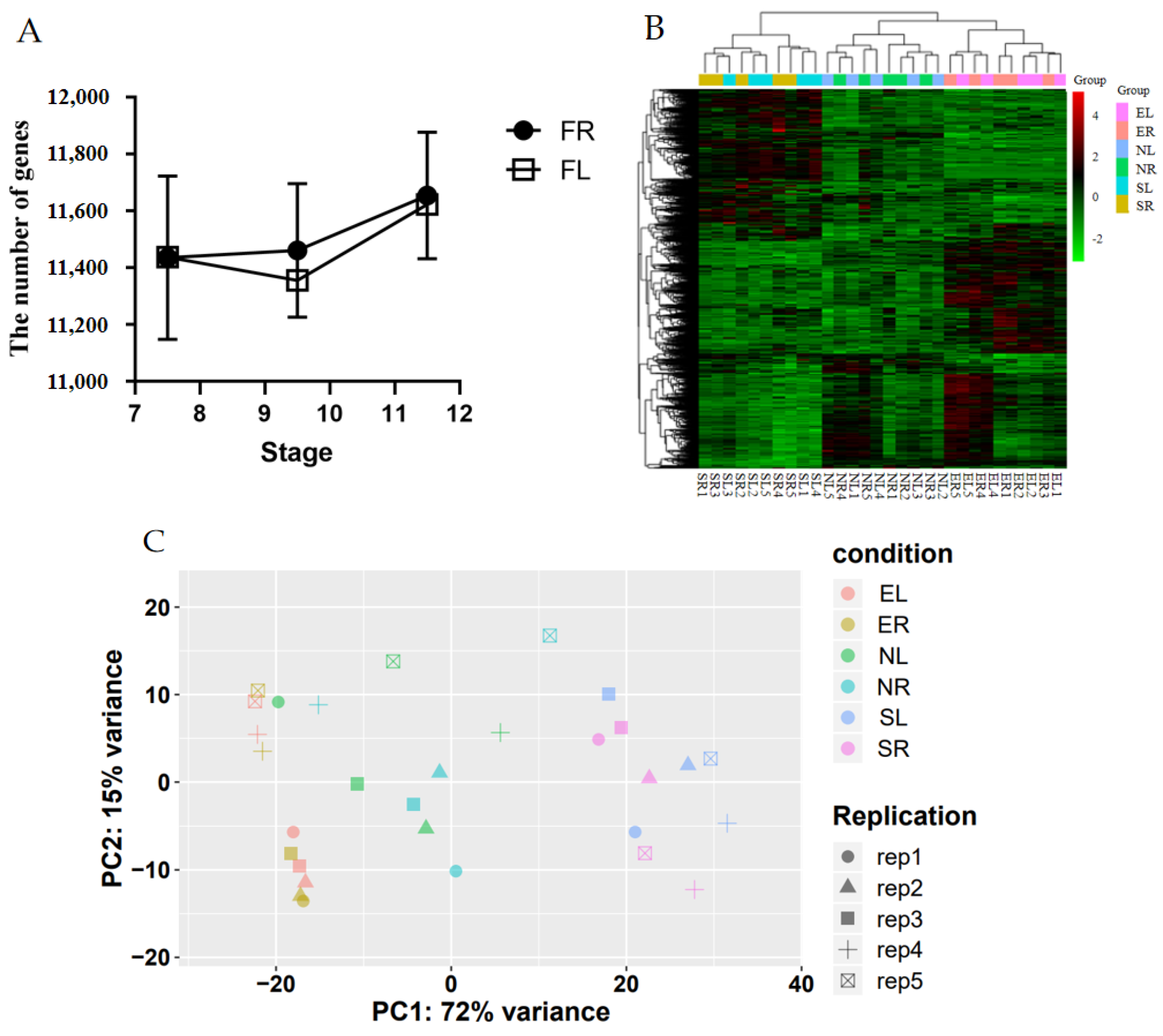
Appendix B.2
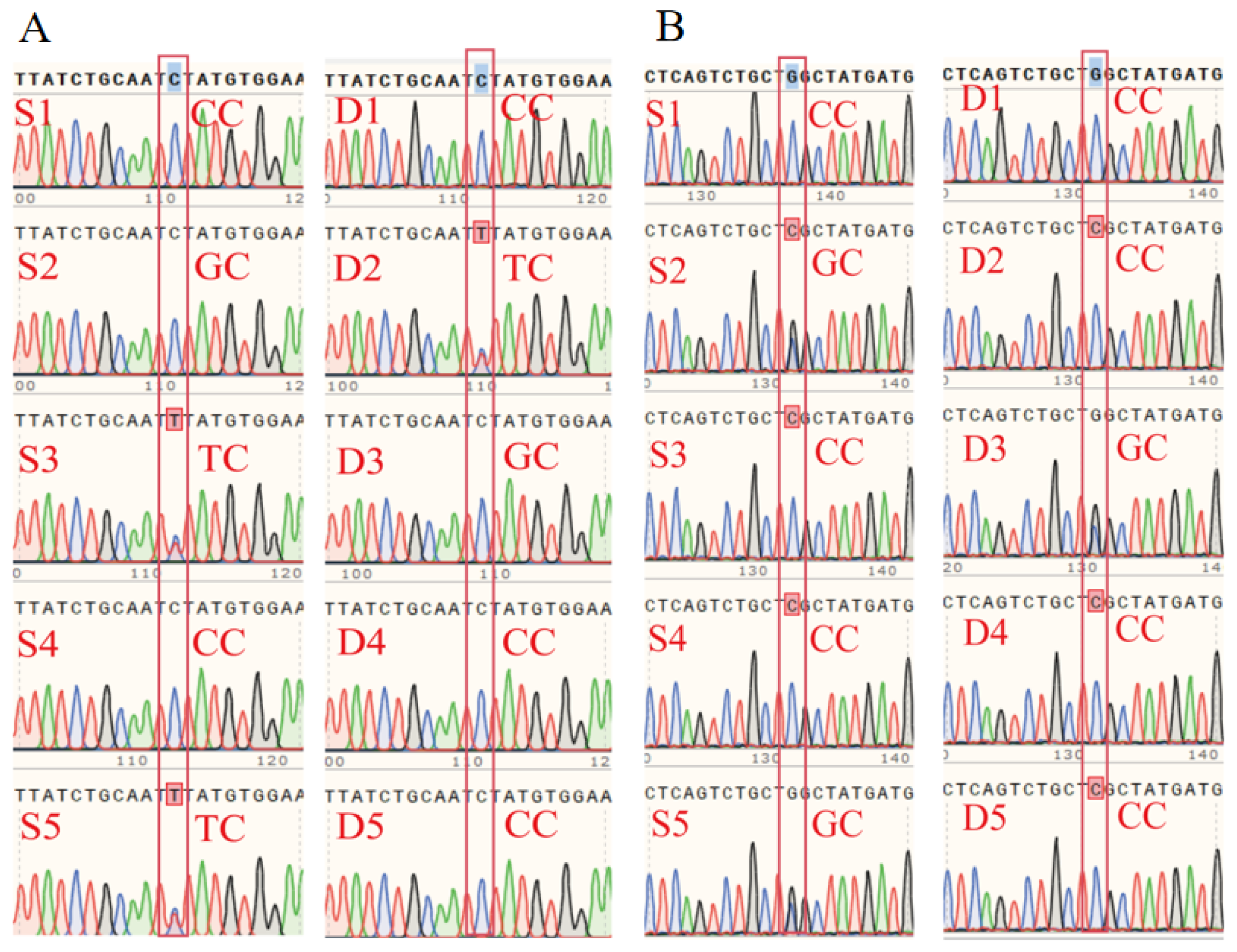
Appendix B.3

References
- Kuroiwa, A. Sex-Determining Mechanism in Avians. Adv. Exp. Med. Biol. 2017, 1001, 19–31. [Google Scholar] [PubMed]
- Josso, N.; di Clemente, N.; Gouédard, L. Anti-Müllerian hormone and its receptors. Mol. Cell. Endocrinol. 2001, 179, 25–32. [Google Scholar] [CrossRef]
- Klattig, J.; Englert, C. The Müllerian duct: Recent insights into its development and regression. Sex. Dev. 2007, 1, 271–278. [Google Scholar] [CrossRef]
- Mullen, R.D.; Behringer, R. Molecular genetics of Müllerian duct formation, regression and differentiation. Sex. Dev. 2014, 8, 281–296. [Google Scholar] [CrossRef]
- Smith, C.A.; Sinclair, A.H. Sex determination: Insights from the chicken. BioEssays 2004, 26, 120–132. [Google Scholar] [CrossRef]
- González-Morán, M.G. Histological and stereological changes in growing and regressing chicken ovaries during development. Anat. Rec. 2011, 294, 893–904. [Google Scholar] [CrossRef]
- Cunha, G.R.; Kurita, T.; Cao, M.; Shen, J.; Robboy, S.; Baskin, L. Molecular mechanisms of development of the human fetal female reproductive tract. Differentiation 2017, 97, 54–72. [Google Scholar] [CrossRef]
- Roly, Z.Y.; Backhouse, B.; Cutting, A.; Tan, T.Y.; Sinclair, A.H.; Ayers, K.L.; Major, A.T.; Smith, C.A. The cell biology and molecular genetics of Müllerian duct development. Wiley Interdiscip. Rev. Dev. Biol. 2018, 7, e310. [Google Scholar] [CrossRef]
- Isaac, A.; Sargent, M.G.; Cooke, J. Control of vertebrate left-right asymmetry by a snail-related zinc finger gene. Science 1997, 275, 1301–1304. [Google Scholar] [CrossRef]
- Rodríguez Esteban, C.; Capdevila, J.; Economides, A.N.; Pascual, J.; Ortiz, A.; Izpisúa Belmonte, J.C. The novel Cer-like protein Caronte mediates the establishment of embryonic left-right asymmetry. Nature 1999, 401, 243–251. [Google Scholar] [CrossRef]
- Welsh, I.C.; Thomsen, M.; Gludish, D.W.; Alfonso-Parra, C.; Bai, Y.; Martin, J.F.; Kurpios, N.A. Integration of left-right Pitx2 transcription and Wnt signaling drives asymmetric gut morphogenesis via Daam2. Dev. Cell 2013, 26, 629–644. [Google Scholar] [CrossRef]
- Vermot, J.; Pourquié, O. Retinoic acid coordinates somitogenesis and left-right patterning in vertebrate embryos. Nature 2005, 435, 215–220. [Google Scholar] [CrossRef]
- Guioli, S.; Lovell-Badge, R. PITX2 controls asymmetric gonadal development in both sexes of the chick and can rescue the degeneration of the right ovary. Development 2007, 134, 4199–4208. [Google Scholar] [CrossRef]
- Beaupain, R. Cell division and cell death during regression of the chick embryo Müllerian ducts. Experientia 1979, 35, 1380–1382. [Google Scholar] [CrossRef]
- Howard, J.A.; Hart, K.N.; Thompson, T.B. Molecular Mechanisms of AMH Signaling. Front. Endocrinol. 2022, 13, 927824. [Google Scholar] [CrossRef]
- Moses, M.M.; Behringer, R.R. A gene regulatory network for Müllerian duct regression. Environ. Epigenetics 2019, 5, dvz017. [Google Scholar] [CrossRef]
- Kang, S.J.; Choi, J.W.; Park, K.J.; Lee, Y.M.; Kim, T.M.; Sohn, S.H.; Lim, J.M.; Han, J.Y. Development of a pheasant interspecies primordial germ cell transfer to chicken embryo: Effect of donor cell sex on chimeric semen production. Theriogenology 2009, 72, 519–527. [Google Scholar] [CrossRef]
- Kim, D.; Paggi, J.M.; Park, C.; Bennett, C.; Salzberg, S.L. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 2019, 37, 907–915. [Google Scholar] [CrossRef]
- Putri, G.H.; Anders, S.; Pyl, P.T.; Pimanda, J.E.; Zanini, F. Analysing high-throughput sequencing data in Python with HTSeq 2.0. Bioinformatics 2022, 38, 2943–2945. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Luo, H.; Zhou, H.; Jiang, S.; He, C.; Xu, K.; Ding, J.; Liu, J.; Qin, C.; Chen, K.; Zhou, W.; et al. Gene Expression Profiling Reveals Potential Players of Sex Determination and Asymmetrical Development in Chicken Embryo Gonads. Int. J. Mol. Sci. 2023, 24, 14597. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef] [PubMed]
- Zhu, P.; He, L.; Li, Y.; Huang, W.; Xi, F.; Lin, L.; Zhi, Q.; Zhang, W.; Tang, Y.T.; Geng, C.; et al. OTG-snpcaller: An optimized pipeline based on TMAP and GATK for SNP calling from ion torrent data. PLoS ONE 2014, 9, e97507. [Google Scholar] [CrossRef]
- Cingolani, P.; Platts, A.; Wang le, L.; Coon, M.; Nguyen, T.; Wang, L.; Land, S.J.; Lu, X.; Ruden, D.M. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly 2012, 6, 80–92. [Google Scholar] [CrossRef]
- Islam, K.N.; Modi, M.M.; Siegfried, K.R. The Zebrafish Meiotic Cohesin Complex Protein Smc1b Is Required for Key Events in Meiotic Prophase I. Front. Cell Dev. Biol. 2021, 9, 714245. [Google Scholar] [CrossRef]
- Lin, J.; Wang, C.; Yang, C.; Fu, S.; Redies, C. Pax3 and Pax7 interact reciprocally and regulate the expression of cadherin-7 through inducing neuron differentiation in the developing chicken spinal cord. J. Comp. Neurol. 2016, 524, 940–962. [Google Scholar] [CrossRef]
- Meno, C.; Takeuchi, J.; Sakuma, R.; Koshiba-Takeuchi, K.; Ohishi, S.; Saijoh, Y.; Miyazaki, J.; ten Dijke, P.; Ogura, T.; Hamada, H. Diffusion of nodal signaling activity in the absence of the feedback inhibitor Lefty2. Dev. Cell 2001, 1, 127–138. [Google Scholar] [CrossRef]
- Hoshino, A.; Koide, M.; Ono, T.; Yasugi, S. Sex-specific and left-right asymmetric expression pattern of Bmp7 in the gonad of normal and sex-reversed chicken embryos. Dev. Growth Differ. 2005, 47, 65–74. [Google Scholar] [CrossRef]
- Valdecantos, P.A.; Bravo Miana, R.D.C.; García, E.V.; García, D.C.; Roldán-Olarte, M.; Miceli, D.C. Expression of bone morphogenetic protein receptors in bovine oviductal epithelial cells: Evidence of autocrine BMP signaling. Anim. Reprod. Sci. 2017, 185, 89–96. [Google Scholar] [CrossRef]
- Hamburger, V.; Hamilton, H.L. A series of normal stages in the development of the chick embryo. Dev. Dyn. 1951, 195, 231–272. [Google Scholar] [CrossRef] [PubMed]
- Gros, J.; Feistel, K.; Viebahn, C.; Blum, M.; Tabin, C.J. Cell movements at Hensen’s node establish left/right asymmetric gene expression in the chick. Science 2009, 324, 941–944. [Google Scholar] [CrossRef]
- Tabin, C.J.; Vogan, K.J. A two-cilia model for vertebrate left-right axis specification. Genes Dev. 2003, 17, 1–6. [Google Scholar] [CrossRef]
- Ishimaru, Y.; Komatsu, T.; Kasahara, M.; Katoh-Fukui, Y.; Ogawa, H.; Toyama, Y.; Maekawa, M.; Toshimori, K.; Chandraratna, R.A.; Morohashi, K.; et al. Mechanism of asymmetric ovarian development in chick embryos. Development 2008, 35, 677–685. [Google Scholar] [CrossRef]
- Intarapat, S.; Stern, C.D. Sexually dimorphic and sex-independent left-right asymmetries in chicken embryonic gonads. PLoS ONE 2013, 8, e69893. [Google Scholar] [CrossRef]
- Rodríguez-León, J.; Rodríguez Esteban, C.; Martí, M.; Santiago-Josefat, B.; Dubova, I.; Rubiralta, X.; Izpisúa Belmonte, J.C. Pitx2 regulates gonad morphogenesis. Proc. Natl. Acad. Sci. USA 2008, 105, 11242–11247. [Google Scholar] [CrossRef]
- Yoshino, T.; Murai, H.; Saito, D. Hedgehog-BMP signalling establishes dorsoventral patterning in lateral plate mesoderm to trigger gonadogenesis in chicken embryos. Nat. Commun. 2016, 7, 12561. [Google Scholar] [CrossRef]
- Ayers, K.L.; Lambeth, L.S.; Davidson, N.M.; Sinclair, A.H.; Oshlack, A.; Smith, C.A. Identification of candidate gonadal sex differentiation genes in the chicken embryo using RNA-seq. BMC Genom. 2015, 16, 704. [Google Scholar] [CrossRef]
- Doi, O.; Hutson, J.M. Pretreatment of chick embryos with estrogen in ovo prevents mullerian duct regression in organ culture. Endocrinology 1988, 122, 2888–2891. [Google Scholar] [CrossRef]
- Elbrecht, A.; Smith, R. Aromatase enzyme activity and sex determination in chickens. Science 1992, 255, 467–470. [Google Scholar] [CrossRef]
- Roly, Z.Y.; Major, A.T.; Fulcher, A.; Estermann, M.A.; Hirst, C.E.; Smith, C.A. Adhesion G-protein-coupled receptor, GPR56, is required for Müllerian duct development in the chick. J. Endocrinol. 2020, 44, 395–413. [Google Scholar] [CrossRef] [PubMed]
- Ha, Y.; Tsukada, A.; Saito, N.; Shimada, K. Changes in mRNA expression of MMP-2 in the Müllerian duct of chicken embryo. Gen. Comp. Endocrinol. 2004, 139, 131–136. [Google Scholar] [CrossRef]
- Ha, Y.; Tsukada, A.; Saito, N.; Zadworny, D.; Shimada, K. Identification of differentially expressed genes involved in the regression and development of the chicken Müllerian duct. Int. J. Dev. Biol. 2008, 52, 1135–1141. [Google Scholar] [CrossRef]
- Roberts, L.M.; Visser, J.A.; Ingraham, H.A. Involvement of a matrix metalloproteinase in MIS-induced cell death during urogenital development. Development 2002, 129, 1487–1496. [Google Scholar] [CrossRef]


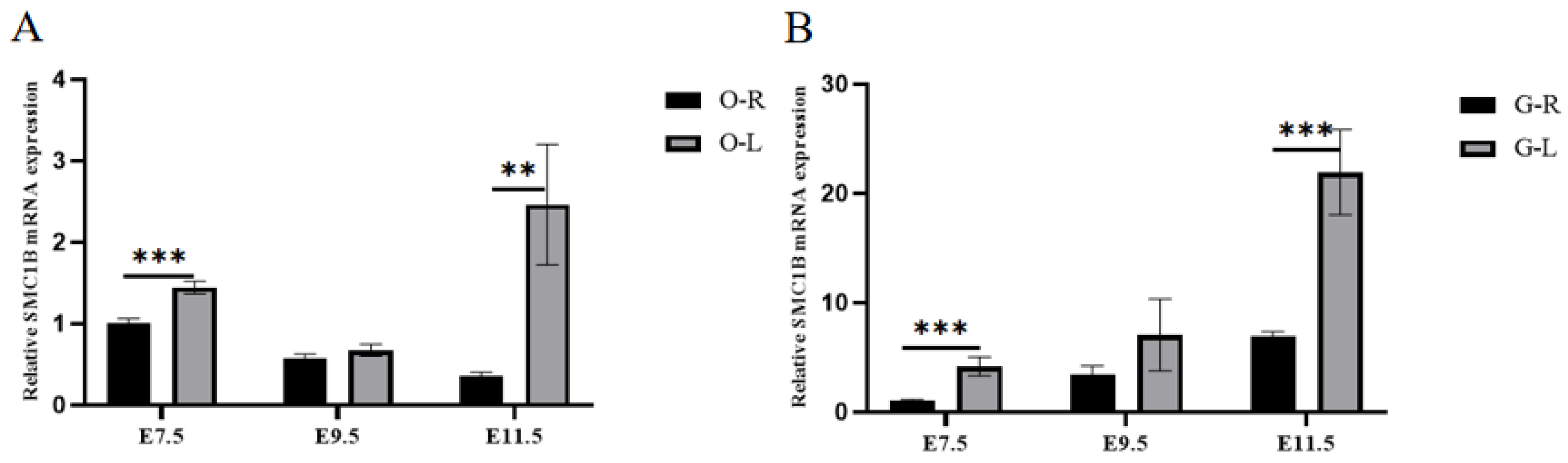
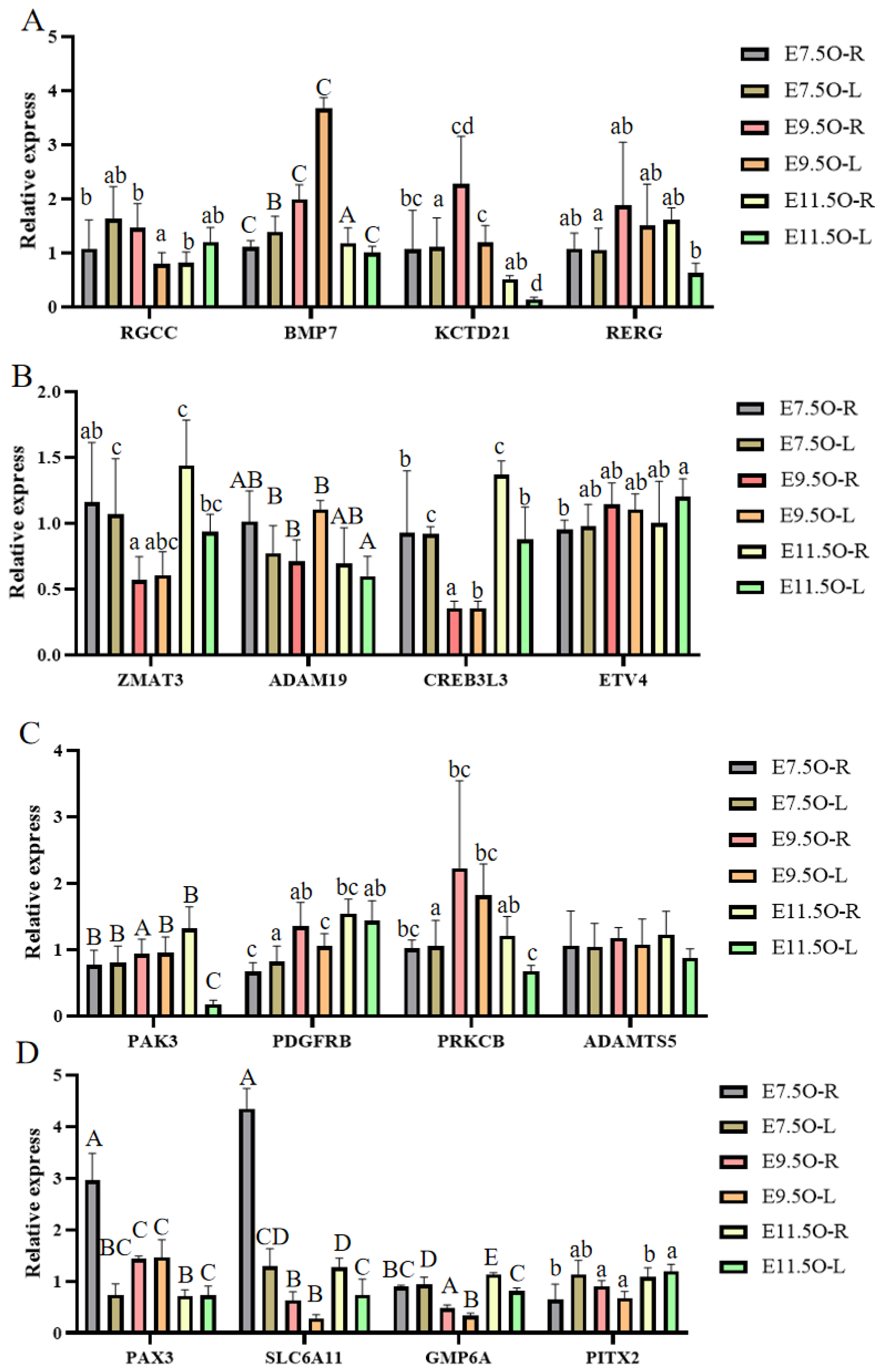
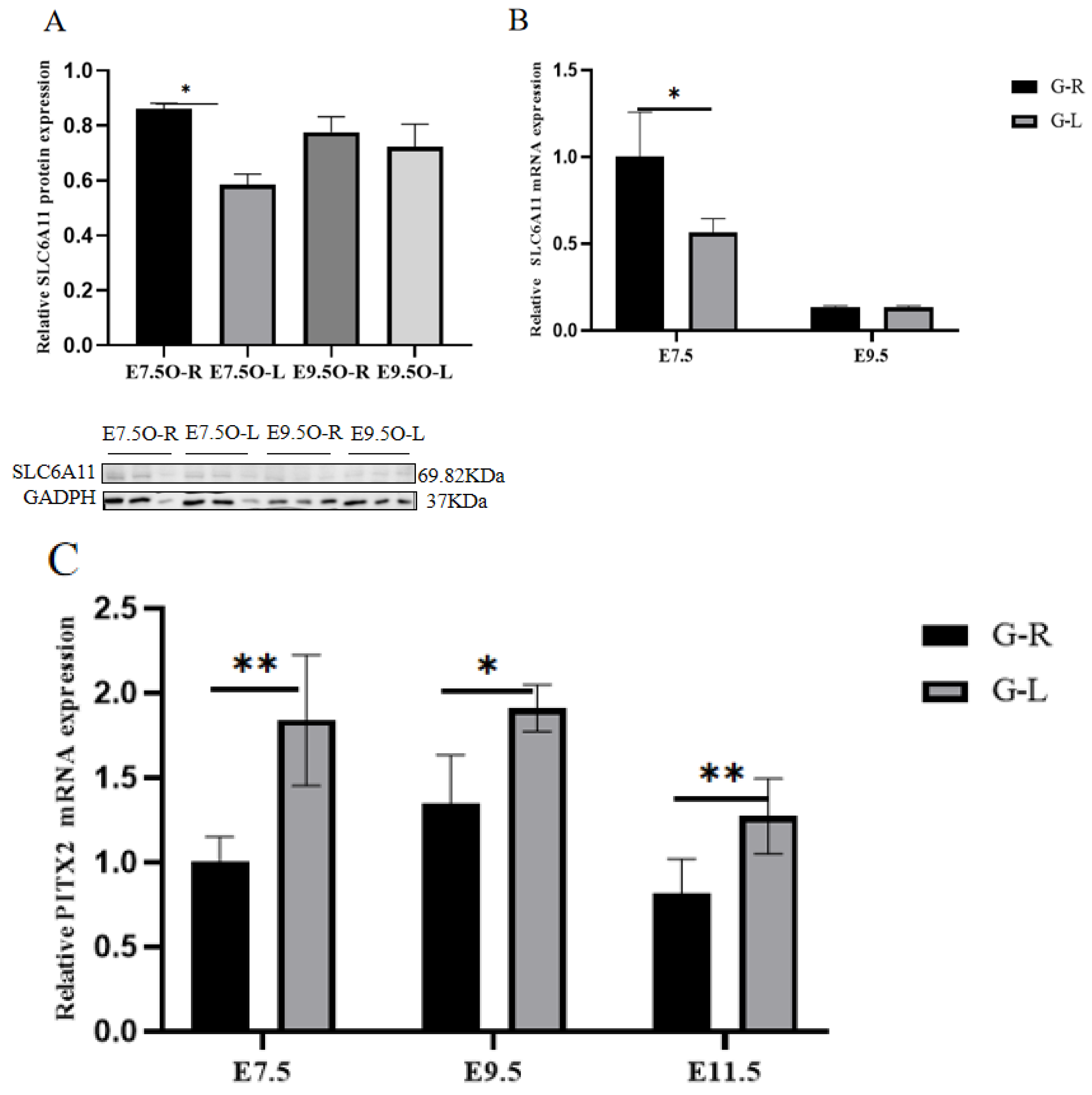
| Gene Name | Gene Sequence | Gene ID |
|---|---|---|
| CHD | CTGCGAGAACGTGGCAACAGAGT | NM_204941.2 |
| ATTGAAATGATCCAGTGCTTC |
| Group 1 | Name | Group 1 | Name | Group 1 | Name |
|---|---|---|---|---|---|
| E7.5O-L vs. E7.5O-R | EYA1 | E9.5O-L vs. E9.5O-R | GJD2 | E11.5O-L vs. E11.5O-R | LUZP2 |
| EBF2 | GPM6A | SLC30A8 | |||
| NPHS2 | RANBP3L | TNFRSF6B | |||
| SPP1 | USP42 | SLC38A3 | |||
| DDEFL1 | PPARG | VTG1 | |||
| E11.5O-R vs. E9.5O-R | SOX10 | E9.5O-R vs. E7.5O-R | SRD5A2 | E11.5O-R vs. E7.5O-R | CYP2AC1 |
| PDK4 | DLX6 | SLC34A1 | |||
| ACTG2 | DLX5 | SRD5A2 | |||
| TUBB3 | FST | ACSM3 | |||
| SLC17A8 | FSTL4 | SLC6A19 | |||
| E11.5O-L vs. E9.5O-L | SOX10 | E9.5O-L vs. E7.5O-L | SRD5A2 | E11.5O-L vs. E7.5O-L | SRD5A2 |
| SLC17A8 | FSTL4 | RHCG | |||
| ECEL1 | DLX5 | CALB1 | |||
| ENSGALG00000051930 | FST | ENSGALG00000051882 | |||
| LAMA3 | CLCNKB | ENSGALG00000039159 |
| Region 1 | Number | Percentage |
|---|---|---|
| INTRON | 13,885 | 62.173% |
| INTERGENIC | 5603 | 25.088% |
| SYNONYMOUS_CODING | 290 | 1.299% |
| NON_SYNONYMOUS_CODING | 114 | 0.510% |
| UPSTREAM | 845 | 3.784% |
| DOWNSTREAM | 905 | 4.052% |
| UTR_3_PRIME | 476 | 2.131% |
| UTR_5_PRIME | 83 | 0.372% |
| START_GAINED | 51 | 0.228% |
| SPLICE_SITE_REGION | 50 | 0.224% |
| INTRAGENIC | 28 | 0.125% |
| SPLICE_SITE_ACCEPTOR | 1 | 0.004% |
| START_LOST | 1 | 0.004% |
| STOP_GAINED | 1 | 0.004% |
| Total | 22,333 | 100.000% |
| Overlap 1 | E7.5-Oviduct | E9.5-Oviduct | E11.5-Oviduct |
|---|---|---|---|
| E4.5-gonad | NPHS2 | / | LUZP2, GRIA1 |
| E5.5-gonad | / | / | LUZP2, GRIA1 |
| E6.5-gonad | ENSGALG00000054136, SMC1B | / | LUZP2, SLC7A9, GRIA1 |
| Co-Pathways 1 | E7.5-Oviduct 2 | E6.5-Gonad 3 |
|---|---|---|
| Oocyte meiosis | SMC1B, CPEB1, CPEB2 | SMC1B |
| Neuroactive ligand–receptor interaction | HTR1E, P2RY8, GRIA1, DRD4, GABRA3 | CCKAR, GLRB, AVPR2 |
| Gene | Site | Region | Type of Base Mutation | WGRS | Small Group Verification Results | Large Group Verification Results | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| Ref 1 | Alt 2 | D 3 (n = 6) | S 4 (n = 6) | D (n = 5) | S (n = 5) | D (n = 18) | S (n = 18) | |||
| PITX2 | 57301142 | UPSTREAM | T | C | T(5) T + C(1) | T(1) T + C(5) | T(5) | T + C(4) T(1) | T(14) C(2) T + C(2) | T + C(10) T(8) |
| ASZ1 | 24814074 | UPSTREAM | G | A | G + A(5) G(1) | G(5) G + A(1) | G + A(3) G(2) | G + A(1) G(4) | / | / |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, R.; Song, X.; Lin, X.; Deng, M.; Liu, Y.; Geng, T.; Gong, D.; Zhao, M.; Liu, L. Combined Genomic and Transcriptomic Screening of Candidate Genes for Asymmetric Oviduct Development in Hens. Agriculture 2025, 15, 1245. https://doi.org/10.3390/agriculture15121245
Li R, Song X, Lin X, Deng M, Liu Y, Geng T, Gong D, Zhao M, Liu L. Combined Genomic and Transcriptomic Screening of Candidate Genes for Asymmetric Oviduct Development in Hens. Agriculture. 2025; 15(12):1245. https://doi.org/10.3390/agriculture15121245
Chicago/Turabian StyleLi, Ruijie, Xiang Song, Xiao Lin, Mingli Deng, Yin Liu, Tuoyu Geng, Daoqing Gong, Minmeng Zhao, and Long Liu. 2025. "Combined Genomic and Transcriptomic Screening of Candidate Genes for Asymmetric Oviduct Development in Hens" Agriculture 15, no. 12: 1245. https://doi.org/10.3390/agriculture15121245
APA StyleLi, R., Song, X., Lin, X., Deng, M., Liu, Y., Geng, T., Gong, D., Zhao, M., & Liu, L. (2025). Combined Genomic and Transcriptomic Screening of Candidate Genes for Asymmetric Oviduct Development in Hens. Agriculture, 15(12), 1245. https://doi.org/10.3390/agriculture15121245







