Assessment of Molecular Diversity and Population Structure of Pakistani Mulberry Accessions Using Retrotransposon-Based DNA Markers
Abstract
1. Introduction
2. Materials and Methods
2.1. Leaf Samples and DNA Extraction
2.2. PCR Amplification for iPBS Markers
2.3. Statistical Analysis
3. Results
3.1. Optimization and Reproducibility of iPBS for 30 Mulberry Accessions
3.2. Genetic Diversity and Heterozygosity Interpretation
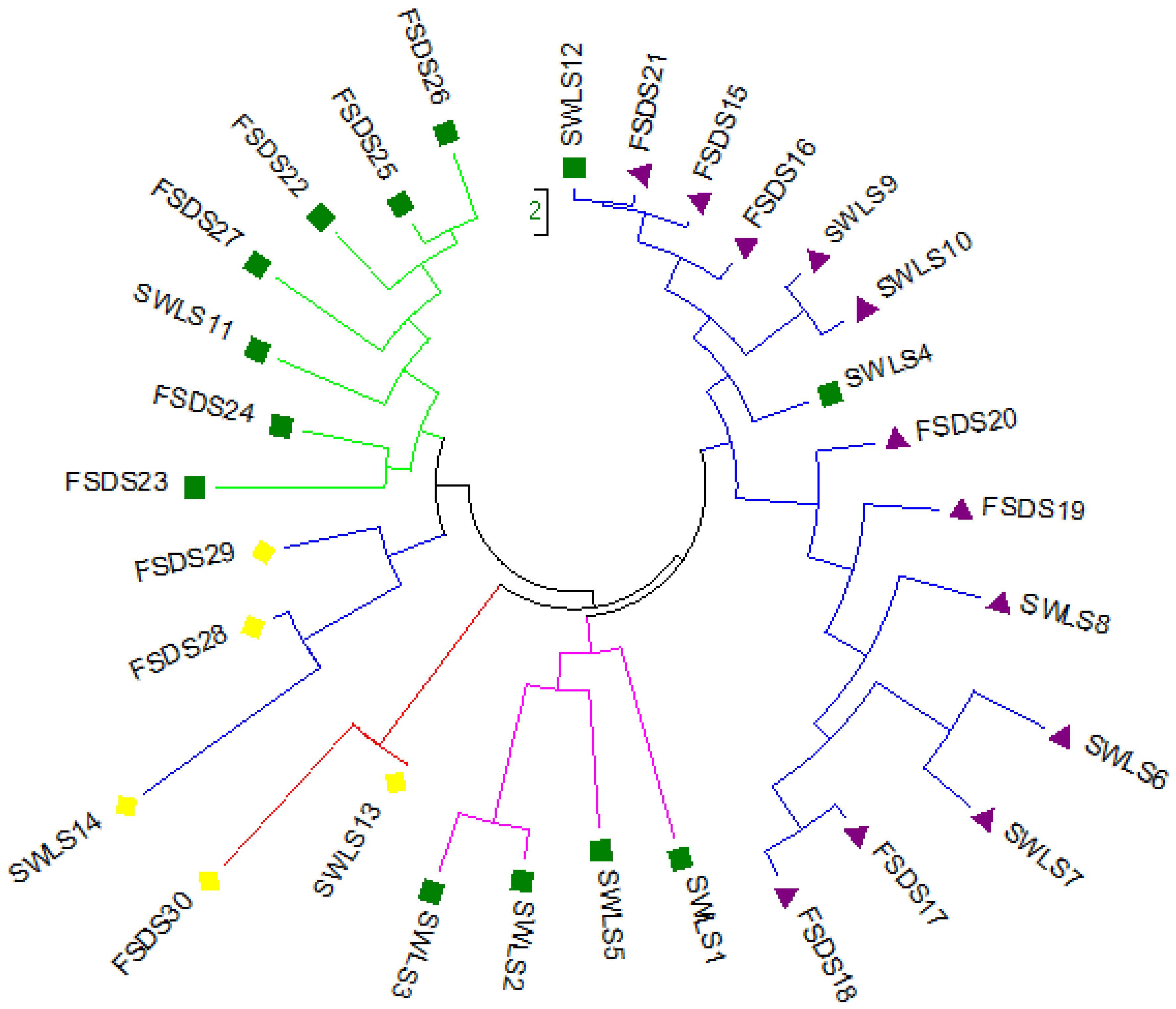
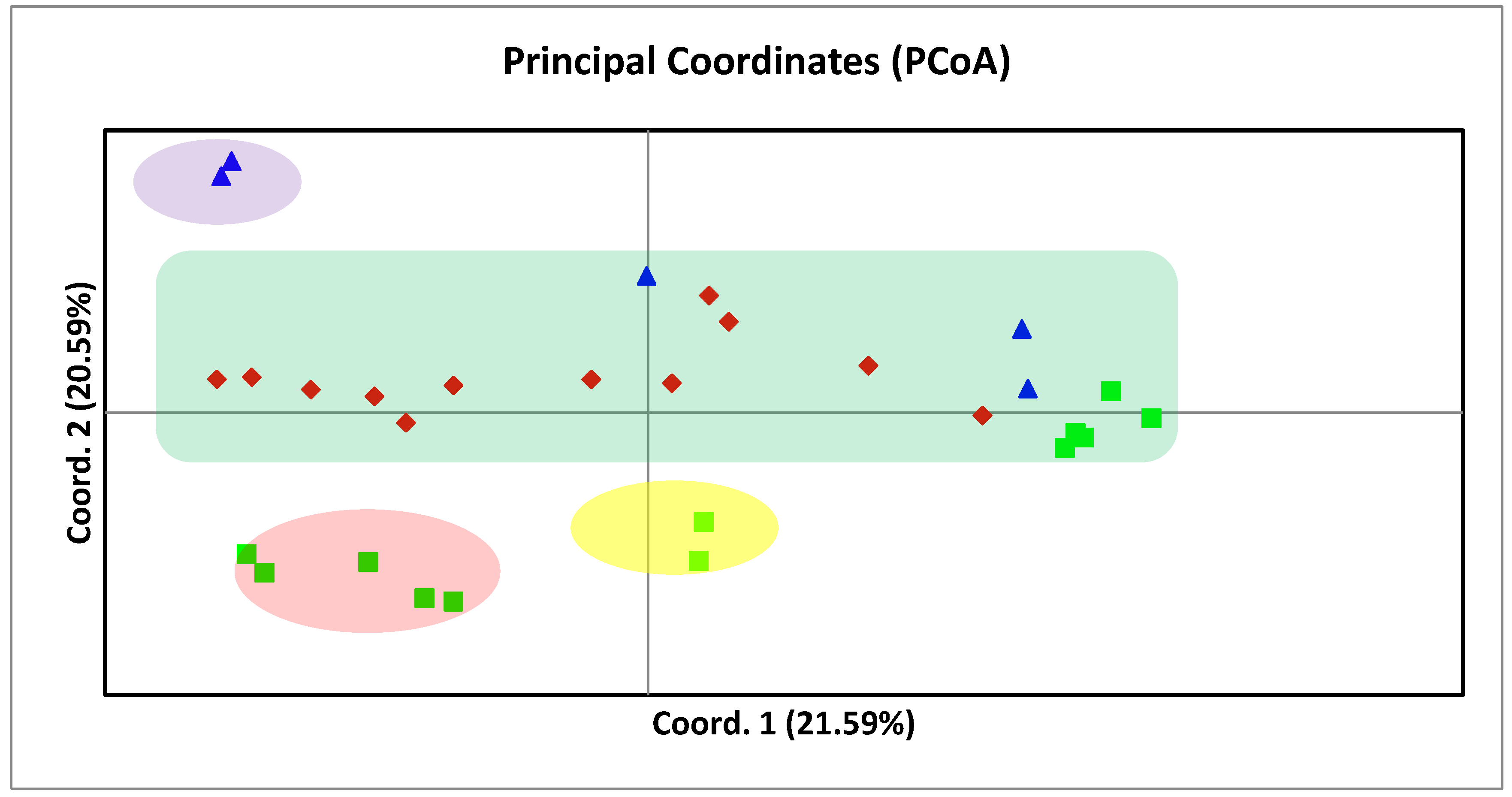
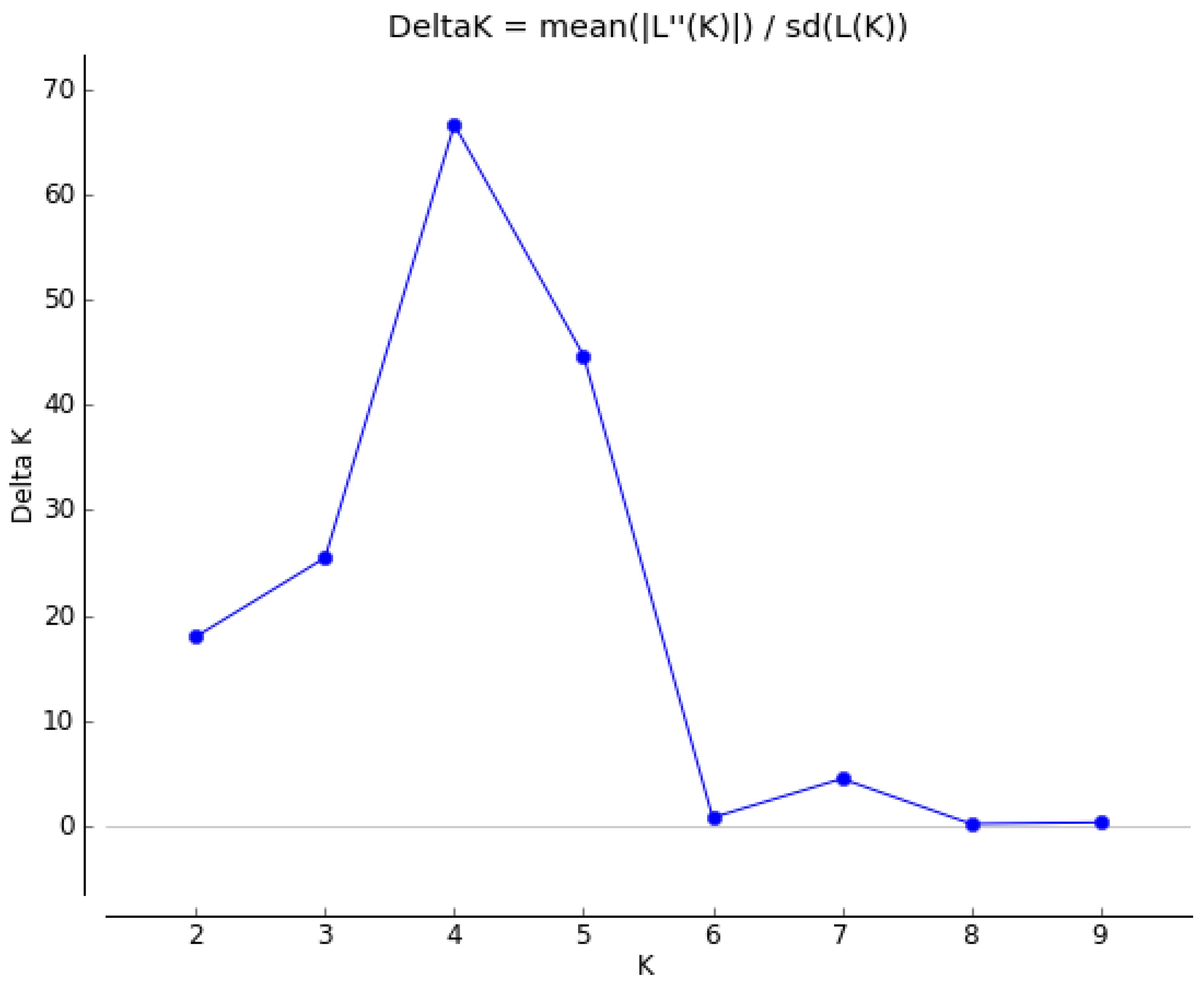
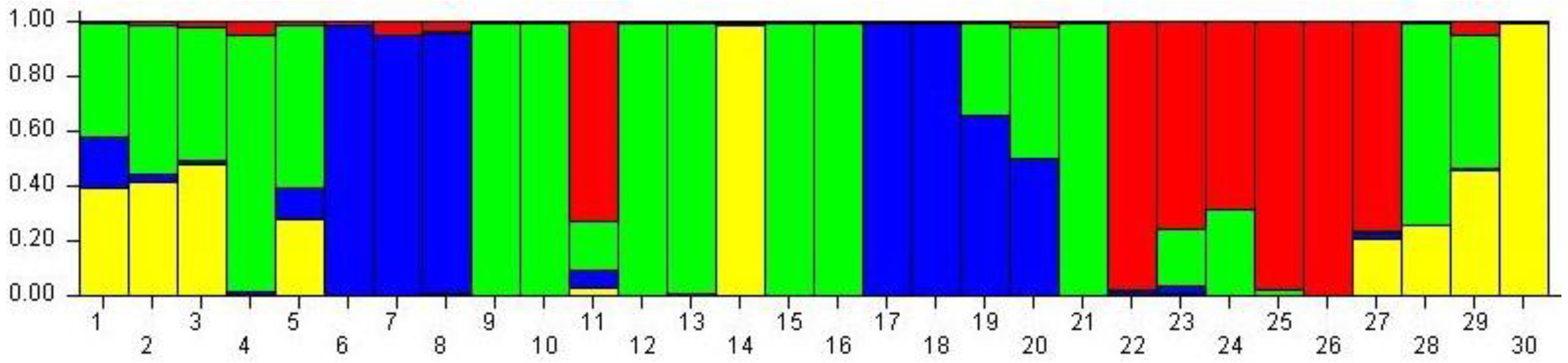
3.3. Principal Coordinate Analysis (PCoA) and Population Structure of 30 Mulberry Accessions
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Kafkas, S.; Özen, M.; Doğan, Y.B.; Özcan, B.; Ercişli, S.; Serçe, S. Molecular characterization of mulberry accessions in Turkey by AFLP markers. Sci. Hortic. 2008, 133, 593–597. [Google Scholar] [CrossRef]
- Gnanesh, B.N.; Tejaswi, A.; Arunakumar, G.S.; Supriya, M.; Manojkumar, H.B.; Tewary, P. Molecular phylogeny, identification and pathogenicity of Rhizopus oryzae associated with root rot of mulberry in India. J. Appl. Microbiol. 2021, 131, 360–374. [Google Scholar] [CrossRef]
- Arunakumar, G.S.; Gnanesh, B.N. Evaluation of artificial inoculation methods to determine resistance reaction to dry root rot and black root rot disease in mulberry (Morus spp.). Arch. Phytopathol. Plant Prot. 2023, 56, 49–65. [Google Scholar] [CrossRef]
- Gnanesh, B.N.; Arunakumar, G.S.; Tejaswi, A.; Supriya, M.; Manojkumar, H.B.; Devi, S.S. Characterization and pathogenicity of Lasiodiplodia theobromae causing black root rot and identification of novel sources of resistance in mulberry collections. Plant Pathol. J. 2022, 38, 272–286. [Google Scholar] [CrossRef]
- Thabti, I.; Elfalleh, W.; Tlili, N.; Ziadi, M.; Campos, M.G.; Ferchichi, A. Phenols, flavonoids, and antioxidant and antibacterial activity of leaves and stem bark of Morus Species. Int. J. Food Prop. 2014, 17, 842–854. [Google Scholar] [CrossRef]
- Gerasopouls, D.; Stravroulakis, G. Quality characteristics of four mulberry (Morus sp.) cultivars in the area of Chania, Greece. J. Sci. Food Agric. 1997, 73, 261–264. [Google Scholar] [CrossRef]
- Memon, A.A.; Memon, N.; Luthria, D.L.; Bhanger, M.I.; Pitafi, A.A. Phenolic acids profiling and antioxidant potential of mulberry (Morus laevigata W., Morus nigra L., Morus alba L.) leaves and fruits grown in Pakistan. Polish J. Food Nutr. Sci. 2010, 60, 25–32. [Google Scholar]
- Van de Peer, Y.; Ashman, T.L.; Soltis, P.S.; Soltis, D.E. Polyploidy: An evolutionary and ecological force in stressful times. Plant Cell. 2021, 33, 11–26. [Google Scholar] [CrossRef] [PubMed]
- Letcher, L.K. The Complete Guide to Edible Wild Plants, Mushrooms, Fruits, and Nuts: How to Find, Identify, and Cook Them, 2nd ed.; FalconGuides: Guilford, CN, USA, 2010; p. 103. ISBN 978-1-59921-887-8. [Google Scholar]
- Hosseini, A.S.; Akramian, M.; Khadivi, A.; Salehi-Arjmand, H. Phenotypic and chemical variation of black mulberry (Morus nigra) genotypes. Ind. Crops Prod. 2018, 117, 260–270. [Google Scholar] [CrossRef]
- Tojo, I. Research of polyploidy and its application in Morus. Jpn. Agric. Res. Q. 1985, 18, 222–228. [Google Scholar]
- Chang, L.Y.; Li, K.T.; Yang, W.J.; Chang, J.C.; Chang, M.W. Phenotypic classification of mulberry (Morus) species in Taiwan using numerical taxonomic analysis through the characterization of vegetative traits and chilling requirements. Sci. Hortic. 2014, 176, 208–217. [Google Scholar] [CrossRef]
- Peris, N.W.; Gacheri, K.M.; Theophillus, M.M.; Lucas, N. Morphological characterization of mulberry (Morus spp.) accessions grown in Kenya. Sustain. Agric. Res. 2013, 3, 10–17. [Google Scholar] [CrossRef]
- Ellstrand, N.C. Gene flow by pollen-implications for plant conservation genetics. Oikos 1992, 63, 77–86. [Google Scholar] [CrossRef]
- Kumar, L.S. DNA markers in plant improvement: An overview. Biotechnol. Adv. 1999, 17, 143–182. [Google Scholar] [CrossRef]
- Aydın, F.; Özer, G.; Alkan, M.; Çakır, İ. The utility of iPBS retrotransposons markers to analyze genetic variation in yeast. Int. J. Food Microbiol. 2020, 325, 108647. [Google Scholar] [CrossRef] [PubMed]
- Mehmood, A.; Jaskani, M.J.; Khan, I.A.; Ahmad, S.; Ahmad, R.; Luo, S.; Ahmad, N.M. Genetic diversity of Pakistani guava (Psidium guajava L.) germplasm and its implications for conservation and breeding. Sci. Hortic. 2014, 172, 221–232. [Google Scholar] [CrossRef]
- Naeem, H.; Awan, F.S.; Dracatos, P.M.; Sajid, M.W.; Saleem, S.; Yousafi, Q.; Zulfiqar, B. Population structure and phylogenetic relationship of Peach [Prunus persica (L.) Batsch] and Nectarine [Prunus persica var. nucipersica (L.) CK Schneid.] based on retrotransposon markers. Genet. Resour. Crop Evol. 2021, 68, 3011–3023. [Google Scholar] [CrossRef]
- Mehmood, A.; Luo, S.; Ahmad, N.M.; Dong, C.; Mahmood, T.; Sajjad, Y.; Sharp, P. Molecular variability and phylogenetic relationships of guava (Psidium guajava L.) cultivars using inter-primer binding site (iPBS) and microsatellite (SSR) markers. Genet. Resour. Crop Evol. 2016, 63, 1345–1361. [Google Scholar] [CrossRef]
- Demirel, U.; Tındaş, İ.; Yavuz, C.; Baloch, F.S.; Çalışkan, M.E. Assessing genetic diversity of potato genotypes using inter-PBS retrotransposon marker system. Plant Genet. Res. 2018, 16, 137–145. [Google Scholar] [CrossRef]
- Murray, M.; Thompson, W.F. Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res. 1980, 8, 4321–4325. [Google Scholar] [CrossRef]
- Janaki, A.E.K. The origin of Black Mulberry. J. Roy. Hort. Soc. 1948, 73, 117–120. [Google Scholar]
- Jiao, F.; Luo, R.; Dai, X.; Liu, H.; Yu, G.; Han, S.; Lu, X.; Su, C.; Chen, Q.; Song, Q.; et al. Chromosome-level reference genome and population genomic analysis provide insights into the evolution and improvement of domesticated mulberry (Morus alba). Mol. Plant. 2020, 13, 1001–1012. [Google Scholar] [CrossRef]
- Zeng, Q.; Chen, H.; Zhang, C.; Han, M.; Li, T.; Qi, X.; Xiang, Z.; He, N. Definition of eight mulberry species in the genus morus by internal transcribed spacer based phylogeny. PLoS ONE 2015, 10, e0135411. [Google Scholar] [CrossRef]
- Kalendar, R.; Antonius, K.; Smýkal, P.; Schulman, A.H. iPBS: A universal method for DNA fingerprinting and retrotransposon isolation. Theor. Appl. Genet. 2010, 121, 1419–1430. [Google Scholar] [CrossRef]
- Peakall, R.; Smouse, P.E. GenAlEx tutorials-part 2: Genetic distance and analysis of molecular variance (AMOVA). Bioinformatics 2012, 28, 2537–2539. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Pritchard, J.K.; Stephens, M.; Donnelly, P. Inference of population structure using multilocus genotype data. Genetics 2000, 155, 945–959. [Google Scholar] [CrossRef] [PubMed]
- Evanno, G.; Regnaut, S.; Goudet, J. Detecting the number of clusters of individuals using the software STRUCTURE: A simulation study. Mol. Ecol. 2005, 14, 2611–2620. [Google Scholar] [CrossRef] [PubMed]
- Earl, D.A. Structure Harvester: A website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv. Genet. Resour. 2012, 4, 359–361. [Google Scholar] [CrossRef]
- Gao, L.Z. Studies on Genetic Variation of Three Wild Rices (Oryza spp.) in China and Their Conservation Biology. Ph.D. Dissertation, Institute of Botany, Chinese Academy of Sciences, Beijing, China, 1997. [Google Scholar]
- Wu, C.J.; Cheng, Z.Q.; Huang, X.Q.; Yin, S.H.; Cao, K.M.; Sun, C.R. Genetic diversity among and within populations of Oryza granulata from Yunnan of China revealed by RAPD and ISSR markers: Implications for conservation of the endangered species. Plant Sci. 2004, 167, 35–42. [Google Scholar] [CrossRef]
- Barut, M.; Nadeem, M.A.; Karaköy, T.; Baloch, F.S. DNA fingerprinting and genetic diversity analysis of world quinoa germplasm using iPBS-retrotransposon marker system. Turk. J. Agric. For. 2020, 44, 479–491. [Google Scholar] [CrossRef]
- He, N.; Zhang, C.; Qi, X.; Zhao, S.; Tao, Y.; Yang, G.; Lee, T.-H.; Wang, X.; Cai, Q.; Li, D.; et al. Drat genome sequence of the mulberry tree Morus notabilis. Nat. Commun. 2013, 4, 2445. [Google Scholar] [CrossRef] [PubMed]
- Sharma, A.; Sharma, R.; Machii, H. Assessment of genetic diversity in a Morus germplasm collection using fluorescence based AFLP markers. Theor. Appl. Genet. 2000, 101, 1049–1055. [Google Scholar] [CrossRef]
- Botton, A.; Barcaccia, G.; Cappellozza, S.; Da Tos, R.; Bonghi, C.; Ramina, A. DNA fingerprinting sheds light on the origin of introduced mulberry (Morus spp.) accessions in Italy. Genet. Resour. Crop Evol. 2005, 52, 181–192. [Google Scholar] [CrossRef]
- Pinar, H.; Yahya, H.N.; Erċışlı, S.; Coskun, O.F.; Yaman, M.; Turgunbaev, K.; Uzun, A. Molecular Characterization of Barberry Genotypes from Turkey and Kyrgyzstan. Erwerbs-Obstbau 2021, 63, 403–407. [Google Scholar] [CrossRef]
- Uzun, A.; Yaman, M.; Pinar, H.; Gok, B.D.; Gazel, I. Leaf and fruit characteristics and genetic diversity of wild fruit cerasus prostrata genotypes collected from the Central Anatolia. Turkey. Acta Sci. Pol. Hortorum Cultus 2021, 20, 53–62. [Google Scholar] [CrossRef]
- Yaman, M. Determination of genetic diversity in european cranberrybush (Viburnum opulus L.) genotypes based on morphological, phytochemical and ISSR markers. Genet. Resour. Crop Evol. 2022, 69, 1889–1899. [Google Scholar] [CrossRef]
- Yildiz, E.; Pinar, H.; Uzun, A.; Yaman, M.; Sumbul, A.; Ercisli, S. Identification of genetic diversity among Juglans regia L. genotypes using molecular, morphological, and fatty acid data. Genet. Resour. Crop Evol. 2021, 68, 1425–1437. [Google Scholar] [CrossRef]
- Pinto, M.V.; Poornima, H.S.; Rukmangada, M.S.; Triveni, R.; Girish, N.V. Association mapping of quantitative resistance to charcoal root rot in mulberry germplasm. PLoS ONE 2018, 13, e0200099. [Google Scholar] [CrossRef]
- Orhan, E.; Ercisli, S.; Yildirim, N.; Agar, G. Genetic variations among mulberry genotypes (Morus alba) as revealed by random amplified polymorphic DNA (RAPD) markers. Plant Syst. Evol. 2007, 265, 251–258. [Google Scholar] [CrossRef]
- Wangari, N.P.; Gacheri, K.M.; Theophilus, M.M.; Lucas, N. Use of SSR markers for genetic diversity studies in mulberry accessions grown in Kenya. Int. J. Biotechnol. Mol. Biol. Res. 2013, 4, 38–44. [Google Scholar] [CrossRef]
- Garcia-Gomez, B.; Gonzalez-Alvarez, H.; Martinez-Mora, C.; Cenis, J.L.; Perez-Hernandez, M.D.C.; Martinez-Zubiaur, Y.; Martinez-Gomez, P. The molecular characterization of an extended mulberry germplasm by SSR markers. Genetika 2019, 51, 389–403. [Google Scholar] [CrossRef]
- Aggarwal, R.K.; Udayakumar, D.; Hendre, P.S.; Sarkar, A.; Singh, L.I. Isolation and Characterization of six novel microsatellite markers for mulberry (Morus indica). Mol. Ecol. 2004, 4, 477–479. [Google Scholar] [CrossRef]
- Wani, S.A.; Bhat, M.A.; Malik, G.N.; Zaki, F.A.; Mir, M.R.; Wani, N.; Bhat, K.M. Genetic diversity and relationship assessment among mulberry (Morus spp.) genotypes by simple sequence repeat (SSR) marker profile. Afr. J. Biotechnol. 2013, 12, 3181–3187. [Google Scholar]
- Das, A.B.; Mohanty, I.C.; Mahapatra, D.; Mohanty, S.; Ray, A. Genetic variation of Indian potato (Solanum tuberosum L.) genotypes using chromosomal and RAPD markers. Crop Breed. Appl. Biotechnol. 2010, 10, 238–246. [Google Scholar] [CrossRef][Green Version]
- Zhao, W.G.; Zhou, Z.H.; Miao, X.X.; Zhang, Y.; Wang, S.B.; Huang, J.H.; Xiang, H.; Pan, Y.L.; Huang, Y.P. A comparison of genetic variation among wild and cultivated Morus species (Moraceae: Morus) as revealed by ISSR and SSR markers. Biodivers. Conserv. 2007, 6, 275–290. [Google Scholar]
- Awasthi, A.K.; Nagaraja, G.M.; Naik, G.V.; Kanginakudru, S.; Thangavelu, K.; Nagaraju, J. Genetic diversity and relationships in mulberry (genus Morus) as revealed by RAPD and ISSR marker assays. BMC Genet. 2004, 5, 1. [Google Scholar] [CrossRef]
- Hamrick, J.L. Genetic diversity and conservation in tropical forest. In Proceedings of the ASEAN-Canada Symposium on Genetic Conservation and Production of Tropical Tree Seed, Chiang Mai, Thailand, 4–16 June 1993; Drysdale, R.M., John, S.E.T., Yapa, A.C., Eds.; ASEAN-Canada Forest Tree Seed Centre: Saraburi, Thailand, 1994; pp. 1–9. [Google Scholar]
- Vijayan, K.; Srivastava, P.P.; Awasthi, A.K. Analysis of phylogenetic relationship among five mulberry (Morus) species using molecular markers. Genome 2004, 47, 439–448. [Google Scholar] [CrossRef]
- Gnanesh, B.N.; Mondal, R.; Arunakumar, G.S.; Manojkumar, H.B.; Singh, P.; Bhavya, M.R.; Sowbhagya, P.; Burji, S.M.; Mogili, T.; Sivaprasad, V. Genome size, genetic diversity, and phenotypic variability imply the effect of genetic variation instead of ploidy on trait plasticity in the cross-pollinated tree species of mulberry. PLoS ONE 2023, 18, e0289766. [Google Scholar] [CrossRef]
| Serial No. | Accessions | Species | Source | Morphological Characters | Ploidy Level |
|---|---|---|---|---|---|
| 1 | SWLS1 | Morus nigra | Sahiwal | Black fruit | 2n = 22x = 308 [22] |
| 2 | SWLS2 | Morus nigra | Sahiwal | Small-sized, black fruit | 2n = 22x = 308 [22] |
| 3 | SWLS3 | Morus nigra | Sahiwal | Small-sized, black fruit | 2n = 22x = 308 [22] |
| 4 | SWLS4 | Morus nigra | Sahiwal | Large-sized, black fruit | 2n = 22x = 308 [22] |
| 5 | SWLS5 | Morus nigra | Sahiwal | Large-sized, black fruit | 2n = 22x = 308 [22] |
| 6 | SWLS6 | Morus alba | Sahiwal | White fruit | 2n = 2x = 28 [23] |
| 7 | SWLS7 | Morus alba | Sahiwal | White fruit | 2n = 2x = 28 [23] |
| 8 | SWLS8 | Morus alba | Sahiwal | White fruit | 2n = 2x = 28 [23] |
| 9 | SWLS9 | Morus alba | Sahiwal | Small-sized, white fruit | 2n = 2x = 28 [23] |
| 10 | SWLS10 | Morus alba | Sahiwal | Small-sized, white fruit | 2n = 2x = 28 [23] |
| 11 | SWLS11 | Morus nigra | Sahiwal | Large-sized, black fruit | 2n = 22x = 308 [22] |
| 12 | SWLS12 | Morus nigra | Sahiwal | Large-sized, black fruit | 2n = 22x = 308 [22] |
| 13 | SWLS13 | Morus rubra | Sahiwal | Small berry-sized reddish-black fruit | 2n = 2x = 28 [24] |
| 14 | SWLS14 | Morus rubra | Sahiwal | Small berry sized, reddish-black fruit | 2n = 2x = 28 [24] |
| 15 | FSDS15 | Morus alba | Faisalabad | Large-sized, white fruit | 2n = 2x = 28 [23] |
| 16 | FSDS16 | Morus alba | Faisalabad | Large-sized, white fruit | 2n = 2x = 28 [23] |
| 17 | FSDS17 | Morus alba | Faisalabad | Large-sized, white fruit | 2n = 2x = 28 [23] |
| 18 | FSDS18 | Morus alba | Faisalabad | Large-sized, white fruit | 2n = 2x = 28 [23] |
| 19 | FSDS19 | Morus alba | Faisalabad | Large-sized, white fruit | 2n = 2x = 28 [23] |
| 20 | FSDS20 | Morus alba | Faisalabad | Large-sized, white fruit | 2n = 2x = 28 [23] |
| 21 | FSDS21 | Morus alba | Faisalabad | Large-sized, white fruit | 2n = 2x = 28 [23] |
| 22 | FSDS22 | Morus nigra | Faisalabad | Large-sized, black fruit | 2n = 22x = 308 [22] |
| 23 | FSDS23 | Morus nigra | Faisalabad | Large-sized, black fruit | 2n = 22x = 308 [22] |
| 24 | FSDS24 | Morus nigra | Faisalabad | Large-sized, black fruit | 2n = 22x = 308 [22] |
| 25 | FSDS25 | Morus nigra | Faisalabad | Large-sized, black fruit | 2n = 22x = 308 [22] |
| 26 | FSDS26 | Morus nigra | Faisalabad | Large-sized, black fruit | 2n = 22x = 308 [22] |
| 27 | FSDS27 | Morus nigra | Faisalabad | Large-sized, black fruit | 2n = 22x = 308 [22] |
| 28 | FSDS28 | Morus rubra | Faisalabad | Small berry-sized, reddish-black fruit | 2n = 2x = 28 [24] |
| 29 | FSDS29 | Morus rubra | Faisalabad | Small berry-sized, reddish-black fruit | 2n = 2x = 28 [24] |
| 30 | FSDS30 | Morus rubra | Faisalabad | Small -sized, reddish-black fruit | 2n = 2x = 28 [24] |
| Serial # | iPBS Primers | Primer Sequence (5′-3′) | Tm (°C) | Size Range (bp) | PM | PIC | I | He | µHe |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 2230 | TCTAGGCGTCTGATACCA | 46 | 200–800 | 75 | 0.29 | 0.34 | 0.21 | 0.22 |
| 2 | 2231 | ACTTGGATGCTGATACCA | 44 | 750–1400 | 56 | 0.40 | 0.46 | 0.31 | 0.32 |
| 3 | 2232 | AGAGAGGCTCGGATACCA | 48 | 400–1200 | 56 | 0.29 | 0.34 | 0.20 | 0.21 |
| 4 | 2238 | ACCTAGCTCATGATGCCA | 46 | 250–900 | 21 | 0.34 | 0.20 | 0.20 | 0.21 |
| 5 | 2239 | ACCTAGGCTCGGATGCCA | 50 | 250–1000 | 51 | 0.31 | 1.26 | 0.19 | 0.20 |
| 6 | 2246 | ACTAGGCTCTGTATACCA | 44 | 400–1000 | 35 | 0.33 | 0.34 | 0.20 | 0.21 |
| 7 | 2249 | AACCGACCTCTGATACCA | 46 | 600–1500 | 26 | 0.32 | 0.34 | 0.20 | 0.20 |
| 8 | 2257 | CTCTCAATGAAAGCACCA | 43 | 600–1000 | 41 | 0.47 | 0.53 | 0.35 | 0.36 |
| 9 | 2277 | GGCGATGATACCA | 46 | 400–1000 | 70 | 0.44 | 0.49 | 0.32 | 0.33 |
| 10 | 2251 | GAACAGGCGATGATACCA | 46 | Did not amplify | |||||
| 11 | 2398 | GAACCCTTGCCGATACCA | 48 | Did not amplify | |||||
| 12 | 2255 | GCGTGTGCTCTCATACCA | 48 | Did not amplify | |||||
| 13 | 2252 | TCATGGCTCATGATACCA | 43 | Did not amplify | |||||
| 14 | 2229 | CGACCTGTTCTGATACCA | 46 | Did not amplify | |||||
| 15 | 2377 | ACGAAGGGACCA | 46 | Did not amplify | |||||
| Population | N | Na | Ne | I | He | uHe | %P |
|---|---|---|---|---|---|---|---|
| Morus nigra | 13 | 1.65 | 1.53 | 0.45 | 0.31 | 0.33 | 82.4% |
| Morus alba | 12 | 1.29 | 1.44 | 0.37 | 0.25 | 0.27 | 64.7% |
| Morus rubra | 5 | 1.26 | 1.50 | 0.39 | 0.27 | 0.34 | 60.8% |
| Mean | 30 | 1.88 | 1.36 | 0.38 | 0.24 | 0.25 | 69.3% |
| Source | df | SS | MS | Est. Var. | % | PhiPT |
|---|---|---|---|---|---|---|
| Among subgroups | 2 | 52.57 | 26.29 | 1.97 | 20 | 0.029 * |
| Within subgroups | 27 | 211.78 | 7.84 | 7.84 | 80 | |
| Total | 29 | 264.37 | 9.81 | 100 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mehmood, A.; Dracatos, P.M.; Arshad, L.; Bibi, S.; Zaheer, A. Assessment of Molecular Diversity and Population Structure of Pakistani Mulberry Accessions Using Retrotransposon-Based DNA Markers. Agriculture 2024, 14, 400. https://doi.org/10.3390/agriculture14030400
Mehmood A, Dracatos PM, Arshad L, Bibi S, Zaheer A. Assessment of Molecular Diversity and Population Structure of Pakistani Mulberry Accessions Using Retrotransposon-Based DNA Markers. Agriculture. 2024; 14(3):400. https://doi.org/10.3390/agriculture14030400
Chicago/Turabian StyleMehmood, Asim, Peter M. Dracatos, Linta Arshad, Shabana Bibi, and Ahmad Zaheer. 2024. "Assessment of Molecular Diversity and Population Structure of Pakistani Mulberry Accessions Using Retrotransposon-Based DNA Markers" Agriculture 14, no. 3: 400. https://doi.org/10.3390/agriculture14030400
APA StyleMehmood, A., Dracatos, P. M., Arshad, L., Bibi, S., & Zaheer, A. (2024). Assessment of Molecular Diversity and Population Structure of Pakistani Mulberry Accessions Using Retrotransposon-Based DNA Markers. Agriculture, 14(3), 400. https://doi.org/10.3390/agriculture14030400






