The Residue Chemistry Transformation Linked to the Fungi Keystone Taxa during Different Residue Tissues Incorporation into Mollisols in Northeast China
Abstract
1. Introduction
2. Materials and Methods
2.1. Experimental Site and Soil Characteristics
2.2. Incubation Experimental Design
2.3. Amplification and Sequencing of Fungal ITS RNA
2.4. 13C Nuclear Magnetic Resonance (NMR) Spectroscopy
2.5. Extracellular Enzyme Activity (EEA) Assays
2.6. Statistical Analyses
3. Results
3.1. The Decomposition of Straw Tissues
3.2. 13C Nuclear Magnetic Resonance (NMR) Spectroscopy
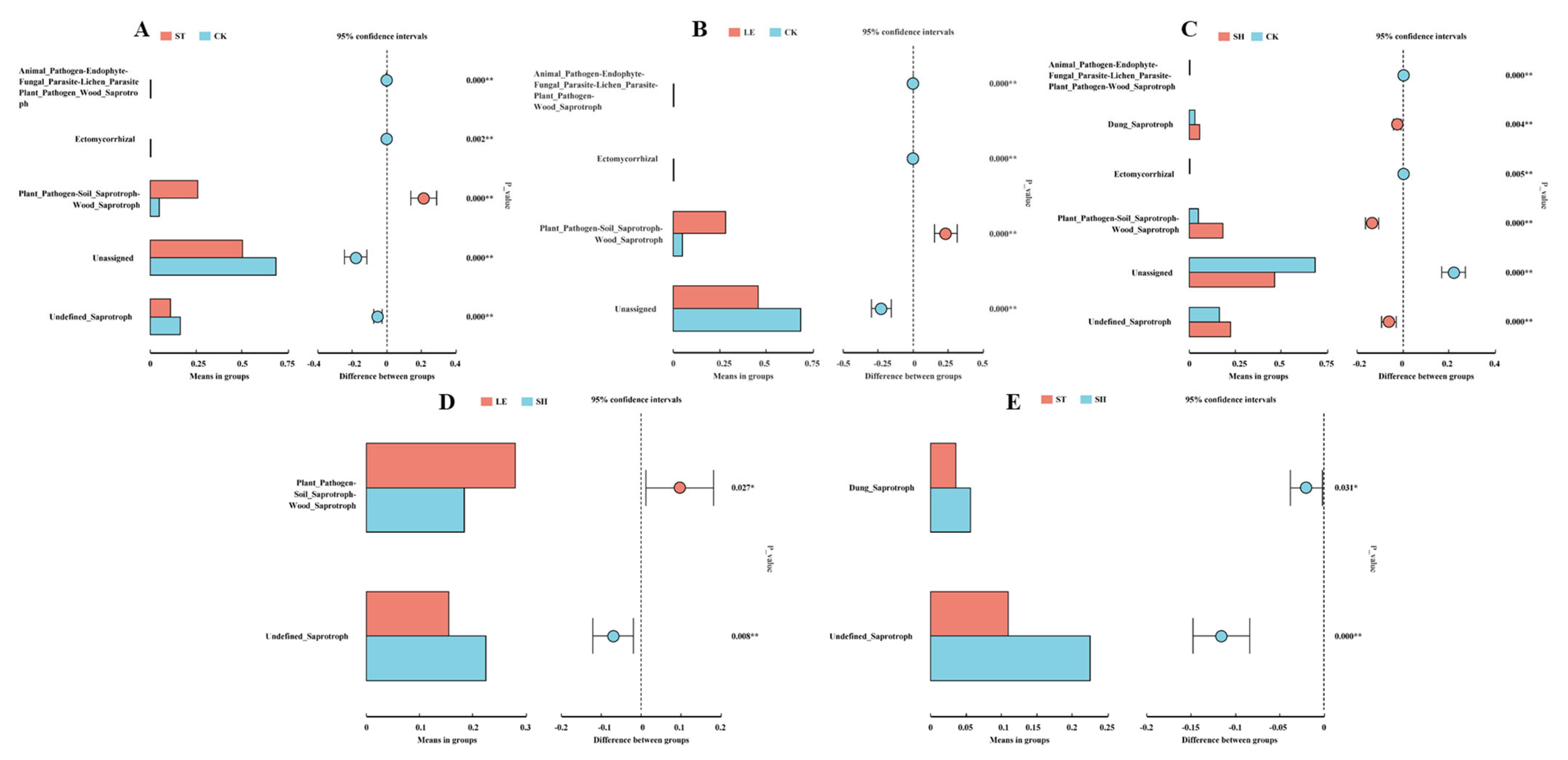
3.3. Soil Community Composition and the Co-Occurrence Network of Fungi
3.4. Changes in EEA Activity with Decomposition
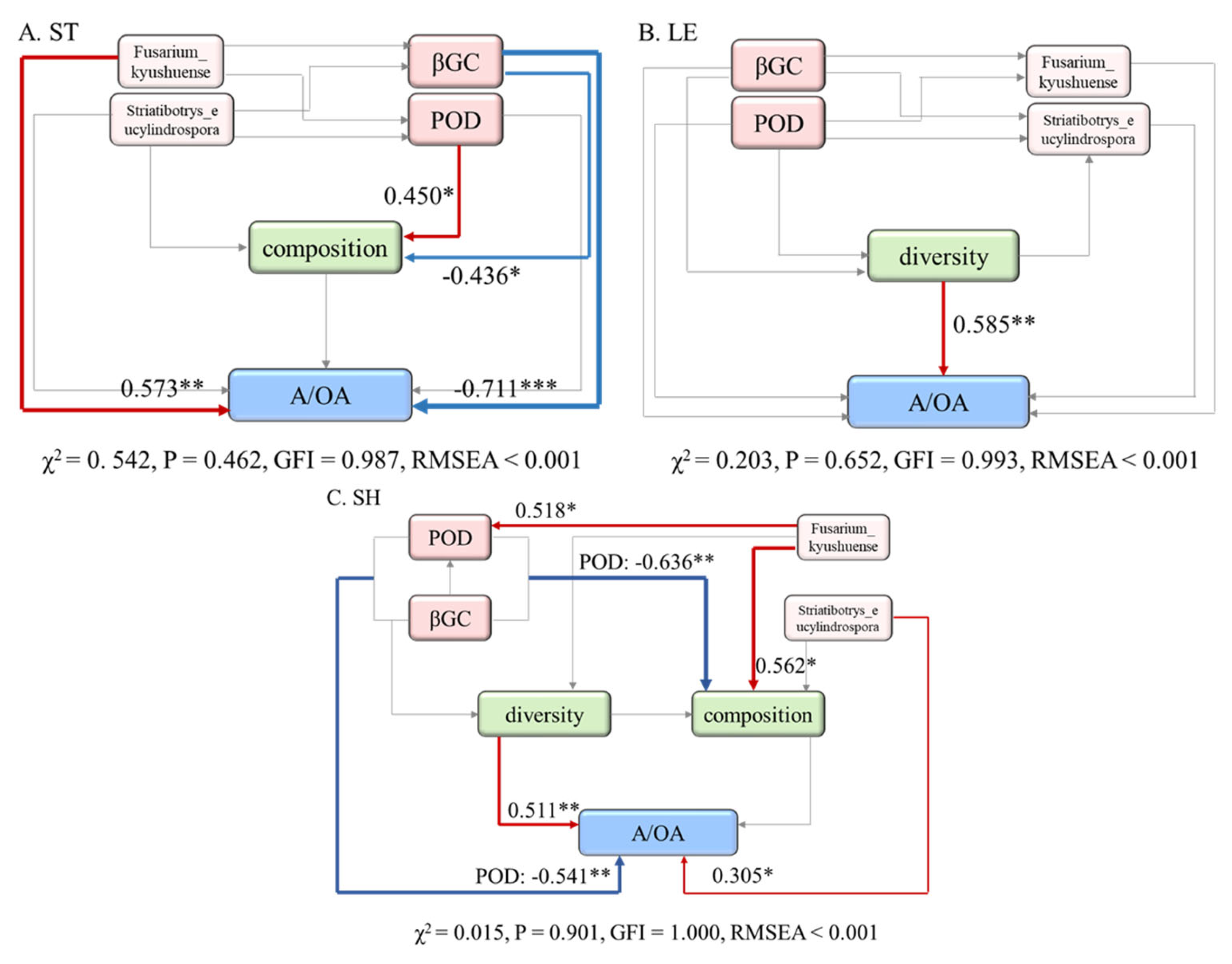
3.5. The Correlation Analysis
4. Discussion
4.1. Difference of Chemical Structure in Straw Tissue
4.2. Difference of Fungi Community Composition
4.3. Linkages between Chemical Structure, EEA Activities and Fungi Composition
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Xu, Y.; Chen, Z.; Fontaine, S.; Wang, W.; Luo, J.; Fan, J.; Ding, W. Dominant effects of organic carbon chemistry on decomposition dynamics of crop residues in a Mollisol. Soil Biol. Biochem. 2017, 115, 221–232. [Google Scholar] [CrossRef]
- Li, D.; Li, Z.; Zhao, B.; Zhang, J. Relationship between the chemical structure of straw and composition of main microbial groups during the decomposition of wheat and maize straws as affected by soil texture. Biol. Fertil. Soils 2020, 56, 11–24. [Google Scholar] [CrossRef]
- Lal, R. Soil carbon sequestration impacts on global climate change and food security. Science 2004, 304, 1623–1627. [Google Scholar] [CrossRef] [PubMed]
- Wahdan, S.F.M.; Ji, L.; Schadler, M.; Wu, Y.-T.; Sansupa, C.; Tanunchai, B.; Buscot, F.; Purahong, W. Future climate conditions accelerate wheat straw decomposition alongside altered microbial community composition, assembly patterns, and interaction networks. ISME J. 2023, 17, 238–251. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.; Keiblinger, I.M.; Leitner, S.; Mentler, A.; Zechmeister-Boltenstem, S. Is there a convergence of deciduous leaf litter stoichiometry, biochemistry and microbial population during decay? Geoderma 2016, 272, 93–100. [Google Scholar] [CrossRef]
- Wang, H.; Liu, S.; Wang, J.; Shi, Z.; Lu, L.; Guo, W.; Jia, H.; Cai, D. Dynamics and speciation of organic carbon during decomposition of leaf litter and fine roots in four subtropical plantations of China. For. Ecol. Manag. 2013, 300, 43–52. [Google Scholar] [CrossRef]
- Tharayil, N.; Suseela, V.; Triebwasser, D.J.; Preston, C.M.; Gerard, P.D.; Dukes, J.S. Changes in the structural composition and reactivity of Acer rubrum leaf litter tannins exposed to warming and altered precipitation: Climatic stress-induced tannins are more reactive. New Phytol. 2011, 191, 132–145. [Google Scholar] [CrossRef] [PubMed]
- Huang, F.; Li, Q.; Xue, L.; Han, J.; Zamanian, K.; Zhao, X. The succession of microbial communities after residue returning in a Solonchak. Plant Soil 2023, 492, 191–208. [Google Scholar] [CrossRef]
- Han, Y.; Yao, S.-H.; Jiang, H.; Ge, X.-l.; Zhang, Y.; Mao, J.; Dou, S.; Zhang, B. Effects of mixing maize straw with soil and placement depths on decomposition rates and products at two cold sites in the mollisol region of China. Soil Tillage Res. 2020, 197, 104519. [Google Scholar] [CrossRef]
- Wang, X.; Sun, B.; Mao, J.; Sui, Y.; Cao, X. Structural Convergence of Maize and Wheat Straw during Two-Year Decomposition under Different Climate Conditions. Environ. Sci. Technol. 2012, 46, 7159–7165. [Google Scholar] [CrossRef]
- Thongjoo, C.; Miyagawa, S.; Kawakubo, N. Effects of soil moisture and temperature on decomposition rates of some waste materials from agriculture and agro-industry. Plant Prod. Sci. 2005, 8, 475–481. [Google Scholar] [CrossRef]
- Seyfried, G.S.; Dalling, J.W.; Yang, W.H. Mycorrhizal type effects on leaf litter decomposition depend on litter quality and environmental context. Biogeochemistry 2021, 155, 21–38. [Google Scholar] [CrossRef]
- Mwafulirwa, L.; Baggs, E.M.; Morley, N.; Paterson, E. Ryegrass root and shoot residues differentially affect short-term priming of soil organic matter and net soil C-balance. Eur. J. Soil Biol. 2019, 93, 103096. [Google Scholar] [CrossRef]
- Su, Y.; Gong, Y.; Han, W.; Li, K.; Liu, X. Dependency of litter decomposition on litter quality, climate change, and grassland type in the alpine grassland of Tianshan Mountains, Northwest China. J. Arid Land 2022, 14, 691–703. [Google Scholar] [CrossRef]
- Pascault, N.; Cecillon, L.; Mathieu, O.; Henault, C.; Sarr, A.; Leveque, J.; Farcy, P.; Ranjard, L.; Maron, P.-A. In Situ Dynamics of Microbial Communities during Decomposition of Wheat, Rape, and Alfalfa Residues. Microb. Ecol. 2010, 60, 816–828. [Google Scholar] [CrossRef]
- Clemente, J.S.; Simpson, M.J.; Simpson, A.J.; Yanni, S.F.; Whalen, J.K. Comparison of soil organic matter composition after incubation with maize leaves, roots, and stems. Geoderma 2013, 192, 86–96. [Google Scholar] [CrossRef]
- Bertrand, I.; Chabbert, B.; Kurek, B.; Recous, S. Can the biochemical features and histology of wheat residues explain their decomposition in soil? Plant Soil 2006, 281, 291–307. [Google Scholar] [CrossRef]
- Liu, J.; Ding, C.; Zhang, W.; Wei, Y.; Zhou, Y.; Zhu, W. Litter mixing promoted decomposition rate through increasing diversities of phyllosphere microbial communities. Front. Microbiol. 2022, 13, 1009091. [Google Scholar] [CrossRef] [PubMed]
- Wickings, K.; Grandy, A.S.; Reed, S.C.; Cleveland, C.C. The origin of litter chemical complexity during decomposition. Ecol. Lett. 2012, 15, 1180–1188. [Google Scholar] [CrossRef]
- Li, D.; Zhao, B.; Olk, D.C.; Zhang, J. Soil texture and straw type modulate the chemical structure of residues during four-year decomposition by regulating bacterial and fungal communities. Appl. Soil Ecol. 2020, 155, 103664. [Google Scholar] [CrossRef]
- Nair, A.; Ngouajio, M. Soil microbial biomass, functional microbial diversity, and nematode community structure as affected by cover crops and compost in an organic vegetable production system. Appl. Soil Ecol. 2012, 58, 45–55. [Google Scholar] [CrossRef]
- Zhao, S.; Fan, F.; Qiu, S.; Xu, X.; He, P.; Ciampitti, I.A. Dynamic of fungal community composition during maize residue decomposition process in north-central China. Appl. Soil Ecol. 2021, 167, 104057. [Google Scholar] [CrossRef]
- Paterson, E.; Sim, A.; Osborne, S.M.; Murray, P.J. Long-term exclusion of plant-inputs to soil reduces the functional capacity of microbial communities to mineralise recalcitrant root-derived carbon sources. Soil Biol. Biochem. 2011, 43, 1873–1880. [Google Scholar] [CrossRef]
- Cho, H.; Kim, M.; Tripathi, B.; Adams, J. Changes in Soil Fungal Community Structure with Increasing Disturbance Frequency. Microb. Ecol. 2017, 74, 62–77. [Google Scholar] [CrossRef] [PubMed]
- Golebiewski, M.; Tarasek, A.; Sikora, M.; Deja-Sikora, E.; Tretyn, A.; Niklinska, M. Rapid Microbial Community Changes During Initial Stages of Pine Litter Decomposition. Microb. Ecol. 2019, 77, 56–75. [Google Scholar] [CrossRef] [PubMed]
- Qiao, Y.-F.; Miao, X.-C.; Burger, M.; Miao, S.-J. Shift of fungal community composition in response to exogenous C application associated with soil properties after 10-year field experiment in black soil of China. J. Soils Sediments 2022, 22, 2281–2289. [Google Scholar] [CrossRef]
- Lopez-Mondejar, R.; Brabcova, V.; Stursova, M.; Davidova, A.; Jansa, J.; Cajthaml, T.; Baldrian, P. Decomposer food web in a deciduous forest shows high share of generalist microorganisms and importance of microbial biomass recycling. ISME J. 2018, 12, 1768–1778. [Google Scholar] [CrossRef] [PubMed]
- Xiao, D.; He, X.; Wang, G.; Xu, X.; Hu, Y.; Chen, X.; Zhang, W.; Su, Y.A.; Wang, K.; Soromotin, A.V.; et al. Network analysis reveals bacterial and fungal keystone taxa involved in straw and soil organic matter mineralization. Appl. Soil Ecol. 2022, 173, 104395. [Google Scholar] [CrossRef]
- Veen, G.F.; ten Hooven, F.C.; Weser, C.; Hannula, S.E. Steering the soil microbiome by repeated litter addition. J. Ecol. 2021, 109, 2499–2513. [Google Scholar] [CrossRef]
- Baumann, K.; Marschner, P.; Smernik, R.J.; Baldock, J.A. Residue chemistry and microbial community structure during decomposition of eucalypt, wheat and vetch residues. Soil Biol. Biochem. 2009, 41, 1966–1975. [Google Scholar] [CrossRef]
- Bonanomi, G.; De Filippis, F.; Cesarano, G.; La Storia, A.; Zotti, M.; Mazzoleni, S.; Incerti, G. Linking bacterial and eukaryotic microbiota to litter chemistry: Combining next generation sequencing with C-13 CPMAS NMR spectroscopy. Soil Biol. Biochem. 2019, 129, 110–121. [Google Scholar] [CrossRef]
- Eichlerova, I.; Homolka, L.; Zifcakova, L.; Lisa, L.; Dobiasova, P.; Baldrian, P. Enzymatic systems involved in decomposition reflects the ecology and taxonomy of saprotrophic fungi. Fungal Ecol. 2015, 13, 10–22. [Google Scholar] [CrossRef]
- Li, Y.; Nie, C.; Liu, Y.; Du, W.; He, P. Soil microbial community composition closely associates with specific enzyme activities and soil carbon chemistry in a long-term nitrogen fertilized grassland. Sci. Total Environ. 2019, 654, 264–274. [Google Scholar] [CrossRef]
- Laza, H.E.; Acosta-Martinez, V.; Cano, A.; Baker, J.; Mahan, J.; Gitz, D.; Emendack, Y.; Slaughter, L.; Lascano, R.; Tissue, D.; et al. Elevated CO2 enhances soil respiration and AMF abundance in a semiarid peanut agroecosystem. Agric. Ecosyst. Environ. 2023, 355, 108592. [Google Scholar] [CrossRef]
- Sinsabaugh, R.L. Phenol oxidase, peroxidase and organic matter dynamics of soil. Soil Biol. Biochem. 2010, 42, 391–404. [Google Scholar] [CrossRef]
- Purahong, W.; Krueger, D.; Buscot, F.; Wubet, T. Correlations between the composition of modular fungal communities and litter decomposition-associated ecosystem functions. Fungal Ecol. 2016, 22, 106–114. [Google Scholar] [CrossRef]
- Peng, Y.; Li, Y.-j.; Song, S.-y.; Chen, Y.-q.; Chen, G.-t.; Tu, L.-h. Nitrogen addition slows litter decomposition accompanied by accelerated manganese release: A five-year experiment in a subtropical evergreen broadleaf forest. Soil Biol. Biochem. 2022, 165, 108511. [Google Scholar] [CrossRef]
- Cusack, D.F.; Silver, W.L.; Torn, M.S.; Burton, S.D.; Firestone, M.K. Changes in microbial community characteristics and soil organic matter with nitrogen additions in two tropical forests. Ecology 2011, 92, 621–632. [Google Scholar] [CrossRef] [PubMed]
- Zheng, H.; Yang, T.; Bao, Y.; He, P.; Yang, K.; Mei, X.; Wei, Z.; Xu, Y.; Shen, Q.; Banerjee, S. Network analysis and subsequent culturing reveal keystone taxa involved in microbial litter decomposition dynamics. Soil Biol. Biochem. 2021, 157, 108230. [Google Scholar] [CrossRef]
- Gonzalez, A.; Germain, R.M.; Srivastava, D.S.; Filotas, E.; Dee, L.E.; Gravel, D.; Thompson, P.L.; Isbell, F.; Wang, S.; Kefi, S.; et al. Scaling-up biodiversity-ecosystem functioning research. Ecol. Lett. 2020, 23, 757–776. [Google Scholar] [CrossRef]
- Banerjee, S.; Kirkby, C.A.; Schmutter, D.; Bissett, A.; Kirkegaard, J.A.; Richardson, A.E. Network analysis reveals functional redundancy and keystone taxa amongst bacterial and fungal communities during organic matter decomposition in an arable soil. Soil Biol. Biochem. 2016, 97, 188–198. [Google Scholar] [CrossRef]
- Banerjee, S.; Schlaeppi, K.; van der Heijden, M.G.A. Keystone taxa as drivers of microbiome structure and functioning. Nat. Rev. Microbiol. 2018, 16, 567–576. [Google Scholar] [CrossRef] [PubMed]
- Liao, H.; Xu, C.; Tan, S.; Wei, Z.; Ling, N.; Yu, G.; Raza, W.; Zhang, R.; Shen, Q.; Xu, Y. Production and characterization of acidophilic xylanolytic enzymes from Penicillium oxalicum GZ-2. Bioresour. Technol. 2012, 123, 117–124. [Google Scholar] [CrossRef] [PubMed]
- Mei, N.; Zhang, X.; Wang, X.; Peng, C.; Gao, H.; Zhu, P.; Gu, Y. Effects of 40 years applications of inorganic and organic fertilization on soil bacterial community in a maize agroecosystem in northeast China. Eur. J. Agron. 2021, 130, 126332. [Google Scholar] [CrossRef]
- Zhang, Q.; Lu, X.; Chen, G.; Luo, N.; Sun, J.; Li, X.; Ngozi, E.A. The priming effect patterns linked to the dominant bacterial keystone taxa during different straw tissues incorporation into Mollisols in Northeast China. Appl. Soil Ecol. 2024, 197, 105330. [Google Scholar] [CrossRef]
- Shrestha, B.M.; Singh, B.R.; Forte, C.; Certini, G. Long-term effects of tillage, nutrient application and crop rotation on soil organic matter quality assessed by NMR spectroscopy. Soil Use Manag. 2015, 31, 358–366. [Google Scholar] [CrossRef]
- Wang, X.; Liang, C.; Mao, J.; Jiang, Y.; Bian, Q.; Liang, Y.; Chen, Y.; Sun, B. Microbial keystone taxa drive succession of plant residue chemistry. ISME J. 2023, 17, 748–757. [Google Scholar] [CrossRef] [PubMed]
- Baldock, J.A.; Masiello, C.A.; Gelinas, Y.; Hedges, J.I. Cycling and composition of organic matter in terrestrial and marine ecosystems. Mar. Chem. 2004, 92, 39–64. [Google Scholar] [CrossRef]
- Grandy, A.S.; Salam, D.S.; Wickings, K.; McDaniel, M.D.; Culman, S.W.; Snapp, S.S. Soil respiration and litter decomposition responses to nitrogen fertilization rate in no-till corn systems. Agric. Ecosyst. Environ. 2013, 179, 35–40. [Google Scholar] [CrossRef]
- Liu, S.; Fan, R.; Yang, X.; Zhang, Z.; Zhang, X.; Liang, A. Decomposition of maize stover varies with maize type and stover management strategies: A microcosm study on a Black soil (Mollisol) in northeast China. J. Environ. Manag. 2019, 234, 226–236. [Google Scholar] [CrossRef]
- Xu, Y.; Fan, J.; Ding, W.; Gunina, A.; Chen, Z.; Bol, R.; Luo, J.; Bolan, N. Characterization of organic carbon in decomposing litter exposed to nitrogen and sulfur additions: Links to microbial community composition and activity. Geoderma 2017, 286, 116–124. [Google Scholar] [CrossRef]
- Lundberg, P.; Ekblad, A.; Nilsson, M. C-13 NMR spectroscopy studies of forest soil microbial activity: Glucose uptake and fatty acid biosynthesis. Soil Biol. Biochem. 2001, 33, 621–632. [Google Scholar] [CrossRef]
- Zhao, H.; Lv, Y.; Wang, X.; Zhang, H.; Yang, X. Tillage impacts on the fractions and compositions of soil organic carbon. Geoderma 2012, 189, 397–403. [Google Scholar] [CrossRef]
- Lorenz, K.; Preston, C.M.; Raspe, S.; Morrison, I.K.; Feger, K.H. Litter decomposition and humus characteristics in Canadian and German spruce ecosystems: Information from tannin analysis and C-13 CPMAS NMR. Soil Biol. Biochem. 2000, 32, 779–792. [Google Scholar] [CrossRef]
- Sanaullah, M.; Chabbi, A.; Maron, P.-A.; Baumann, K.; Tardy, V.; Blagodatskaya, E.; Kuzyakov, Y.; Rumpel, C. How do microbial communities in top-and subsoil respond to root litter addition under field conditions? Soil Biol. Biochem. 2016, 103, 28–38. [Google Scholar] [CrossRef]
- Zhao, S.; Qiu, S.; Xu, X.; Ciampitti, I.A.; Zhang, S.; He, P. Change in straw decomposition rate and soil microbial community composition after straw addition in different long-term fertilization soils. Appl. Soil Ecol. 2019, 138, 123–133. [Google Scholar] [CrossRef]
- Herzog, C.; Hartmann, M.; Frey, B.; Stierli, B.; Rumpel, C.; Buchmann, N.; Brunner, I. Microbial succession on decomposing root litter in a drought-prone Scots pine forest. ISME J. 2019, 13, 2346–2362. [Google Scholar] [CrossRef] [PubMed]
- Pascault, N.; Ranjard, L.; Kaisermann, A.; Bachar, D.; Christen, R.; Terrat, S.; Mathieu, O.; Leveque, J.; Mougel, C.; Henault, C.; et al. Stimulation of Different Functional Groups of Bacteria by Various Plant Residues as a Driver of Soil Priming Effect. Ecosystems 2013, 16, 810–822. [Google Scholar] [CrossRef]
- Song, A.; Zhang, J.; Xu, D.; Wang, E.; Bi, J.; Asante-Badu, B.; Njyenawe, M.C.; Sun, M.; Xue, P.; Wang, S.; et al. Keystone microbial taxa drive the accelerated decompositions of cellulose and lignin by long-term resource enrichments. Sci. Total Environ. 2022, 842, 156814. [Google Scholar] [CrossRef]
- Voriskova, J.; Baldrian, P. Fungal community on decomposing leaf litter undergoes rapid successional changes. ISME J. 2013, 7, 477–486. [Google Scholar] [CrossRef]
- Jin, X.; Wang, Z.; Wu, F.; Li, X.; Zhou, X. Litter Mixing Alters Microbial Decomposer Community to Accelerate Tomato Root Litter Decomposition. Microbiol. Spectr. 2022, 10, e0018622. [Google Scholar] [CrossRef] [PubMed]
- Espana, M.; Rasche, F.; Kandeler, E.; Brune, T.; Rodriguez, B.; Bending, G.D.; Cadisch, G. Assessing the effect of organic residue quality on active decomposing fungi in a tropical Vertisol using N-15-DNA stable isotope probing. Fungal Ecol. 2011, 4, 115–119. [Google Scholar] [CrossRef]
- Purahong, W.; Wubet, T.; Lentendu, G.; Schloter, M.; Pecyna, M.J.; Kapturska, D.; Hofrichter, M.; Krueger, D.; Buscot, F. Life in leaf litter: Novel insights into community dynamics of bacteria and fungi during litter decomposition. Mol. Ecol. 2016, 25, 4059–4074. [Google Scholar] [CrossRef] [PubMed]
- Osono, T. Ecology of ligninolytic fungi associated with leaf litter decomposition. Ecol. Res. 2007, 22, 955–974. [Google Scholar] [CrossRef]
- Cenini, V.L.; Fornara, D.A.; McMullan, G.; Ternan, N.; Carolan, R.; Crawley, M.J.; Clement, J.-C.; Lavorel, S. Linkages between extracellular enzyme activities and the carbon and nitrogen content of grassland soils. Soil Biol. Biochem. 2016, 96, 198–206. [Google Scholar] [CrossRef]
- Wilhelm, R.C.; Cardenas, E.; Leung, H.; Szeitz, A.; Jensen, L.D.; Mohn, W.W. Long-Term Enrichment of Stress-Tolerant Cellulolytic Soil Populations following Timber Harvesting Evidenced by Multi-Omic Stable Isotope Probing. Front. Microbiol. 2017, 8, 537. [Google Scholar] [CrossRef] [PubMed]
- Clocchiatti, A.; Hannula, S.E.; van den Berg, M.; Korthals, G.; de Boer, W. The hidden potential of saprotrophic fungi in arable soil: Patterns of short-term stimulation by organic amendments. Appl. Soil Ecol. 2020, 147, 103434. [Google Scholar] [CrossRef]
- Miao, Y.; Lin, Y.; Chen, Z.; Zheng, H.; Niu, Y.; Kuzyakov, Y.; Liu, D.; Ding, W. Fungal key players of cellulose utilization: Microbial networks in aggregates of long-term fertilized soils disentangled using C-13-DNA-stable isotope probing. Sci. Total Environ. 2022, 832, 155051. [Google Scholar] [CrossRef] [PubMed]
- Miao, Y.; Li, J.; Li, Y.; Niu, Y.; He, T.; Liu, D.; Ding, W. Long-Term Compost Amendment Spurs Cellulose Decomposition by Driving Shifts in Fungal Community Composition and Promoting Fungal Diversity and Phylogenetic Relatedness. mBio 2022, 13, e0032322. [Google Scholar] [CrossRef]
- Koechli, C.; Campbell, A.N.; Pepe-Ranney, C.; Buckley, D.H. Assessing fungal contributions to cellulose degradation in soil by using high-throughput stable isotope probing. Soil Biol. Biochem. 2019, 130, 150–158. [Google Scholar] [CrossRef]
- Rodrigues, P.d.O.; Alves Gurgel, L.V.; Pasquini, D.; Badotti, F.; Goes-Neto, A.; Baffi, M.A. Lignocellulose-degrading enzymes production by solid-state fermentation through fungal consortium among Ascomycetes and Basidiomycetes. Renew. Energy 2020, 145, 2683–2693. [Google Scholar] [CrossRef]
- Liao, H.; Li, S.; Wei, Z.; Shen, Q.; Xu, Y. Insights into high-efficiency lignocellulolytic enzyme production by Penicillium oxalicum GZ-2 induced by a complex substrate. Biotechnol. Biofuels 2014, 7, 162. [Google Scholar] [CrossRef] [PubMed]
- Oates, N.C.; Abood, A.; Schirmacher, A.M.; Alessi, A.M.; Bird, S.M.; Bennett, J.P.; Leadbeater, D.R.; Li, Y.; Dowle, A.A.; Liu, S.; et al. A multi-omics approach to lignocellulolytic enzyme discovery reveals a new ligninase activity from Parascedosporium putredinis NO1. Proc. Natl. Acad. Sci. USA 2021, 118, e2008888118. [Google Scholar] [CrossRef] [PubMed]
- Cui, J.; Song, D.; Dai, X.; Xu, X.; He, P.; Wang, X.; Liang, G.; Zhou, W.; Zhu, P. Effects of long-term cropping regimes on SOC stability, soil microbial community and enzyme activities in the Mollisol region of Northeast China. Appl. Soil Ecol. 2021, 164, 103941. [Google Scholar] [CrossRef]
- de Vries, F.T.; Manning, P.; Tallowin, J.R.B.; Mortimer, S.R.; Pilgrim, E.S.; Harrison, K.A.; Hobbs, P.J.; Quirk, H.; Shipley, B.; Cornelissen, J.H.C.; et al. Abiotic drivers and plant traits explain landscape-scale patterns in soil microbial communities. Ecol. Lett. 2012, 15, 1230–1239. [Google Scholar] [CrossRef] [PubMed]
- Adetunji, A.T.; Lewu, F.B.; Mulidzi, R.; Ncube, B. The biological activities of beta-glucosidase, phosphatase and urease as soil quality indicators: A review. J. Soil Sci. Plant Nutr. 2017, 17, 794–807. [Google Scholar] [CrossRef]
- Kellner, H.; Luis, P.; Pecyna, M.J.; Barbi, F.; Kapturska, D.; Krueger, D.; Zak, D.R.; Marmeisse, R.; Vandenbol, M.; Hofrichter, M. Widespread Occurrence of Expressed Fungal Secretory Peroxidases in Forest Soils. PLoS ONE 2014, 9, e95557. [Google Scholar] [CrossRef]
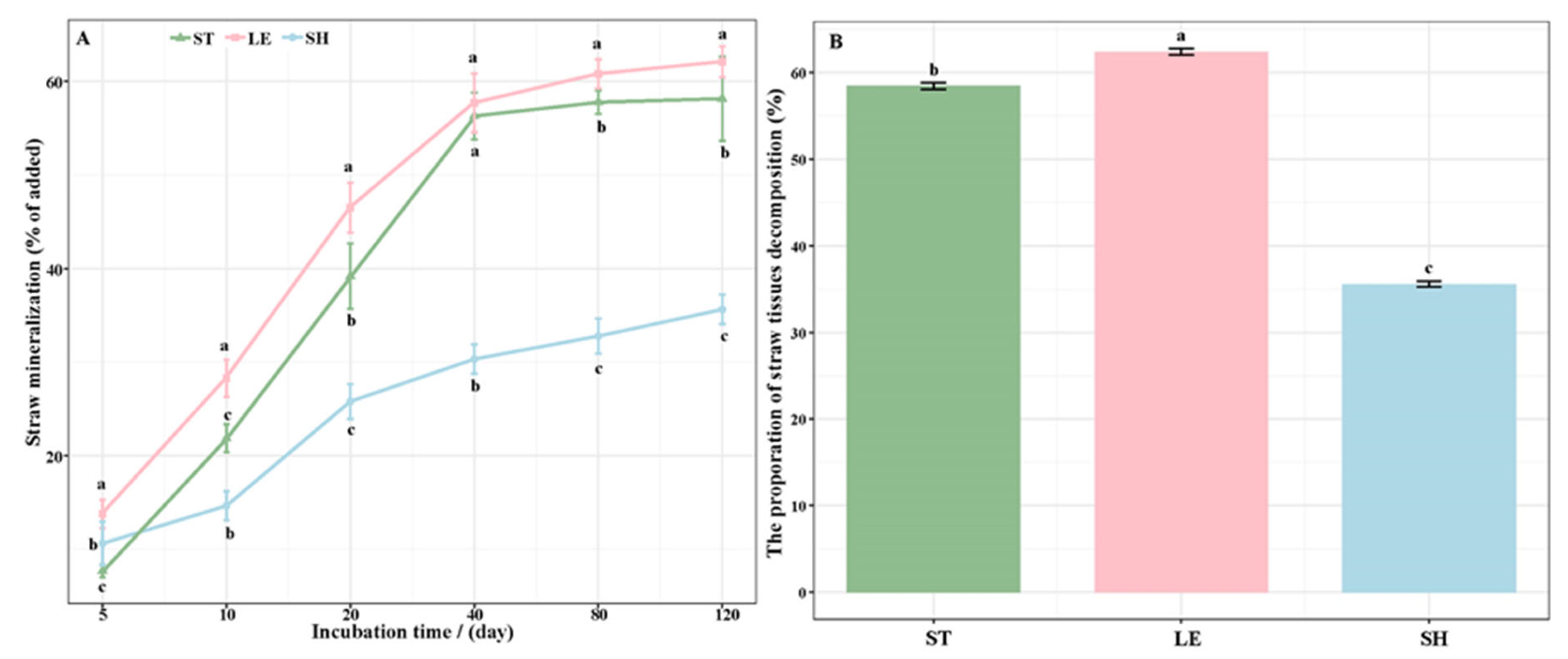
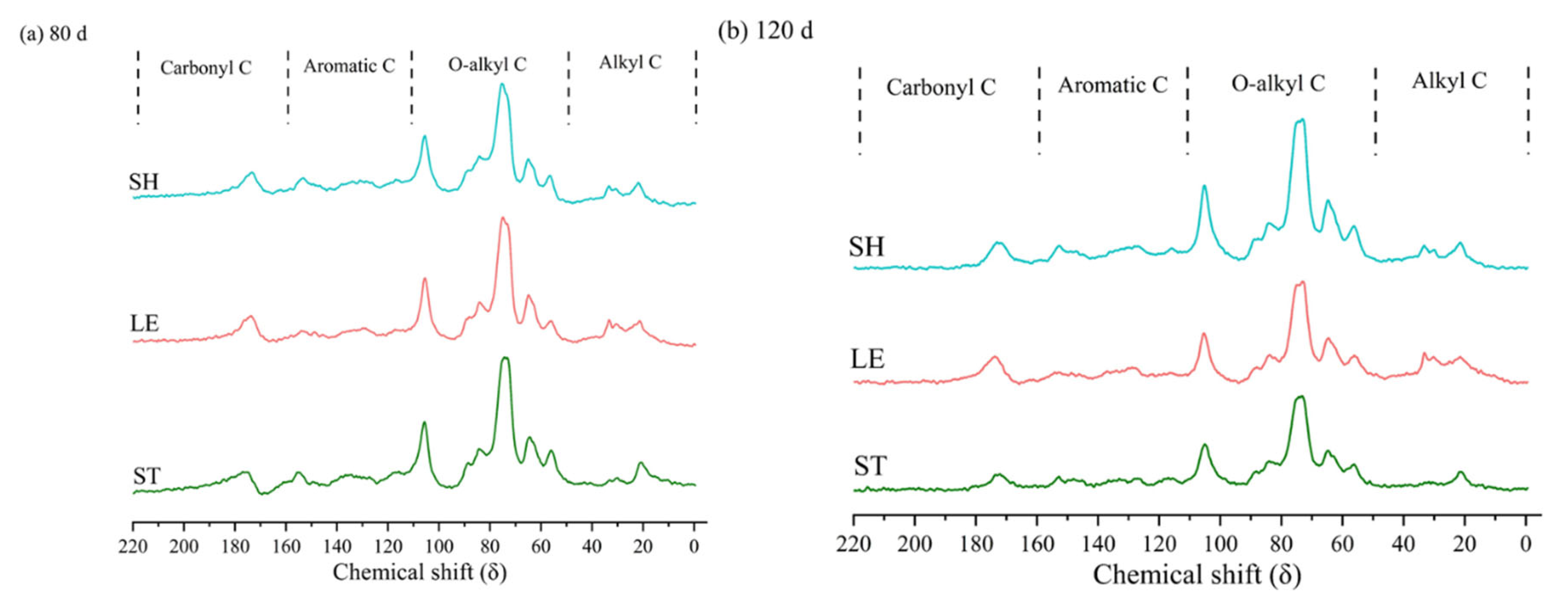
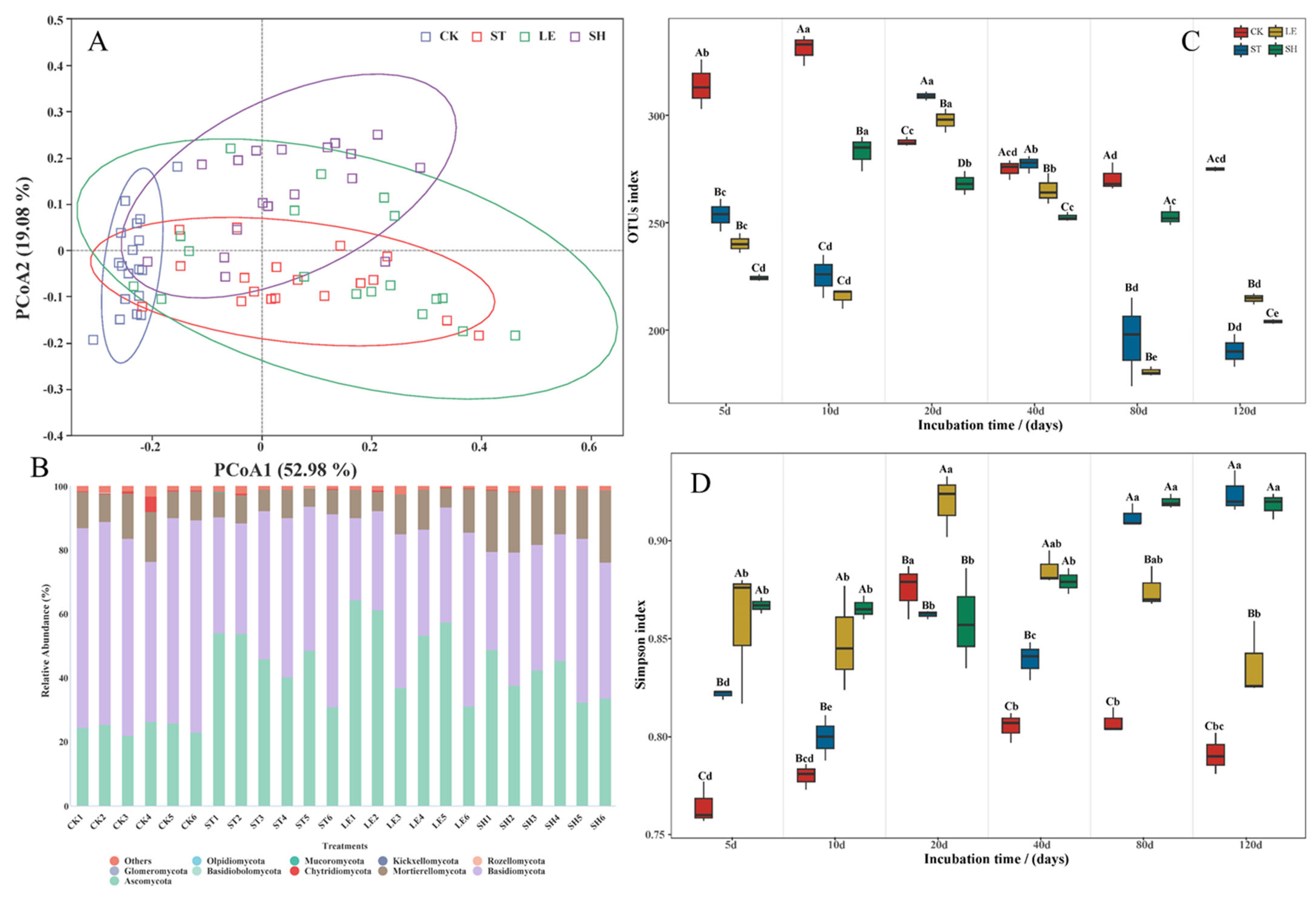
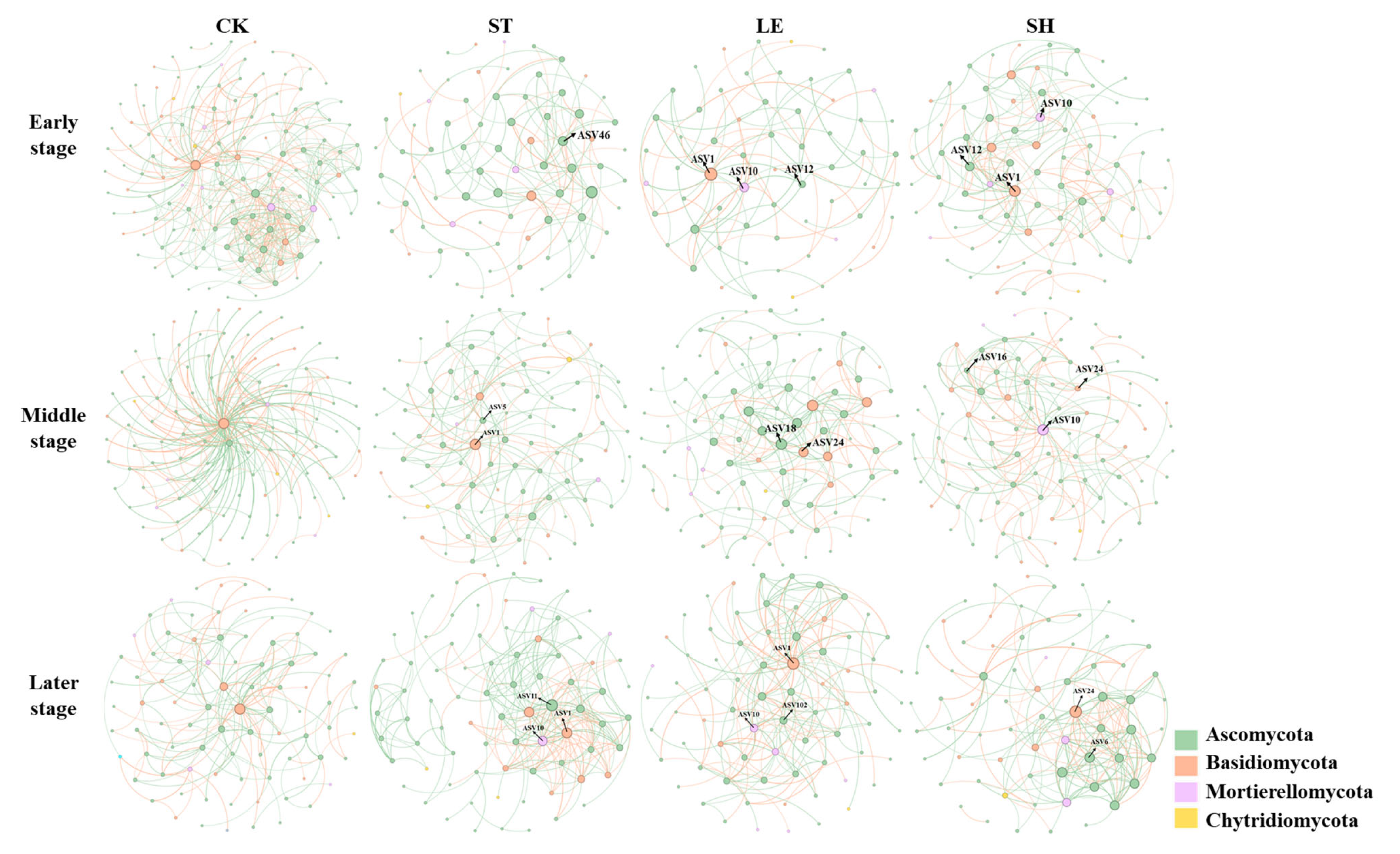
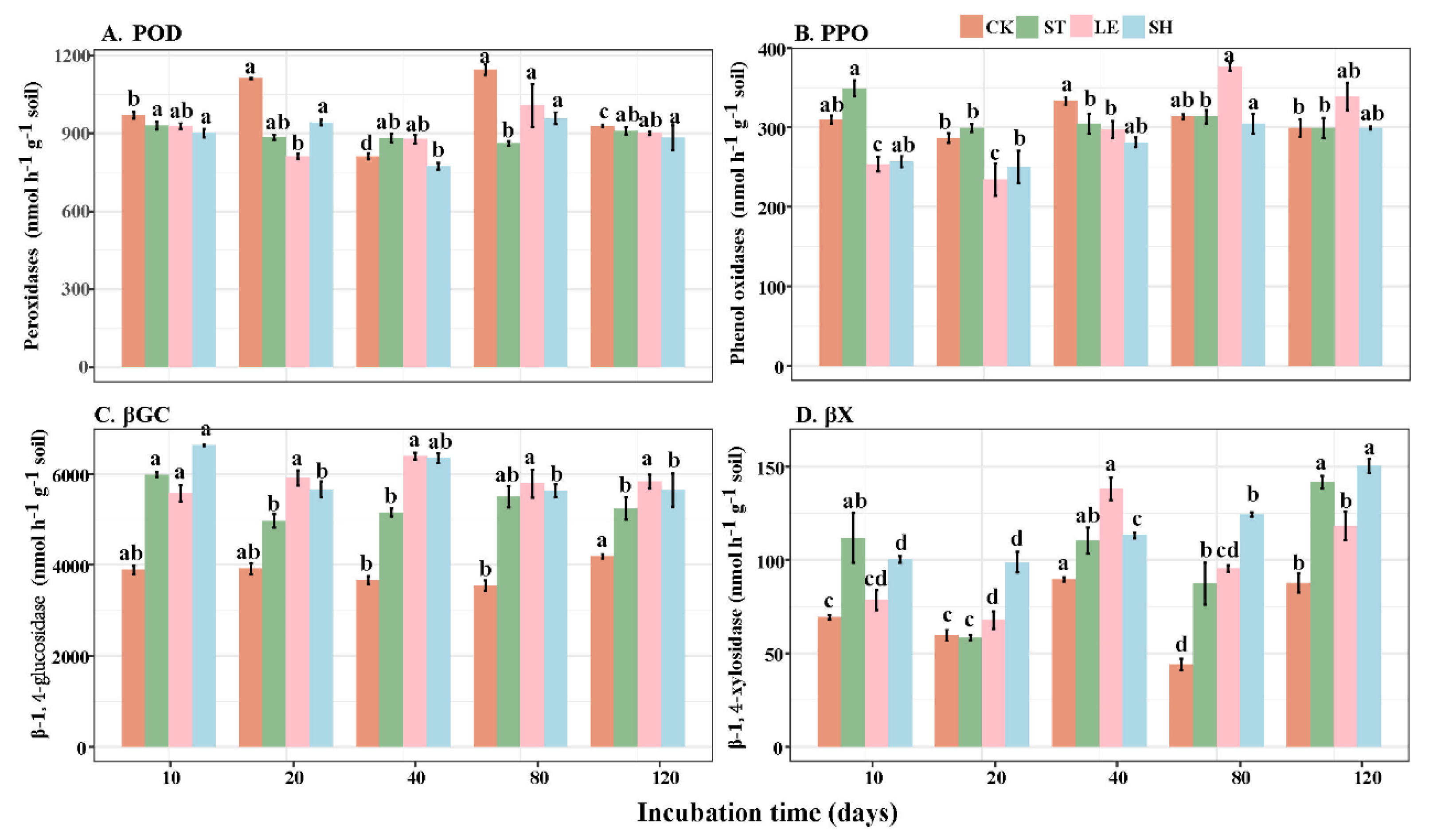
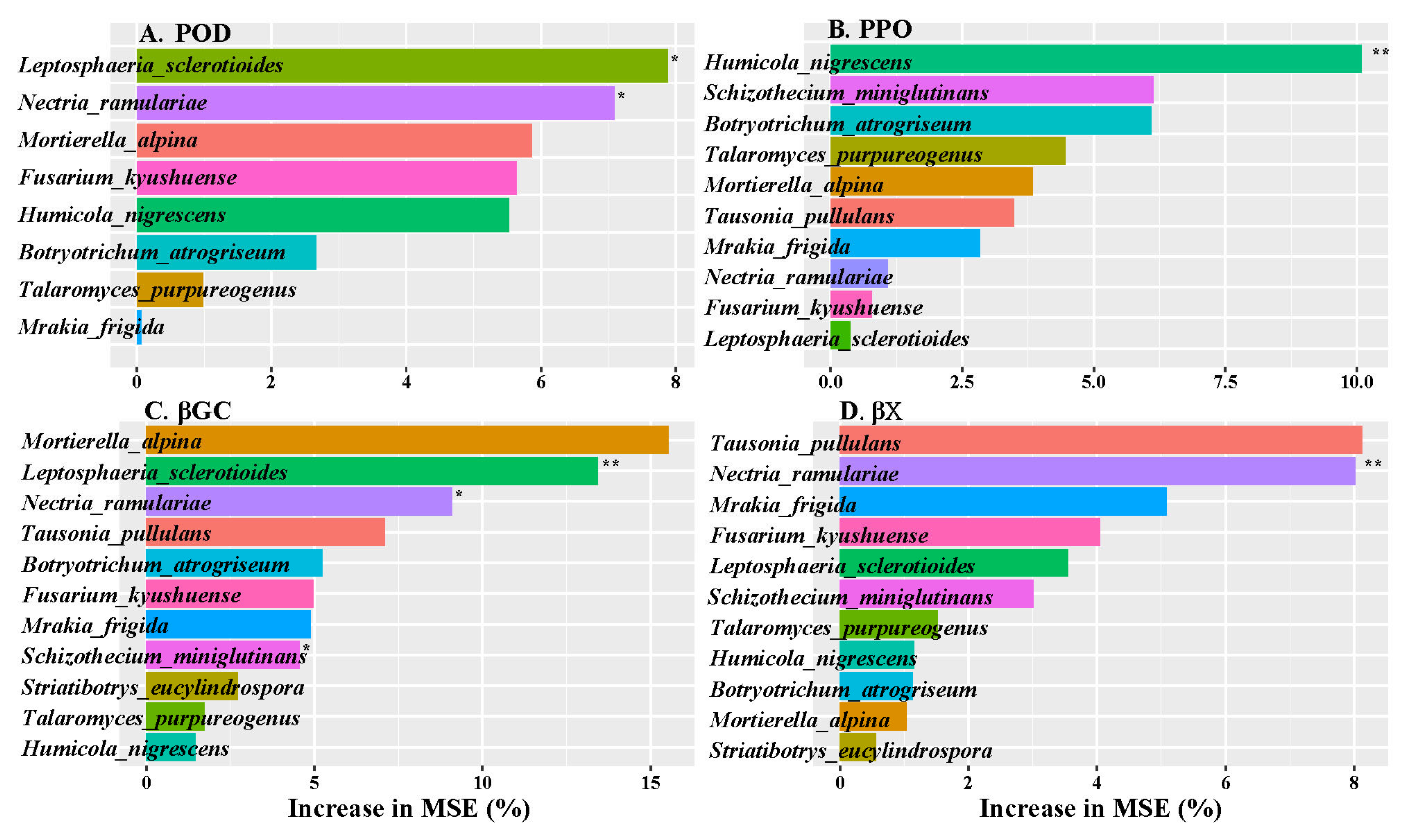

| Alkyl C (0–45 ppm) | O-Alkyl C (45–110 ppm) | Aromatic C (110–160 ppm) | Carbonyl C (160–220 ppm) | A/ OA | AL/AR | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| O/N-alkyl C | di-O-alkyl C | Total | Aryl C/O-aryl C | Phenolic C | Total | ||||||
| 80d | ST | 11.59 | 55.26 | 10.30 | 65.56 | 7.75 | 6.18 | 13.93 | 8.92 | 0.18 | 3.38 |
| LE | 10.17 | 57.49 | 12.40 | 69.89 | 2.68 | 4.96 | 7.64 | 12.30 | 0.15 | 4.02 | |
| SH | 5.42 | 50.05 | 14.43 | 64.48 | 10.31 | 7.55 | 17.86 | 12.24 | 0.08 | 2.32 | |
| 120d | ST | 13.66 | 47.58 | 11.36 | 58.94 | 11.69 | 3.98 | 15.68 | 11.72 | 0.23 | 2.65 |
| LE | 16.25 | 52.01 | 11.17 | 63.18 | 6.66 | 3.16 | 9.83 | 10.74 | 0.26 | 3.86 | |
| SH | 10.68 | 50.05 | 14.42 | 64.47 | 6.70 | 7.38 | 17.48 | 7.38 | 0.16 | 3.02 | |
| Biomolecule | C-Type | 80d | 120d | ||||
|---|---|---|---|---|---|---|---|
| ST | LE | SH | ST | LE | SH | ||
| Carbohydrate | O-alkyl, di-O-alkyl | 65.56 | 69.89 | 64.48 | 58.94 | 63.18 | 64.47 |
| Protein | alkyl, N-alkyl, carbonyl | 20.51 | 22.47 | 17.66 | 25.38 | 26.99 | 18.06 |
| Lignin | alkyl, N-alkyl, O-alkyl, aryl, O-aryl | 80.78 | 75.30 | 73.33 | 76.92 | 78.09 | 78.20 |
| Lipid | alkyl | 11.59 | 10.17 | 5.42 | 13.66 | 16.25 | 10.68 |
| Treatment | Node | Edge | Positive | Negative | Average Degree (avgK) | Diameter | Density | Modularity | Average Clustering Coefficient (avgCC) | Average Path Distance (GD) | Betweenness_Centralization | Degree_Centralization | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ST | Early | 157 | 119 | 56.3 | 43.7 | 2.644 | 15 | 0.0097 | 0.706 | 0.3626 | 5.4129 | 0.0621 | 0.0544 |
| Middle | 203 | 175 | 60.6 | 39.4 | 3.103 | 10 | 0.0085 | 0.631 | 0.2849 | 4.0872 | 0.0749 | 0.0855 | |
| Later | 193 | 247 | 66.8 | 33.2 | 5.277 | 10 | 0.0133 | 0.451 | 0.4929 | 3.6832 | 0.0427 | 0.1325 | |
| LE | Early | 157 | 85 | 55.3 | 44.7 | 2.366 | 11 | 0.0069 | 0.621 | 0.2897 | 4.3958 | 0.0449 | 0.0764 |
| Middle | 211 | 183 | 61.8 | 38.3 | 3.208 | 14 | 0.0083 | 0.62 | 0.4331 | 4.8536 | 0.0480 | 0.0584 | |
| Later | 129 | 187 | 52.9 | 47.1 | 4.456 | 7 | 0.0227 | 0.439 | 0.3850 | 3.0074 | 0.0774 | 0.1727 | |
| SH | Early | 169 | 179 | 55.9 | 44.1 | 3.612 | 7 | 0.0126 | 0.616 | 0.3757 | 3.0385 | 0.0266 | 0.0886 |
| Middle | 198 | 182 | 52.2 | 47.8 | 3.241 | 11 | 0.0093 | 0.608 | 0.2917 | 3.9736 | 0.0919 | 0.1125 | |
| Later | 126 | 164 | 60.4 | 39.6 | 4.245 | 9 | 0.0208 | 0.431 | 0.5655 | 3.7670 | 0.0625 | 0.1312 | |
| CK | Early | 238 | 424 | 53.5 | 46.5 | 5.507 | 8 | 0.0150 | 0.56 | 0.4635 | 3.1973 | 0.1237 | 0.1369 |
| Middle | 145 | 218 | 57.3 | 42.7 | 3.559 | 7 | 0.0209 | 0.441 | 0.0994 | 2.6351 | 0.4478 | 0.5000 | |
| Later | 151 | 162 | 56.2 | 43.8 | 3.239 | 8 | 0.0143 | 0.621 | 0.2421 | 3.9261 | 0.0999 | 0.1324 | |
| Treatments | Keystone Taxa | Degree | Kingdom | Phylum | Class | Order | Family | Genus | Species | VIP Score of EEA | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| ST | early | ASV46 | 8 | Fungi | Ascomycota | Sordariomycetes | Hypocreales | Nectriaceae | Nectria | Nectria_ramulariae | 1.58713 |
| middle | ASV1 | 19 | Fungi | Basidiomycota | Tremellomycetes | Cystofilobasidiales | Mrakiaceae | Tausonia | Tausonia_pullulans | 2.01394 | |
| ASV5 | 9 | Fungi | Ascomycota | Sordariomycetes | Sordariales | Lasiosphaeriaceae | Schizothecium | Schizothecium_miniglutinans | 1.93246 | ||
| later | ASV1 | 24 | Fungi | Basidiomycota | Tremellomycetes | Cystofilobasidiales | Mrakiaceae | Tausonia | Tausonia_pullulans | 1.93993 | |
| ASV10 | 22 | Fungi | Mortierellomycota | Mortierellomycetes | Mortierellales | Mortierellaceae | Mortierella | Mortierella_alpina | 1.99996 | ||
| ASV11 | 28 | Fungi | Ascomycota | Eurotiomycetes | Eurotiales | Trichocomaceae | Talaromyces | Talaromyces_purpureogenus | 1.62333 | ||
| LE | early | ASV1 | 13 | Fungi | Basidiomycota | Tremellomycetes | Cystofilobasidiales | Mrakiaceae | Tausonia | Tausonia_pullulans | 1.33279 |
| ASV10 | 9 | Fungi | Mortierellomycota | Mortierellomycetes | Mortierellales | Mortierellaceae | Mortierella | Mortierella_alpina | 1.53937 | ||
| ASV12 | 6 | Fungi | Ascomycota | Dothideomycetes | Pleosporales | Leptosphaeriaceae | Leptosphaeria | Leptosphaeria_sclerotioides | 1.51172 | ||
| middle | ASV18 | 14 | Fungi | Ascomycota | Sordariomycetes | Sordariales | Chaetomiaceae | Botryotrichum | Botryotrichum_atrogriseum | 1.92881 | |
| ASV24 | 12 | Fungi | Basidiomycota | Tremellomycetes | Cystofilobasidiales | Mrakiaceae | Mrakia | Mrakia_frigida | 1.83323 | ||
| later | ASV1 | 25 | Fungi | Basidiomycota | Tremellomycetes | Cystofilobasidiales | Mrakiaceae | Tausonia | Tausonia_pullulans | 1.93993 | |
| ASV10 | 15 | Fungi | Mortierellomycota | Mortierellomycetes | Mortierellales | Mortierellaceae | Mortierella | Mortierella_alpina | 1.99996 | ||
| ASV102 | 14 | Fungi | Ascomycota | Sordariomycetes | Hypocreales | Stachybotryaceae | Striatibotrys | Striatibotrys_eucylindrospora | 1.82102 | ||
| SH | early | ASV1 | 17 | Fungi | Basidiomycota | Tremellomycetes | Cystofilobasidiales | Mrakiaceae | Tausonia | Tausonia_pullulans | 1.33279 |
| ASV10 | 12 | Fungi | Mortierellomycota | Mortierellomycetes | Mortierellales | Mortierellaceae | Mortierella | Mortierella_alpina | 1.53937 | ||
| ASV12 | 13 | Fungi | Ascomycota | Dothideomycetes | Pleosporales | Leptosphaeriaceae | Leptosphaeria | Leptosphaeria_sclerotioides | 1.51172 | ||
| middle | ASV10 | 24 | Fungi | Mortierellomycota | Mortierellomycetes | Mortierellales | Mortierellaceae | Mortierella | Mortierella_alpina | 2.20626 | |
| ASV16 | 9 | Fungi | Ascomycota | Sordariomycetes | Sordariales | Chaetomiaceae | Humicola | Humicola_nigrescens | 2.05356 | ||
| ASV24 | 9 | Fungi | Basidiomycota | Tremellomycetes | Cystofilobasidiales | Mrakiaceae | Mrakia | Mrakia_frigida | 1.83323 | ||
| later | ASV6 | 15 | Fungi | Ascomycota | Sordariomycetes | Hypocreales | Nectriaceae | Fusarium | Fusarium_kyushuense | 1.49026 | |
| ASV24 | 19 | Fungi | Basidiomycota | Tremellomycetes | Cystofilobasidiales | Mrakiaceae | Mrakia | Mrakia_frigida | 1.19719 | ||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, Q.; Li, X.; Chen, G.; Luo, N.; Sun, J.; Ngozi, E.A.; Lu, X. The Residue Chemistry Transformation Linked to the Fungi Keystone Taxa during Different Residue Tissues Incorporation into Mollisols in Northeast China. Agriculture 2024, 14, 792. https://doi.org/10.3390/agriculture14060792
Zhang Q, Li X, Chen G, Luo N, Sun J, Ngozi EA, Lu X. The Residue Chemistry Transformation Linked to the Fungi Keystone Taxa during Different Residue Tissues Incorporation into Mollisols in Northeast China. Agriculture. 2024; 14(6):792. https://doi.org/10.3390/agriculture14060792
Chicago/Turabian StyleZhang, Qilin, Xiujun Li, Guoshuang Chen, Nana Luo, Jing Sun, Ezemaduka Anastasia Ngozi, and Xinrui Lu. 2024. "The Residue Chemistry Transformation Linked to the Fungi Keystone Taxa during Different Residue Tissues Incorporation into Mollisols in Northeast China" Agriculture 14, no. 6: 792. https://doi.org/10.3390/agriculture14060792
APA StyleZhang, Q., Li, X., Chen, G., Luo, N., Sun, J., Ngozi, E. A., & Lu, X. (2024). The Residue Chemistry Transformation Linked to the Fungi Keystone Taxa during Different Residue Tissues Incorporation into Mollisols in Northeast China. Agriculture, 14(6), 792. https://doi.org/10.3390/agriculture14060792






