A Genome-Wide Comparative Analysis of AUX1/LAX, PIN, and ABCB Genes Reveals Their Roles in Cucumber Fruit Curving
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials and Treatment
2.2. Characterization and Phylogenetic Analysis of Auxin Transporters
2.3. Gene Structure, Genome Distribution, and Duplication Analysis
2.4. Analysis of Cis-Acting Elements of Auxin Transporters
2.5. Co-Expression Analysis of Auxin Transporters
2.6. Real-Time Quantitative PCR(RT-qPCR) Analysis
3. Results
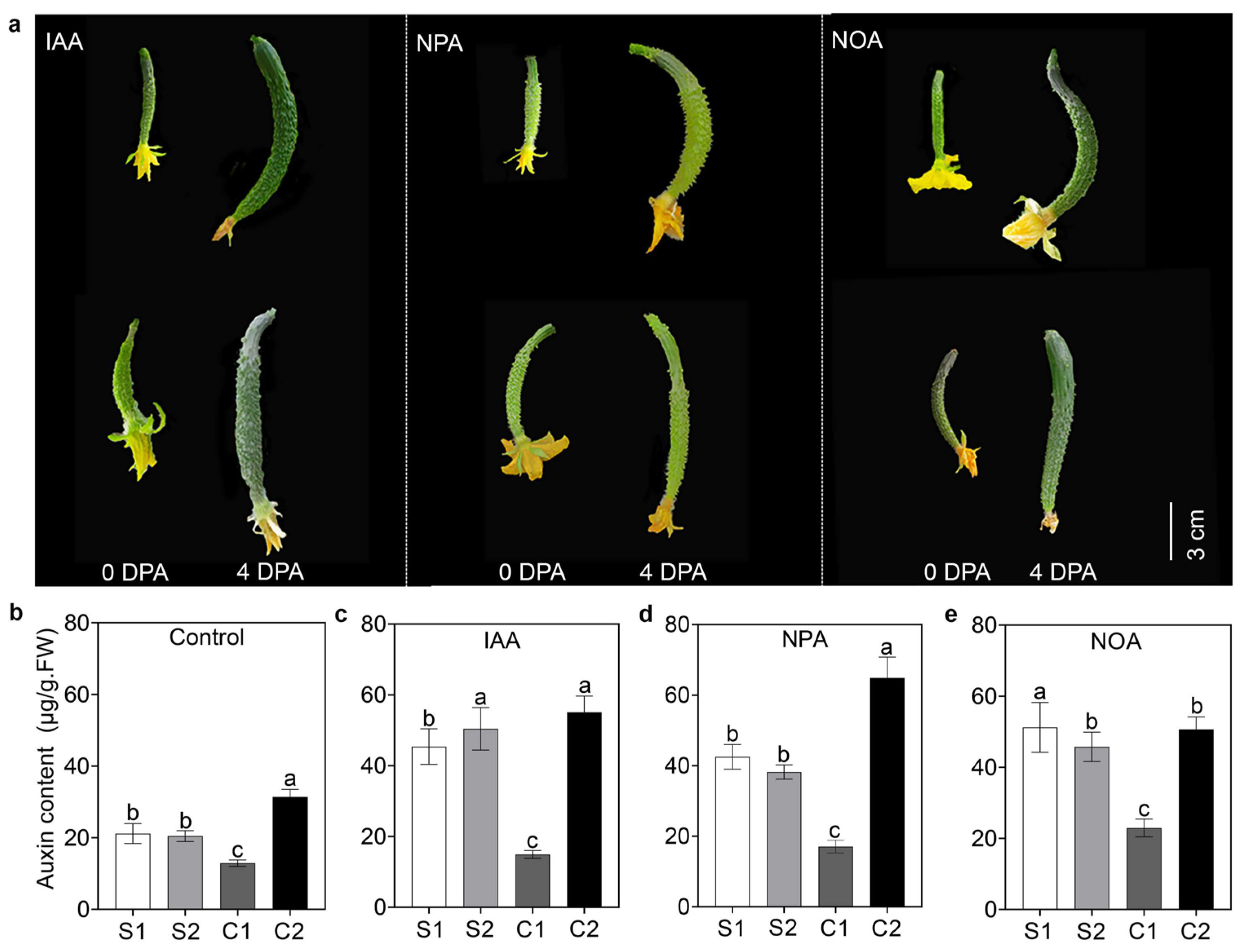
3.1. Auxin Transport Contributed to Asymmetric Auxin Distribution and Resulted in Curved Fruit
3.2. Genome-Wide Identification of Auxin Transporters
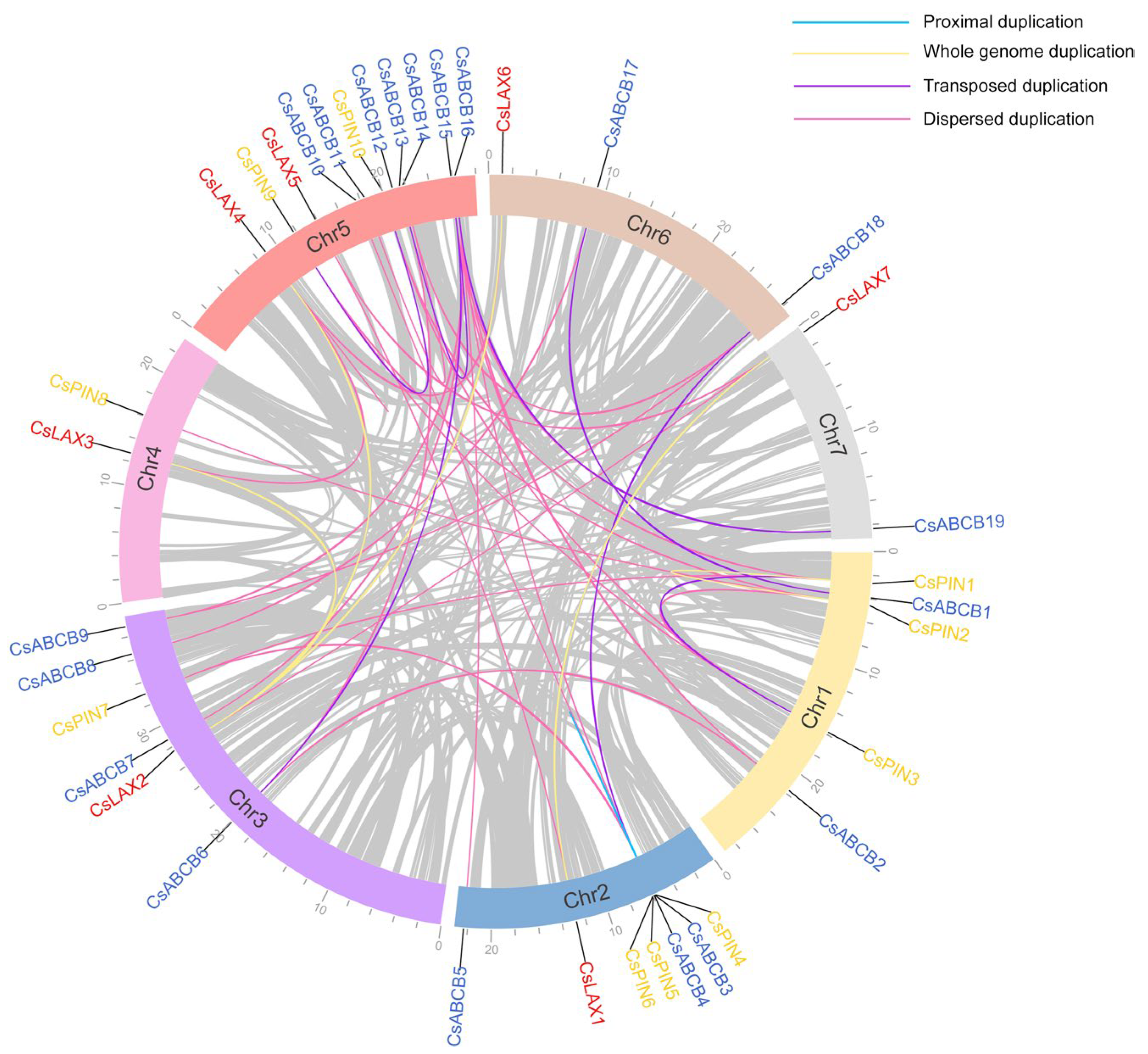
3.3. Distribution and Duplication Analysis of Auxin Transporters
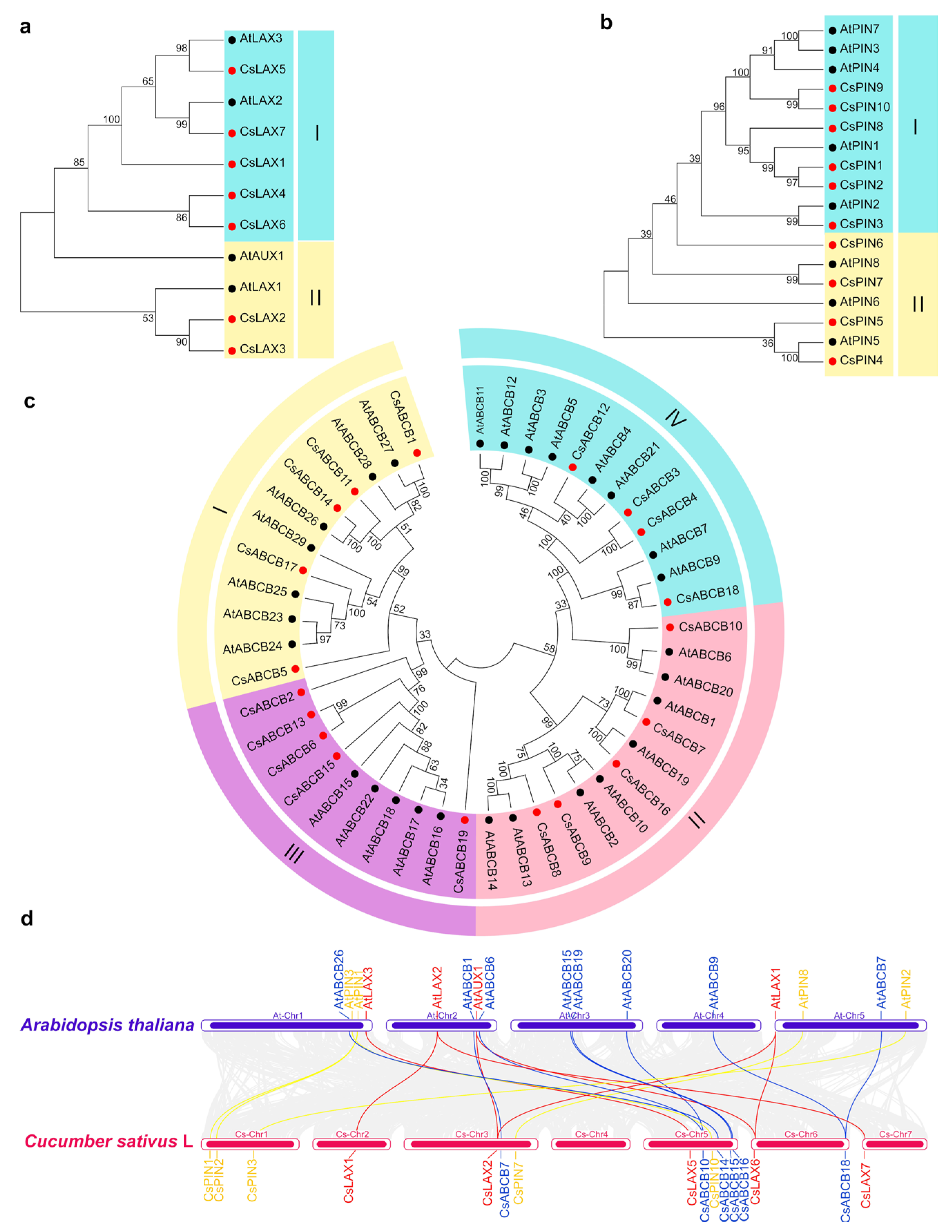
3.4. Analyses of Phylogenetic Relationships, Synteny Relationships, Conservative Motif, and Gene Structure
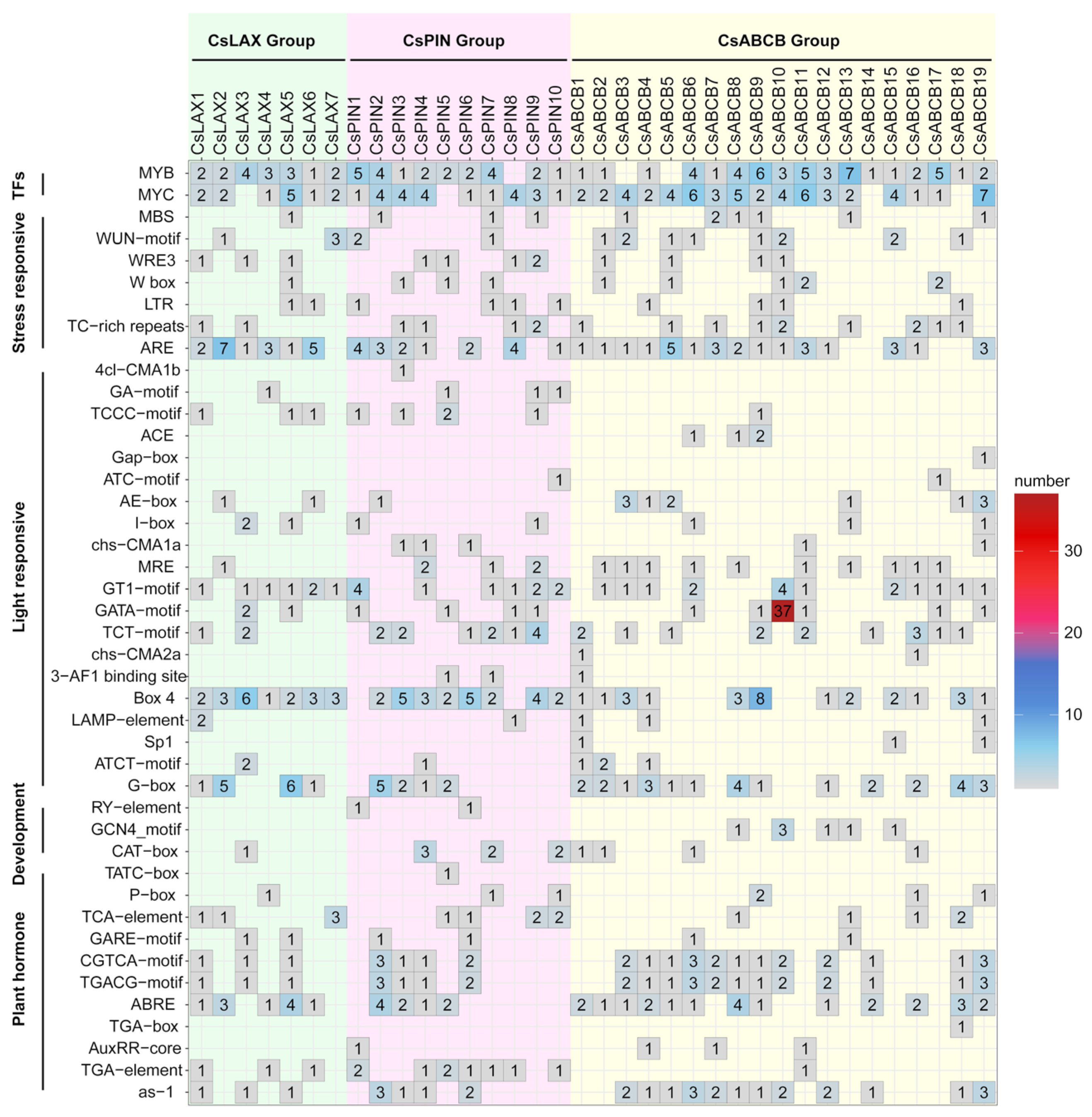
3.5. Analysis of Cis-Regulatory Elements in the Promoter Regions of Auxin Transporters
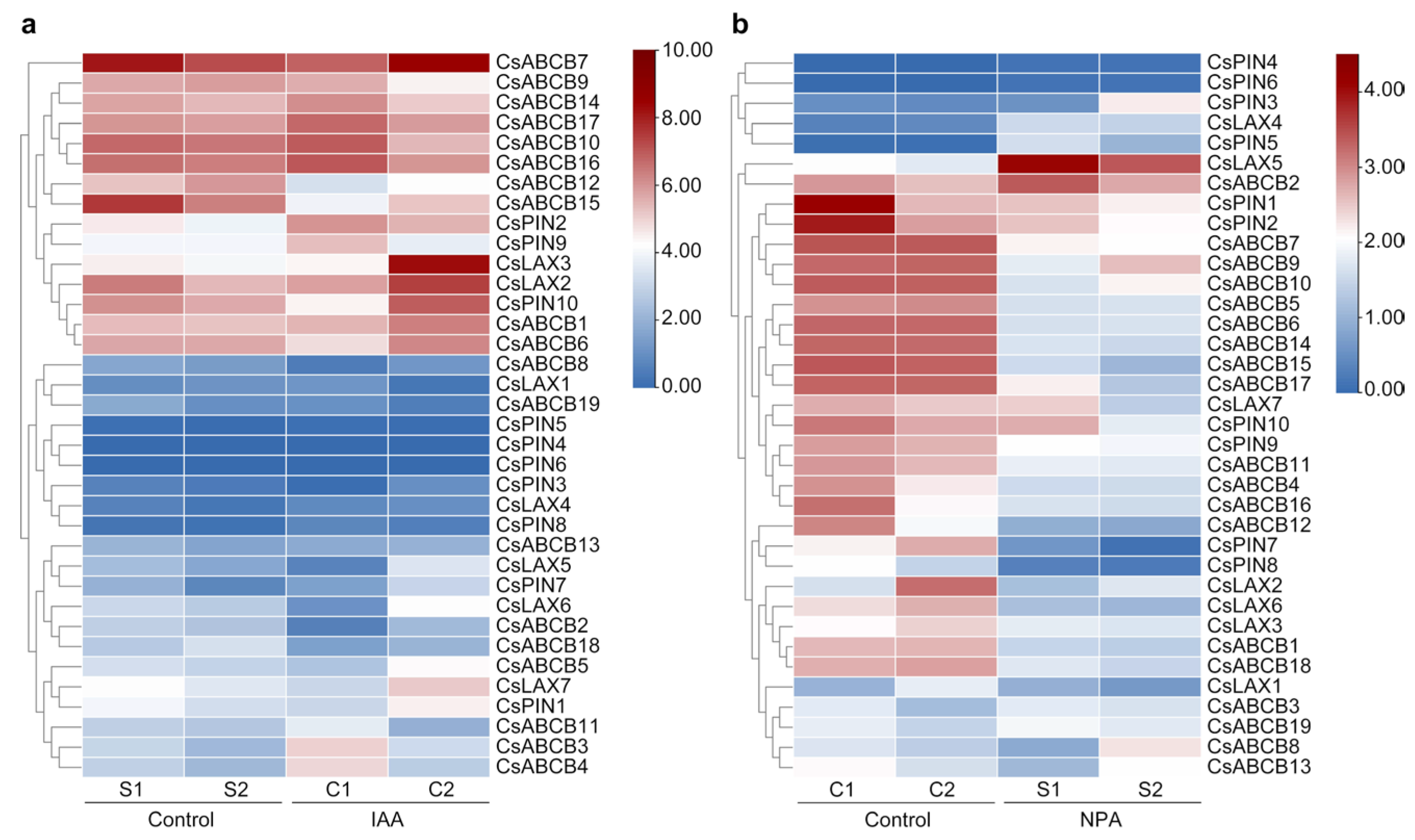
3.6. Differential Expression of Auxin Transporters Control Fruit Curving
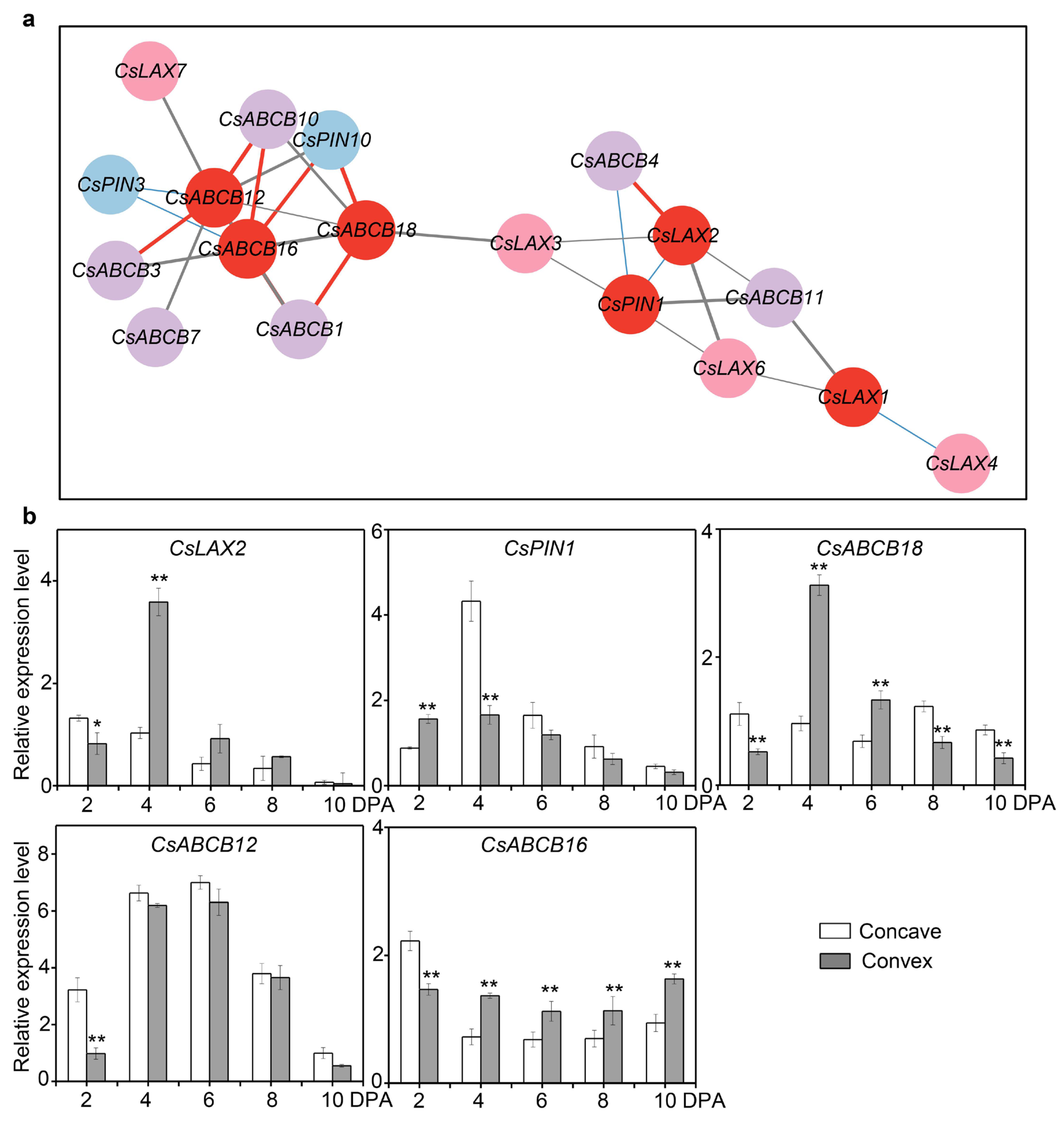
3.7. Potential Roles of Auxin Transporters in Fruit Curving
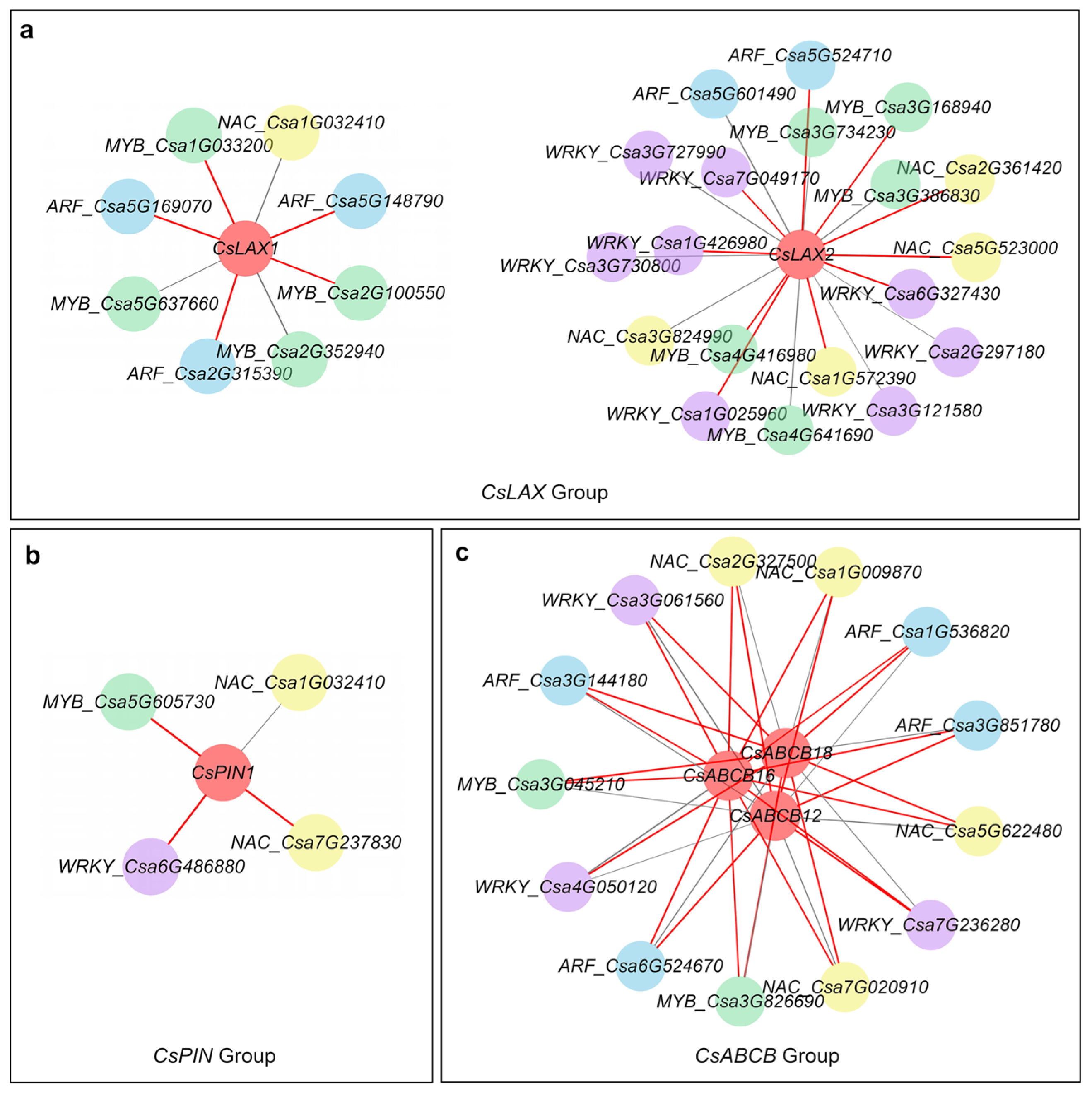
3.8. Co-Expression Analysis of TFs and Transporters
4. Discussion
4.1. Comprehensive Analysis of Auxin Transporters
4.2. Potential Roles of Auxin Transporters
4.3. A Model of Fruit Curving in Cucumber
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Hohm, T.; Demarsy, E.; Quan, C.; Allenbach Petrolati, L.; Preuten, T.; Vernoux, T.; Bergmann, S.; Fankhauser, C. Plasma membrane H+-ATP ase regulation is required for auxin gradient formation preceding phototropic growth. Mol. Syst. Biol. 2014, 10, 751. [Google Scholar] [CrossRef] [PubMed]
- Xiao, G.; Zhang, Y. Adaptive growth: Shaping auxin-mediated root system architecture. Trends Plant Sci. 2020, 25, 121–123. [Google Scholar] [CrossRef] [PubMed]
- Peng, Y.; Zhang, D.; Qiu, Y.; Xiao, Z.; Ji, Y.; Li, W.; Xia, Y.; Wang, Y.; Guo, H. Growth asymmetry precedes differential auxin response during apical hook initiation in Arabidopsis. J. Integr. Plant Biol. 2022, 64, 5–22. [Google Scholar] [CrossRef] [PubMed]
- Dindas, J.; Scherzer, S.; Roelfsema, M.R.G.; von Meyer, K.; Müller, H.M.; Al-Rasheid, K.A.S.; Palme, K.; Dietrich, P.; Becker, D.; Bennett, M.J.; et al. AUX1-mediated root hair auxin influx governs SCFTIR1/AFB-type Ca2+ signaling. Nat. Commun. 2018, 9, 1174. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.; Wang, D.; Zhang, C.; Ye, M.; Kong, N.; Ma, H.; Chen, Q. Comprehensive Analysis and Expression Profiling of PIN, AUX/LAX, and ABCB Auxin Transporter Gene Families in Solanum tuberosum under Phytohormone Stimuli and Abiotic Stresses. Biology 2021, 10, 127. [Google Scholar] [CrossRef] [PubMed]
- An, F.; Zhang, X.; Zhu, Z.; Ji, Y.; He, W.; Jiang, Z.; Li, M.; Guo, H. Coordinated regulation of apical hook development by gibberellins and ethylene in etiolated Arabidopsis seedlings. Cell Res. 2012, 22, 915–927. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.; Maokai, Y.; Cheng, H.; Guo, M.; Liu, Y.; Wang, L.; Chao, S.; Zhang, M.; Lai, L.; Qin, Y. Characterization of auxin transporter AUX, PIN and PILS gene families in pineapple and evaluation of expression profiles during reproductive development and under abiotic stresses. PeerJ 2021, 9, e11410. [Google Scholar] [CrossRef]
- Vandenbussche, F.; Petrášek, J.; Žádníková, P.; Hoyerová, K.; Pešek, B.; Raz, V.; Swarup, R.; Bennett, M.; Zažímalová, E.; Benková, E. The auxin influx carriers AUX1 and LAX3 are involved in auxin-ethylene interactions during apical hook development in Arabidopsis thaliana seedlings. Development 2010, 137, 597–606. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Tidy, A.; Abu Bakar, N.; Carrier, D.; Kerr, I.D.; Hodgman, C.; Bennett, M.J.; Swarup, R. Mechanistic insight into the role of AUXIN RESISTANCE4 in trafficking of AUXIN1 and LIKE AUX1-2. Plant Physiol. 2024, 194, 422–433. [Google Scholar] [CrossRef]
- Liu, H.; Luo, Q.; Tan, C.; Song, J.; Zhang, T.; Men, S. Biosynthesis- and transport-mediated dynamic auxin distribution during seed development controls seed size in Arabidopsis. Plant J. 2023, 113, 1259–1277. [Google Scholar] [CrossRef]
- Zhang, Y.; Berman, A.; Shani, E. Plant Hormone Transport and Localization: Signaling Molecules on the Move. Annu. Rev. Plant Biol. 2023, 74, 453–479. [Google Scholar] [CrossRef] [PubMed]
- Bennett, T.; Brockington, S.F.; Rothfels, C.; Graham, S.W.; Stevenson, D.; Kutchan, T.; Rolf, M.; Thomas, P.; Wong, G.K.-S.; Leyser, O.; et al. Paralogous Radiations of PIN Proteins with Multiple Origins of Noncanonical PIN Structure. Mol. Biol. Evol. 2014, 31, 2042–2060. [Google Scholar] [CrossRef] [PubMed]
- Hammes, U.Z.; Murphy, A.S.; Schwechheimer, C. Auxin transporters—A biochemical view. Cold Spring Harbor Perspect. Biol. 2022, 14, a039875. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Rodriguez, L.; Li, L.; Zhang, X.; Friml, J. Functional innovations of PIN auxin transporters mark crucial evolutionary transitions during rise of flowering plants. Sci. Adv. 2020, 6, eabc8895. [Google Scholar] [CrossRef] [PubMed]
- Žádníková, P.; Petrášek, J.; Marhavý, P.; Raz, V.; Vandenbussche, F.; Ding, Z.; Schwarzerová, K.; Morita, M.T.; Tasaka, M.; Hejátko, J. Role of PIN-mediated auxin efflux in apical hook development of Arabidopsis thaliana. Development 2010, 137, 607–617. [Google Scholar] [CrossRef]
- Wang, H.; Ouyang, Q.; Yang, C.; Zhang, Z.; Hou, D.; Liu, H.; Xu, H. Mutation of OsPIN1b by CRISPR/Cas9 Reveals a Role for Auxin Transport in Modulating Rice Architecture and Root Gravitropism. Int. J. Mol. Sci. 2022, 23, 8965. [Google Scholar] [CrossRef] [PubMed]
- Ung, K.L.; Winkler, M.; Schulz, L.; Kolb, M.; Janacek, D.P.; Dedic, E.; Stokes, D.L.; Hammes, U.Z.; Pedersen, B.P. Structures and mechanism of the plant PIN-FORMED auxin transporter. Nature 2022, 609, 605–610. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Xia, J.; Hong, J.; Zhang, C.; Wei, H.; Ying, W.; Sun, C.; Sun, L.; Mao, Y.; Gao, Y.; et al. Structural insights into auxin recognition and efflux by Arabidopsis PIN1. Nature 2022, 609, 611–615. [Google Scholar] [CrossRef] [PubMed]
- Geisler, M.; Aryal, B.; Di Donato, M.; Hao, P. A critical view on ABC transporters and their interacting partners in auxin transport. Plant Cell Physiol. 2017, 58, 1601–1614. [Google Scholar] [CrossRef]
- Que, F.; Zhu, Y.; Liu, Q.; Wei, Q.; Ramakrishnan, M. Genome-Wide Identification, Expansion, Evolution, and Expression Analysis Reveals ABCB Genes Important for Secondary Cell Wall Development in Moso Bamboo (Phyllostachys edulis). Agronomy 2023, 13, 1828. [Google Scholar] [CrossRef]
- Yang, Y.; Huang, Q.; Wang, X.; Mei, J.; Sharma, A.; Tripathi, D.K.; Yuan, H.; Zheng, B. Genome-wide identification and expression profiles of ABCB gene family in Chinese hickory (Carya cathayensis Sarg.) during grafting. Plant Physiol. Biochem. 2021, 168, 477–487. [Google Scholar] [CrossRef] [PubMed]
- Kubeš, M.; Yang, H.; Richter, G.L.; Cheng, Y.; Młodzińska, E.; Wang, X.; Blakeslee, J.J.; Carraro, N.; Petrášek, J.; Zažímalová, E. The Arabidopsis concentration-dependent influx/efflux transporter ABCB4 regulates cellular auxin levels in the root epidermis. Plant J. 2012, 69, 640–654. [Google Scholar] [CrossRef]
- Cho, M.; Lee, S.H.; Cho, H.-T. P-Glycoprotein4 displays auxin efflux transporter–like action in Arabidopsis root hair cells and tobacco cells. Plant Cell 2007, 19, 3930–3943. [Google Scholar] [CrossRef]
- Wu, G.; Cameron, J.N.; Ljung, K.; Spalding, E.P. A role for ABCB19-mediated polar auxin transport in seedling photomorphogenesis mediated by cryptochrome 1 and phytochrome B. Plant J. Cell Mol. Biol. 2010, 62, 179–191. [Google Scholar] [CrossRef] [PubMed]
- Mazzella, M.A.; Casal, J.J.; Muschietti, J.P.; Fox, A.R. Hormonal networks involved in apical hook development in darkness and their response to light. Front. Plant Sci. 2014, 5, 52. [Google Scholar] [CrossRef]
- Jenness, M.K.; Tayengwa, R.; Bate, G.A.; Tapken, W.; Zhang, Y.; Pang, C.; Murphy, A.S. Loss of multiple ABCB auxin transporters recapitulates the major twisted dwarf 1 phenotypes in Arabidopsis thaliana. Front. Plant Sci. 2022, 13, 840260. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Nasser, V.; Pisanty, O.; Omary, M.; Wulff, N.; Di Donato, M.; Tal, I.; Hauser, F.; Hao, P.; Roth, O. A transportome-scale amiRNA-based screen identifies redundant roles of Arabidopsis ABCB6 and ABCB20 in auxin transport. Nat. Commun. 2018, 9, 4204. [Google Scholar] [CrossRef]
- Chen, J.; Hu, Y.; Hao, P.; Tsering, T.; Xia, J.; Zhang, Y.; Roth, O.; Njo, M.F.; Sterck, L.; Hu, Y. ABCB-mediated shootward auxin transport feeds into the root clock. EMBO Rep. 2023, 24, e56271. [Google Scholar] [CrossRef]
- Li, S.; Wang, C.; Zhou, X.; Liu, D.; Liu, C.; Luan, J.; Qin, Z.; Xin, M. The curvature of cucumber fruits is associated with spatial variation in auxin accumulation and expression of a YUCCA biosynthesis gene. Hortic. Res. 2020, 7, 35. [Google Scholar] [CrossRef]
- Jahn, L.; Hofmann, U.; Ludwig-Müller, J. Indole-3-Acetic Acid Is Synthesized by the Endophyte Cyanodermella asteris via a Tryptophan-Dependent and -Independent Way and Mediates the Interaction with a Non-Host Plant. Int. J. Mol. Sci. 2021, 22, 2651. [Google Scholar] [CrossRef]
- Weiler, E.; Jourdan, P.; Conrad, W. Levels of indole-3-acetic acid in intact and decapitated coleoptiles as determined by a specific and highly sensitive solid-phase enzyme immunoassay. Planta 1981, 153, 561–571. [Google Scholar] [CrossRef] [PubMed]
- Horton, P.; Park, K.-J.; Obayashi, T.; Fujita, N.; Harada, H.; Adams-Collier, C.; Nakai, K. WoLF PSORT: Protein localization predictor. Nucleic Acids Res. 2007, 35, W585–W587. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef] [PubMed]
- He, Z.; Zhang, H.; Gao, S.; Lercher, M.J.; Chen, W.-H.; Hu, S. Evolview v2: An online visualization and management tool for customized and annotated phylogenetic trees. Nucleic Acids Res. 2016, 44, W236–W241. [Google Scholar] [CrossRef] [PubMed]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.; Li, W.W.; Noble, W.S. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 2009, 37, W202–W208. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Wu, Y.; Li, J.; Wang, X.; Zeng, Z.; Xu, J.; Liu, Y.; Feng, J.; Chen, H.; He, Y. TBtools-II: A “one for all, all for one” bioinformatics platform for biological big-data mining. Mol. Plant 2023, 16, 1733–1742. [Google Scholar] [CrossRef] [PubMed]
- Hallgren, J.; Tsirigos, K.D.; Pedersen, M.D.; Almagro Armenteros, J.J.; Marcatili, P.; Nielsen, H.; Krogh, A.; Winther, O. DeepTMHMM predicts alpha and beta transmembrane proteins using deep neural networks. BioRxiv 2022. [Google Scholar] [CrossRef]
- Wang, Y.; Tang, H.; Debarry, J.D.; Tan, X.; Li, J.; Wang, X.; Lee, T.-h.; Jin, H.; Marler, B.; Guo, H. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012, 40, e49. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Xin, M.; Zhou, X.; Liu, C.; Li, S.; Liu, D.; Xu, Y.; Qin, Z. The novel ethylene-responsive factor CsERF025 affects the development of fruit bending in cucumber. Plant Mol. Biol. 2017, 95, 519–531. [Google Scholar] [CrossRef]
- Smoot, M.E.; Ono, K.; Ruscheinski, J.; Wang, P.-L.; Ideker, T. Cytoscape 2.8: New features for data integration and network visualization. Bioinformatics 2011, 27, 431–432. [Google Scholar] [CrossRef]
- Li, Y.; Yang, S.; Shi, M.; Zhang, S.; Wu, S.; Chen, Y.; Li, W.; Tian, W.-M. HbARF2 and HbARF16. 3 function as negative regulators for the radial trunk growth of rubber tree. Ind. Crops Prod. 2020, 158, 112978. [Google Scholar] [CrossRef]
- Gou, H.; Nai, G.; Lu, S.; Ma, W.; Chen, B.; Mao, J. Genome-wide identification and expression analysis of PIN gene family under phytohormone and abiotic stresses in Vitis vinifera L. Physiol. Mol. Biol. Plants 2022, 28, 1905–1919. [Google Scholar] [CrossRef]
- Villaécija-Aguilar, J.A.; Körösy, C.; Maisch, L.; Hamon-Josse, M.; Petrich, A.; Magosch, S.; Chapman, P.; Bennett, T.; Gutjahr, C. KAI2 promotes Arabidopsis root hair elongation at low external phosphate by controlling local accumulation of AUX1 and PIN2. Curr. Biol. 2022, 32, 228–236. e223. [Google Scholar] [CrossRef] [PubMed]
- Jia, Z.; Giehl, R.F.H.; Hartmann, A.; Estevez, J.M.; Bennett, M.J.; von Wirén, N. A spatially concerted epidermal auxin signaling framework steers the root hair foraging response under low nitrogen. Curr. Biol. 2023, 33, 3926–3941.e3925. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Jiang, L.; Che, G.; Pan, Y.; Li, Y.; Hou, Y.; Zhao, W.; Zhong, Y.; Ding, L.; Yan, S. A functional allele of CsFUL1 regulates fruit length through repressing CsSUP and inhibiting auxin transport in cucumber. Plant Cell 2019, 31, 1289–1307. [Google Scholar] [CrossRef]
- Dong, X.; Ma, C.; Xu, T.; Reid, M.S.; Jiang, C.-Z.; Li, T. Auxin response and transport during induction of pedicel abscission in tomato. Hortic. Res. 2021, 8, 192. [Google Scholar] [CrossRef]
- Teale, W.D.; Pasternak, T.; Dal Bosco, C.; Dovzhenko, A.; Kratzat, K.; Bildl, W.; Schwörer, M.; Falk, T.; Ruperti, B.; V Schaefer, J.; et al. Flavonol-mediated stabilization of PIN efflux complexes regulates polar auxin transport. EMBO J. 2021, 40, e104416. [Google Scholar] [CrossRef]
- Mellor, N.L.; Voß, U.; Ware, A.; Janes, G.; Barrack, D.; Bishopp, A.; Bennett, M.J.; Geisler, M.; Wells, D.M.; Band, L.R. Systems approaches reveal that ABCB and PIN proteins mediate co-dependent auxin efflux. Plant Cell 2022, 34, 2309–2327. [Google Scholar] [CrossRef]
- Hajný, J.; Tan, S.; Friml, J. Auxin canalization: From speculative models toward molecular players. Curr. Opin. Plant Biol. 2022, 65, 102174. [Google Scholar] [CrossRef]
- Hu, J.; Su, H.; Cao, H.; Wei, H.; Fu, X.; Jiang, X.; Song, Q.; He, X.; Xu, C.; Luo, K. AUXIN RESPONSE FACTOR7 integrates gibberellin and auxin signaling via interactions between DELLA and AUX/IAA proteins to regulate cambial activity in poplar. Plant Cell 2022, 34, 2688–2707. [Google Scholar] [CrossRef]
- Wang, H.-Z.; Yang, K.-Z.; Zou, J.-J.; Zhu, L.-L.; Xie, Z.D.; Morita, M.T.; Tasaka, M.; Friml, J.; Grotewold, E.; Beeckman, T. Transcriptional regulation of PIN genes by FOUR LIPS and MYB88 during Arabidopsis root gravitropism. Nat. Commun. 2015, 6, 8822. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y.; Zhang, Y.; Sun, Y.; Xie, Z.; Luo, Y.; Long, Q.; Feng, J.; Liu, X.; Wang, B.; He, D. Natural variations of OsAUX5, a target gene of OsWRKY78, control the neutral essential amino acid content in rice grains. Mol. Plant 2023, 16, 322–336. [Google Scholar] [CrossRef] [PubMed]
- Gu, L.; Hou, Y.; Sun, Y.; Chen, X.; Wang, G.; Wang, H.; Zhu, B.; Du, X. The maize WRKY transcription factor ZmWRKY64 confers cadmium tolerance in Arabidopsis and maize (Zea mays L.). Plant Cell Rep. 2024, 43, 44. [Google Scholar] [CrossRef]
- Béziat, C.; Barbez, E.; Feraru, M.I.; Lucyshyn, D.; Kleine-Vehn, J. Light triggers PILS-dependent reduction in nuclear auxin signalling for growth transition. Nat. Plants 2017, 3, 17105. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lu, K.; Zhang, L.; Fan, L.; Zhou, X.; Li, S. A Genome-Wide Comparative Analysis of AUX1/LAX, PIN, and ABCB Genes Reveals Their Roles in Cucumber Fruit Curving. Agriculture 2024, 14, 657. https://doi.org/10.3390/agriculture14050657
Lu K, Zhang L, Fan L, Zhou X, Li S. A Genome-Wide Comparative Analysis of AUX1/LAX, PIN, and ABCB Genes Reveals Their Roles in Cucumber Fruit Curving. Agriculture. 2024; 14(5):657. https://doi.org/10.3390/agriculture14050657
Chicago/Turabian StyleLu, Ke, La Zhang, Lianxue Fan, Xiuyan Zhou, and Shengnan Li. 2024. "A Genome-Wide Comparative Analysis of AUX1/LAX, PIN, and ABCB Genes Reveals Their Roles in Cucumber Fruit Curving" Agriculture 14, no. 5: 657. https://doi.org/10.3390/agriculture14050657
APA StyleLu, K., Zhang, L., Fan, L., Zhou, X., & Li, S. (2024). A Genome-Wide Comparative Analysis of AUX1/LAX, PIN, and ABCB Genes Reveals Their Roles in Cucumber Fruit Curving. Agriculture, 14(5), 657. https://doi.org/10.3390/agriculture14050657





