Genomic-Mediated Breeding Strategies for Global Warming in Chickpeas (Cicer arietinum L.)
Abstract
1. Introduction
2. Impact on Morpho-Physiological, Biochemical, and Molecular Parameters Due to Drought and Heat Stresses
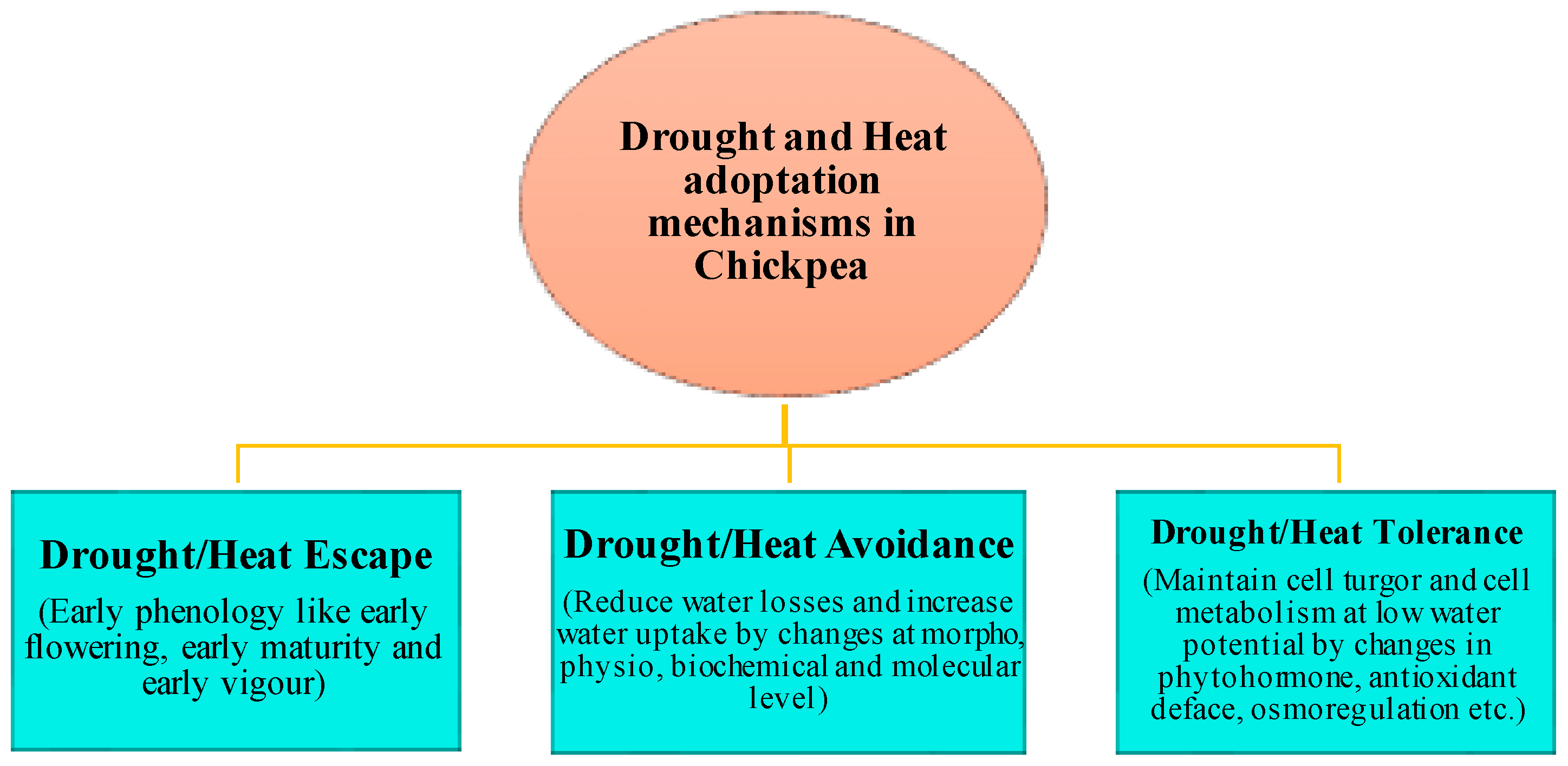
3. Current Knowledge of Different Mechanisms Responsible for the Adaption of Chickpea Plants under Drought and Heat Conditions
4. Genetic Knowledge of Traits Responsible to Chickpea Drought and Heat
5. Selection Indices and the Identification of Donors for Drought Tolerance
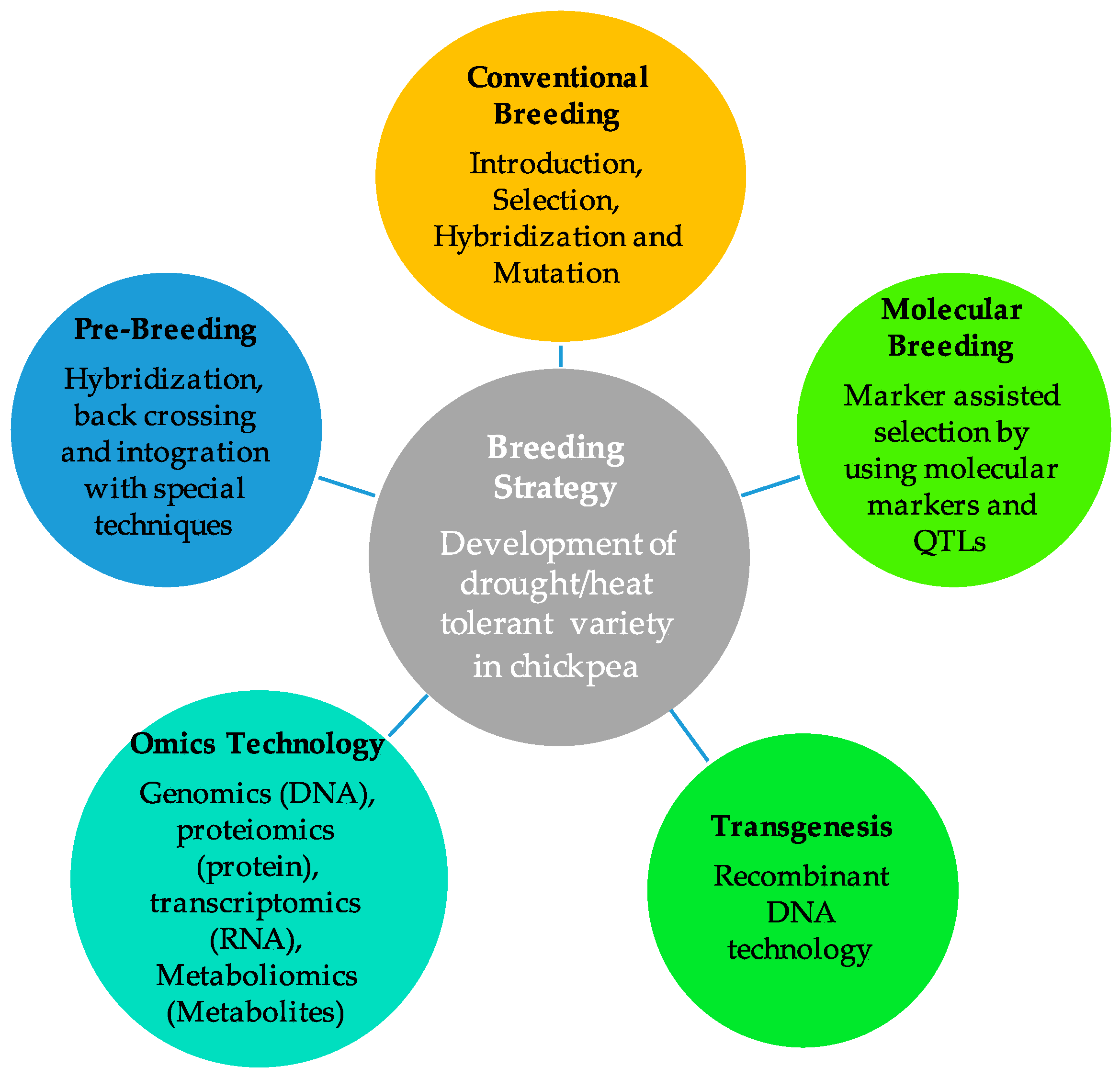
6. Breeding Tactics for Enhancing Drought Tolerance in Chickpeas
6.1. Breeding of Drought- and Heat-Tolerant High-Yield Varieties Using Traditional Breeding Approaches
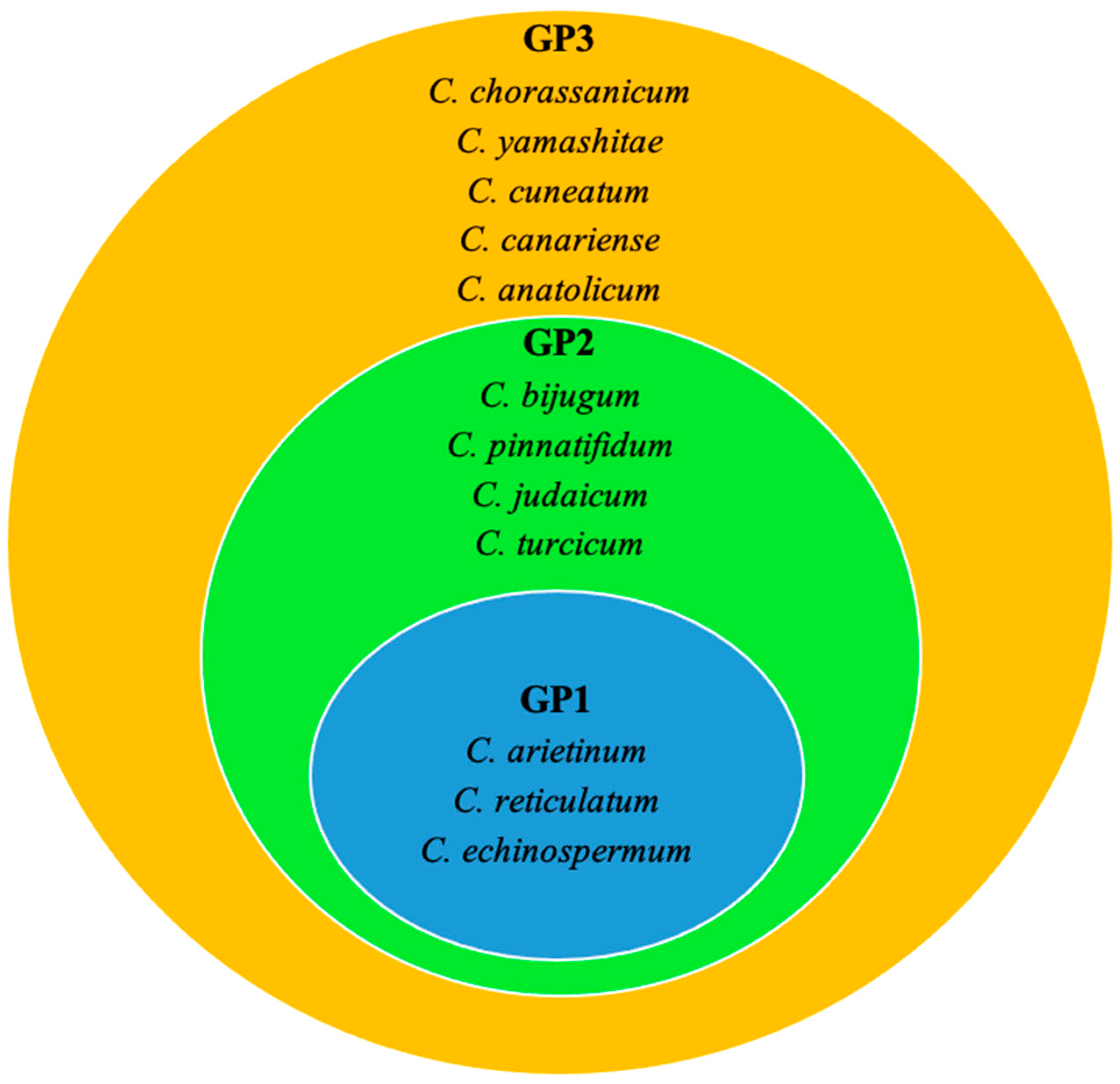
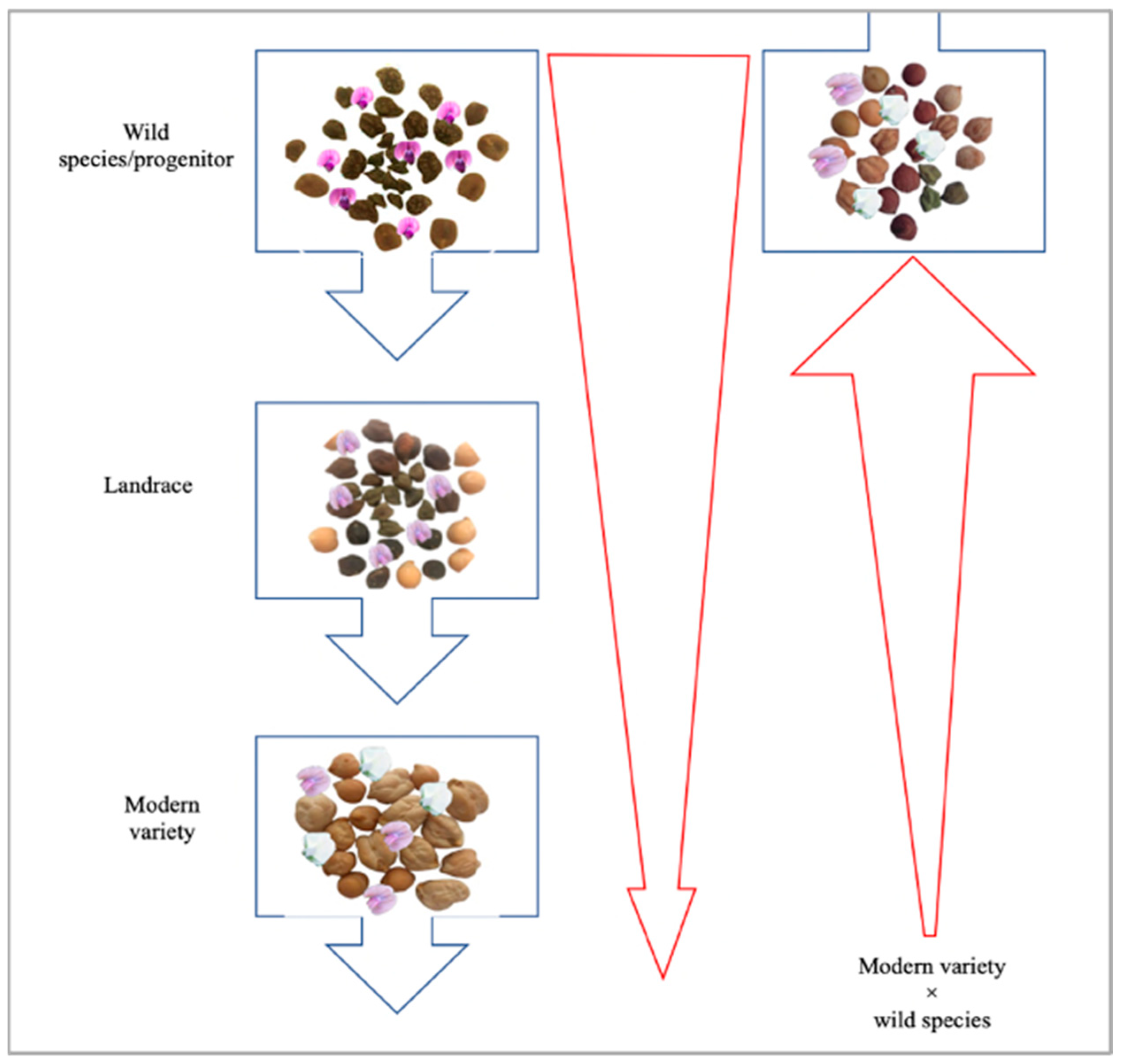
6.2. Exploitation of Wild Relatives through Pre-Breeding for Drought and Heat Tolerance
6.3. Exploitation of QTLs/Genes for Drought and Heat Tolerance Employing Marker-Assisted Breeding
6.4. OMICS-Based Technology and Transgenesis Approach for Drought and Heat Tolerance in Chickpeas
7. Candidate Genes Related to Drought and Heat Tolerance
8. Limitations and Future Directions
- Heat and drought tolerance are complex in nature because multiple traits are involved to control the tolerance to these stresses;
- Low heritability of the traits imparting drought and heat tolerance due to high G × E interactions limit the breeding of high-yield cultivars with heat and drought tolerance;
- There is still a lack of knowledge on the major physiological/biochemical/morphological traits imparting heat and drought tolerance. This could be due to the unavailability of precise screening methods for these traits;
- The molecular mechanism underlying these traits is not known properly and, which limits the scope for molecular breeding for these traits;
- The traits imparting drought and heat tolerance are controlled by many genes, and a network of genes is involved to control these traits. Therefore, major QTLs/genes are not well-characterized for these traits;
- Only a small percentage of accessions have been screened for their performance under water-limited conditions and high-temperature conditions despite the availability of a range of chickpea germplasm.
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Avnery, S.; Mauzerall, D.L.; Liu, J.; Horowitz, L.W. Global Crop Yield Reductions Due to Surface Ozone Exposure: 1. Year 2000 Crop Production Losses and Economic Damage. Atmos. Environ. 2011, 45, 2284–2296. [Google Scholar] [CrossRef]
- Teixeira, E.I.; Fischer, G.; Van Velthuizen, H.; Walter, C.; Ewert, F. Global Hot-Spots of Heat Stress on Agricultural Crops Due to Climate Change. Agric. For. Meteorol. 2013, 170, 206–215. [Google Scholar] [CrossRef]
- Challinor, A.J.; Watson, J.; Lobell, D.B.; Howden, S.M.; Smith, D.R.; Chhetri, N. A Meta-Analysis of Crop Yield under Climate Change and Adaptation. Nat. Clim. Chang. 2014, 4, 287–291. [Google Scholar] [CrossRef]
- Jha, U.C.; Chaturvedi, S.K.; Bohra, A.; Basu, P.S.; Khan, M.S.; Barh, D. Abiotic Stresses, Constraints and Improvement Strategies in Chickpea. Plant Breed. 2014, 133, 163–178. [Google Scholar] [CrossRef]
- Toker, C.; Lluch, C.; Tejera, N.A.; Serraj, R.; Siddique, K.H.M. 23 Abiotic Stresses. In Chickpea Breeding and Management; CABI: Wallingford, UK, 2007; pp. 474–496. [Google Scholar]
- Toker, C.; Mutlu, N. Breeding for Abiotic Stresses. In Biology and Breeding of Food Legumes; CABI: Wallingford, UK, 2011; pp. 241–261. [Google Scholar]
- Canci, H.; Toker, C. Evaluation of Yield Criteria for Drought and Heat Resistance in Chickpea (Cicer arietinum L.). J. Agron. Crop Sci. 2009, 195, 47–54. [Google Scholar] [CrossRef]
- Maphosa, L.; Richards, M.F.; Norton, S.L.; Nguyen, G.N. Breeding for Abiotic Stress Adaptation in Chickpea (Cicer arietinum L.): A Comprehensive Review. Crop Breed. Genet. Genom. 2020, 2, e200015. [Google Scholar] [CrossRef]
- Kumari, P.; Rastogi, A.; Yadav, S. Effects of Heat Stress and Molecular Mitigation Approaches in Orphan Legume, Chickpea. Mol. Biol. Rep. 2020, 47, 4659–4670. [Google Scholar] [CrossRef]
- Sani, S.G.A.S.; Chang, P.L.; Zubair, A.; Carrasquilla-Garcia, N.; Cordeiro, M.; Penmetsa, R.V.; Munis, M.F.H.; Nuzhdin, S.V.; Cook, D.R.; von Wettberg, E.J. Genetic Diversity, Population Structure, and Genetic Correlation with Climatic Variation in Chickpea (Cicer arietinum) Landraces from Pakistan. Plant Genome 2018, 11, 170067. [Google Scholar] [CrossRef]
- Gaur, P.; Srinivasan, S.; Varshney, R. Drought and Heat Tolerance in Chickpea. Legume Perspect. 2014, 3, 15–17. [Google Scholar]
- Harlan, J.R.; de Wet, J.M. Toward a Rational Classification of Cultivated Plants. Taxon 1971, 20, 509–517. [Google Scholar] [CrossRef]
- Kahraman, A.; Pandey, A.; Khan, M.K.; Lindsay, D.; Moenga, S.; Vance, L.; Bergmann, E.; Carrasquilla-Garcia, N.; Shin, M.-G.; Chang, P.L. Distinct Subgroups of Cicer echinospermum Are Associated with Hybrid Sterility and Breakdown in Interspecific Crosses with Cultivated Chickpea. Crop Sci. 2017, 57, 3101–3111. [Google Scholar] [CrossRef]
- Mallikarjuna, N. Ovule and Embryo Culture to Obtain Hybrids from Interspecific Incompatible Pollinations in Chickpea. Euphytica 1999, 110, 1–6. [Google Scholar] [CrossRef]
- Toker, C.; Berger, J.; Eker, T.; Sari, D.; Sari, H.; Gokturk, R.S.; Kahraman, A.; Aydin, B.; von Wettberg, E.J. Cicer turcicum: A New Cicer Species and Its Potential to Improve Chickpea. Front. Plant Sci. 2021, 12, 662891. [Google Scholar] [CrossRef] [PubMed]
- Tanksley, S.D.; McCouch, S.R. Seed Banks and Molecular Maps: Unlocking Genetic Potential from the Wild. Science 1997, 277, 1063–1066. [Google Scholar] [CrossRef] [PubMed]
- Coyne, C.J.; Kumar, S.; von Wettberg, E.J.; Marques, E.; Berger, J.D.; Redden, R.J.; Ellis, T.N.; Brus, J.; Zablatzká, L.; Smỳkal, P. Potential and Limits of Exploitation of Crop Wild Relatives for Pea, Lentil, and Chickpea Improvement. Legume Sci. 2020, 2, e36. [Google Scholar] [CrossRef]
- Collard, B.C.-Y.; Ades, P.K.; Pang, E.C.K.; Brouwer, J.B.; Taylor, P.W.J. Prospecting for Sources of Resistance to Ascochyta Blight in Wild Cicer Species. Australas. Plant Pathol. 2001, 30, 271–276. [Google Scholar] [CrossRef]
- Sharma, M.; Ghosh, R. An Update on Genetic Resistance of Chickpea to Ascochyta Blight. Agronomy 2016, 6, 18. [Google Scholar] [CrossRef]
- Chrigui, N.; Sari, D.; Sari, H.; Eker, T.; Cengiz, M.F.; Ikten, C.; Toker, C. Introgression of Resistance to Leafminer (Liriomyza cicerina Rondani) from Cicer reticulatum Ladiz. to C. arietinum L. and Relationships between Potential Biochemical Selection Criteria. Agronomy 2020, 11, 57. [Google Scholar] [CrossRef]
- Eker, T.; Erler, F.; Adak, A.; Imrek, B.; Guven, H.; Tosun, H.S.; Sari, D.; Sari, H.; Upadhyaya, H.D.; Toker, C. Screening of Chickpea Accessions for Resistance against the Pulse Beetle, Callosobruchus chinensis L. (Coleoptera: Bruchidae). J. Stored Prod. Res. 2018, 76, 51–57. [Google Scholar] [CrossRef]
- Eker, T.; Erler, F.; Sari, H.; Sari, D.; Berger, J.; Toker, C. Deployment of Cicer echinospermum PH Davis for Resistance to Callosobruchus chinensis L. J. Plant Dis. Prot. 2022, 129, 843–851. [Google Scholar] [CrossRef]
- Toker, C. Preliminary Screening and Selection for Cold Tolerance in Annual Wild Cicer Species. Genet. Resour. Crop Evol. 2005, 52, 1–5. [Google Scholar] [CrossRef]
- Bakir, M.; Sari, D.; Sari, H.; Waqas, M.; Atif, R.M. Chickpea Wild Relatives: Potential Hidden Source for the Development of Climate Resilient Chickpea Varieties. In Wild Germplasm for Genetic Improvement in Crop Plants; Elsevier: Amsterdam, The Netherlands, 2021; pp. 269–297. ISBN 978-0-12-822137-2. [Google Scholar]
- Ramgopal, D.; Srivastava, R.K.; Pande, S.; Rathore, A.; Jadhav, D.R.; Sharma, M.; Gaur, P.M.; Mallikarjuna, N. Introgression of Botrytis Grey Mould Resistance Genes from Cicer reticulatum (Bgmr1cr) and C. echinospermum (Bgmr1ce) to Chickpea (C. arietinum). Plant Genet. Resour. 2013, 11, 212–216. [Google Scholar] [CrossRef]
- Singh, M.; Rani, S.; Malhotra, N.; Katna, G.; Sarker, A. Transgressive Segregations for Agronomic Improvement Using Interspecific Crosses between C. arietinum L. × C. reticulatum Ladiz. and C. arietinum L. × C. echinospermum Davis Species. PLoS ONE 2018, 13, e0203082. [Google Scholar] [CrossRef]
- Talip, M.; Adak, A.; Kahraman, A.; Berger, J.; Sari, D.; Sari, H.; Penmetsa, R.V.; Von Wettberg, E.J.; Cook, D.R.; Toker, C. Agro-Morphological Traits of Cicer reticulatum Ladizinsky in Comparison to C. echinospermum PH Davis in Terms of Potential to Improve Cultivated Chickpea (C. arietinum L.). Genet. Resour. Crop Evol. 2018, 65, 951–962. [Google Scholar] [CrossRef]
- Cowling, W.A.; Buirchell, B.J.; Falk, D.E. A Model for Incorporating Novel Alleles from the Primary Gene Pool into Elite Crop Breeding Programs While Reselecting Major Genes for Domestication or Adaptation. Crop Pasture Sci. 2009, 60, 1009–1015. [Google Scholar] [CrossRef]
- Smỳkal, P.; Nelson, M.N.; Berger, J.D.; Von Wettberg, E.J. The Impact of Genetic Changes during Crop Domestication. Agronomy 2018, 8, 119. [Google Scholar] [CrossRef]
- Gross, B.L.; Olsen, K.M. Genetic Perspectives on Crop Domestication. Trends Plant Sci. 2010, 15, 529–537. [Google Scholar] [CrossRef] [PubMed]
- Olsen, K.M.; Wendel, J.F. A Bountiful Harvest: Genomic Insights into Crop Domestication Phenotypes. Annu. Rev. Plant Biol. 2013, 64, 47–70. [Google Scholar] [CrossRef]
- Berger, J.D.; Turner, N.C. The Ecology of Chickpea. In Chickpea Breeding and Management; CABI: Wallingford, UK, 2007; pp. 47–71. [Google Scholar]
- Singh, R.; Sharma, P.; Varshney, R.K.; Sharma, S.K.; Singh, N.K. Chickpea Improvement: Role of Wild Species and Genetic Markers. Biotechnol. Genet. Eng. Rev. 2008, 25, 267–314. [Google Scholar] [CrossRef]
- Lamichaney, A.; Katiyar, P.K.; Laxmi, V.; Pratap, A. Variation in Pre-Harvest Sprouting Tolerance and Fresh Seed Germination in Mungbean (Vigna radiata L.) Genotypes. Plant Genet. Resour. 2018, 16, 437–445. [Google Scholar] [CrossRef]
- Lamichaney, A.; Pratap, A.; KATIYAR, P.K.; Singh, N.P. Genotypic Variability Studies and Identification of Pre-Harvest Sprouting Tolerant Wild Vigna. Indian J. Agric. Sci. 2021, 91, 335–339. [Google Scholar] [CrossRef]
- Lamichaney, A.; Hazra, K.K.; Katiyar, P.K.; Parihar, A.K.; Gupta, D.S.; Kumar, A.; Singh, F. Influence of Seed and Pod Biophysical Characters on Pre-Harvest Sprouting Tolerance in Urdbean (Vigna mungo L.). Acta Physiol. Plant. 2023, 45, 48. [Google Scholar] [CrossRef]
- Serraj, R.; Bidinger, F.R.; Chauhan, Y.S.; Seetharama, N.; Nigam, S.N.; Saxena, N.P. Management of Drought in ICRISAT Cereal and Legume Mandate Crops. In Water Productivity in Agriculture: Limits and Opportunities for Improvement; CABI: Wallingford, UK, 2003; pp. 127–144. [Google Scholar]
- Toker, C.; Canci, H.; Yildirim, T. Evaluation of Perennial Wild Cicer Species for Drought Resistance. Genet. Resour. Crop Evol. 2007, 54, 1781–1786. [Google Scholar] [CrossRef]
- Siemens, H.W. Einführung in Die Allgemeine Konstitutions-Und Vererbungspathologie; Springer: Berlin/Heidelberg, Germany, 1921. [Google Scholar]
- Donald, C.T. The Breeding of Crop Ideotypes. Euphytica 1968, 17, 385–403. [Google Scholar] [CrossRef]
- Trethowan, R.M. Defining a Genetic Ideotype for Crop Improvement. In Crop Breeding: Methods and Protocols; Humana: New York, NY, USA, 2014; pp. 1–20. [Google Scholar]
- Khanna-Chopra, R.; Sinha, S.K. Chickpea: Physiological Aspects of Growth and Yield. In The Chickpea; CABI: Wallingford, UK, 1987; pp. 163–187. [Google Scholar]
- Singh, K.B. Chickpea Breeding. In The Chickpea; CABI: Wallingford, UK, 1987; pp. 127–162. [Google Scholar]
- Saxena, N.P.; Johansen, C. Chickpea Ideotypes for Genetic Enhancement of Yield and Yield Stability in South Asia. In Proceedings of the Chickpea in the Nineties: Proceedings of the Second International Workshop on Chickpea Improvement, Patancheru, India, 4–8 December 1989; pp. 4–8. [Google Scholar]
- Saxena, N.P.; Saxena, M.C.; Johansen, C. Chickpea Ideotypes for Low and High-Input Conditions. In Recent Advantages in Pulses Research; Indian Society of Pulses Research and Development, IIPR: Kanpur, India, 1997; pp. 217–231. [Google Scholar]
- Bonfil, D.J.; Goren, O.; Mufradi, I.; Lichtenzveig, J.; Abbo, S. Development of Early-Flowering Kabuli Chickpea with Compound and Simple Leaves. Plant Breed. 2007, 126, 125–129. [Google Scholar] [CrossRef]
- Boyer, J.S. Plant Productivity and Environment. Science 1982, 218, 443–448. [Google Scholar] [CrossRef] [PubMed]
- Kumar, J.; Abbo, S. Genetics of Flowering Time in Chickpea and Its Bearing on Productivity in Semiarid Environments. Adv. Agron. 2001, 72, 107–138. [Google Scholar]
- Dixit, G.P. Anonymous Annual Report of AICRP on Chickpea; IIPR: Kanpur, India, 2022. [Google Scholar]
- Fleury, D.; Jefferies, S.; Kuchel, H.; Langridge, P. Genetic and Genomic Tools to Improve Drought Tolerance in Wheat. J. Exp. Bot. 2010, 61, 3211–3222. [Google Scholar] [CrossRef]
- Kushwah, A.; Bhatia, D.; Singh, I.; Thudi, M.; Singh, G.; Bindra, S.; Vij, S.; Gill, B.S.; Bharadwaj, C.; Singh, S.; et al. Identification of Stable Heat Tolerance QTLs Using Inter-Specific Recombinant Inbred Line Population Derived from GPF 2 and ILWC 292. PLoS ONE 2021, 16, e0254957. [Google Scholar] [CrossRef]
- Devasirvatham, V.; Gaur, P.M.; Mallikarjuna, N.; Tokachichu, R.N.; Trethowan, R.M.; Tan, D.K. Effect of High Temperature on the Reproductive Development of Chickpea Genotypes under Controlled Environments. Funct. Plant Biol. 2012, 39, 1009–1018. [Google Scholar] [CrossRef]
- Chaudhary, S.; Devi, P.; Bhardwaj, A.; Jha, U.C.; Sharma, K.D.; Prasad, P.V.; Siddique, K.H.; Bindumadhava, H.; Kumar, S.; Nayyar, H. Identification and Characterization of Contrasting Genotypes/Cultivars for Developing Heat Tolerance in Agricultural Crops: Current Status and Prospects. Front. Plant Sci. 2020, 11, 587264. [Google Scholar] [CrossRef] [PubMed]
- Asati, R.; Tripathi, M.K.; Tiwari, S.; Yadav, R.K.; Tripathi, N. Molecular Breeding and Drought Tolerance in Chickpea. Life 2022, 12, 1846. [Google Scholar] [CrossRef]
- Sabaghpour, S.H.; Akbar, M.A.; Ali, S.; Masood, K.; Malhotra, R.S. Study on Chickpea Drought Tolerance Lines under Dryland Condition of Iran. Available online: https://www.indianjournals.com/ijor.aspx?target=ijor:ijocs&volume=1&issue=1and2&article=011 (accessed on 27 July 2023).
- Turner, N.C.; Abbo, S.; Berger, J.D.; Chaturvedi, S.; French, R.J.; Ludwig, C.; Mannur, D.; Singh, S.; Yadava, H. Osmotic Adjustment in Chickpea (Cicer arietinum L.) Results in No Yield Benefit under Terminal Drought. J. Exp. Bot. 2007, 58, 187–194. [Google Scholar] [CrossRef]
- Mafakheri, A.; Siosemardeh, A.; Bahramnejad, B.; Struik, P.C.; Sohrabi, Y. Effect of Drought Stress and Subsequent Recovery on Protein, Carbohydrate Contents, Catalase and Peroxidase Activities in Three Chickpea (‘Cicer arietinum’) Cultivars. Aust. J. Crop Sci. 2011, 5, 1255–1260. [Google Scholar]
- Rahbarian, R.; Nejad, R.K.; Ganjeali, A.; Bagheri, A.; Najafi, F. Drought Stress Effects on Photosynthesis, Chlorophyll Fluorescence and Water Relations in Tolerant and Susceptible Chickpea (Cicer arietinum L.) Genotypes. Acta Biol. Cracoviensia 2011, 1, 47–56. [Google Scholar] [CrossRef]
- Sharma, R.A. Influence of Drought Stress on the Emergence and Growth of Chickpea Seedlings. Int. Chickpea Newsl. 1985, 12, 15–16. [Google Scholar]
- Weber, H.; Borisjuk, L.; Wobus, U. Molecular Physiology of Legume Seed Development. Annu. Rev. Plant Biol. 2005, 56, 253–279. [Google Scholar] [CrossRef]
- Vessal, S.; Palta, J.A.; Atkins, C.A.; Siddique, K.H. Development of an Assay to Evaluate Differences in Germination Rate among Chickpea Genotypes under Limited Water Content. Funct. Plant Biol. 2011, 39, 60–70. [Google Scholar] [CrossRef]
- Pouresmael, M.; Khavari-Nejad, R.A.; Mozafari, J.; Najafi, F.; Moradi, F. Efficiency of Screening Criteria for Drought Tolerance in Chickpea. Arch. Agron. Soil Sci. 2013, 59, 1675–1693. [Google Scholar] [CrossRef]
- Chaves, M.M.; Maroco, J.P.; Pereira, J.S. Understanding Plant Responses to Drought—From Genes to the Whole Plant. Funct. Plant Biol. 2003, 30, 239–264. [Google Scholar] [CrossRef]
- Chaves, M.M.; Flexas, J.; Pinheiro, C. Photosynthesis under Drought and Salt Stress: Regulation Mechanisms from Whole Plant to Cell. Ann. Bot. 2009, 103, 551–560. [Google Scholar] [CrossRef]
- Mohammadi, A.; Habibi, D.; Rohami, M.; Mafakheri, S. Effect of Drought Stress on Antioxidant Enzymes Activity of Some Chickpea Cultivars. Am.-Euras. J. Agric. Environ. Sci. 2011, 11, 782–785. [Google Scholar]
- Davies, S.L.; Turner, N.C.; Siddique, K.H.M.; Leport, L.; Plummer, J.A. Seed Growth of Desi and Kabuli Chickpea (Cicer arietinum L.) in a Short-Season Mediterranean-Type Environment. Aust. J. Exp. Agric. 1999, 39, 181–188. [Google Scholar] [CrossRef]
- Leport, L.; Turner, N.C.; French, R.J.; Barr, M.D.; Duda, R.; Davies, S.L.; Tennant, D.; Siddique, K.H.M. Physiological Responses of Chickpea Genotypes to Terminal Drought in a Mediterranean-Type Environment. Eur. J. Agron. 1999, 11, 279–291. [Google Scholar] [CrossRef]
- Leport, L.; Turner, N.C.; Davies, S.L.; Siddique, K.H.M. Variation in Pod Production and Abortion among Chickpea Cultivars under Terminal Drought. Eur. J. Agron. 2006, 24, 236–246. [Google Scholar] [CrossRef]
- He, J.; Du, Y.-L.; Wang, T.; Turner, N.C.; Yang, R.-P.; Jin, Y.; Xi, Y.; Zhang, C.; Cui, T.; Fang, X.-W. Conserved Water Use Improves the Yield Performance of Soybean (Glycine max (L.) Merr.) under Drought. Agric. Water Manag. 2017, 179, 236–245. [Google Scholar] [CrossRef]
- Siddique, K.H.M.; Loss, S.P.; Regan, K.L.; Jettner, R.L. Adaptation and Seed Yield of Cool Season Grain Legumes in Mediterranean Environments of South-Western Australia. Aust. J. Agric. Res. 1999, 50, 375–388. [Google Scholar] [CrossRef]
- Aslam, M.M.; Raja, S.; Saeed, S.; Farhat, F.; Tariq, A.; Rai, H.M.; Javaid, A.; Shahzadi, I.; Asim, M.; Zulfiqar, S. Revisiting the Crucial Role of Reactive Oxygen Species and Antioxidant Defense in Plant Under Abiotic Stress. In Antioxidant Defense in Plants: Molecular Basis of Regulation; Springer: Berlin/Heidelberg, Germany, 2022; pp. 397–419. [Google Scholar]
- Mhadhbi, H.; Fotopoulos, V.; Djebali, N.; Polidoros, A.N.; Aouani, M.E. Behaviours of Medicago Truncatula–Sinorhizobium Meliloti Symbioses under Osmotic Stress in Relation with the Symbiotic Partner Input: Effects on Nodule Functioning and Protection. J. Agron. Crop Sci. 2009, 195, 225–231. [Google Scholar] [CrossRef]
- Labidi, N.; Mahmoudi, H.; Dorsaf, M.; Slama, I.; Abdelly, C. Assessment of Intervarietal Differences in Drought Tolerance in Chickpea Using Both Nodule and Plant Traits as Indicators. J. Plant Breed. Crop Sci. 2009, 1, 80–86. [Google Scholar]
- Singh, N.T.; Dhaliwal, G.S. Effect of Soil Temperature on Seedling Emergence in Different Crops. Plant Soil 1972, 37, 441–444. [Google Scholar] [CrossRef]
- Kumar, S.; Thakur, P.; Kaushal, N.; Malik, J.A.; Gaur, P.; Nayyar, H. Effect of Varying High Temperatures during Reproductive Growth on Reproductive Function, Oxidative Stress and Seed Yield in Chickpea Genotypes Differing in Heat Sensitivity. Arch. Agron. Soil Sci. 2013, 59, 823–843. [Google Scholar] [CrossRef]
- Kaushal, N.; Awasthi, R.; Gupta, K.; Gaur, P.; Siddique, K.H.; Nayyar, H. Heat-Stress-Induced Reproductive Failures in Chickpea (Cicer arietinum) Are Associated with Impaired Sucrose Metabolism in Leaves and Anthers. Funct. Plant Biol. 2013, 40, 1334–1349. [Google Scholar] [CrossRef]
- Devasirvatham, V.; Gaur, P.M.; Mallikarjuna, N.; Raju, T.N.; Trethowan, R.M.; Tan, D.K. Reproductive Biology of Chickpea Response to Heat Stress in the Field Is Associated with the Performance in Controlled Environments. Field Crops Res. 2013, 142, 9–19. [Google Scholar] [CrossRef]
- Eker, T.; Sari, H.; Sari, D.; Canci, H.; Arslan, M.; Aydinoglu, B.; Ozay, H.; Toker, C. Advantage of Multiple Pods and Compound Leaf in Kabuli Chickpea under Heat Stress Conditions. Agronomy 2022, 12, 557. [Google Scholar] [CrossRef]
- Devasirvatham, V.; Gaur, P.M.; Raju, T.N.; Trethowan, R.M.; Tan, D.K.Y. Field Response of Chickpea (Cicer arietinum L.) to High Temperature. Field Crops Res. 2015, 172, 59–71. [Google Scholar] [CrossRef]
- Jha, U.C.; Devi, P.; Prakash, V.; Kumar, S.; Parida, S.K.; Paul, P.J.; Prasad, P.V.; Sharma, K.D.; Siddique, K.H.; Nayyar, H. Response of Physiological, Reproductive Function and Yield Traits in Cultivated Chickpea (Cicer arietinum L.) under Heat Stress. Front. Plant Sci. 2022, 13, 880519. [Google Scholar]
- Srinivasan, A.; Takeda, H.; Senboku, T. Heat Tolerance in Food Legumes as Evaluated by Cell Membrane Thermostability and Chlorophyll Fluorescence Techniques. Euphytica 1996, 88, 35–45. [Google Scholar] [CrossRef]
- Kalaji, M.H.; Goltsev, V.N.; Żuk-Gołaszewska, K.; Zivcak, M.; Brestic, M. Chlorophyll Fluorescence: Understanding Crop Performance—Basics and Applications; CRC Press: Boca Raton, FL, USA, 2017. [Google Scholar]
- Havaux, M.; Tardy, F.; Ravenel, J.; Chanu, D.; Parot, P. Thylakoid Membrane Stability to Heat Stress Studied by Flash Spectroscopic Measurements of the Electrochromic Shift in Intact Potato Leaves: Influence of the Xanthophyll Content. Plant Cell Environ. 1996, 19, 1359–1368. [Google Scholar] [CrossRef]
- Wang, J.; Gan, Y.T.; Clarke, F.; McDonald, C.L. Response of Chickpea Yield to High Temperature Stress during Reproductive Development. Crop Sci. 2006, 46, 2171–2178. [Google Scholar] [CrossRef]
- Basu, P.S.; Ali, M.; Chaturvedi, S.K. Terminal Heat Stress Adversely Affects Chickpea Productivity in Northern India-Strategies to Improve Thermotolerance in the Crop under Climate Change. In W3 Workshop Proceedings: Impact of Climate Change on Agriculture; International Society for Photogrammetry and Remote Sensing: New Delhi, India, 2009; Volume 23, pp. 189–193. [Google Scholar]
- Mittler, R.; Vanderauwera, S.; Gollery, M.; Van Breusegem, F. Reactive Oxygen Gene Network of Plants. Trends Plant Sci. 2004, 9, 490–498. [Google Scholar] [CrossRef]
- Blum, A. Plant Breeding for Stress Environments; CRC Press: Boca Raton, FL, USA, 1988; p. 223. [Google Scholar]
- Ludlow, M.M.; Muchow, R.C. A Critical Evaluation of Traits for Improving Crop Yields in Water-Limited Environments. Adv. Agron. 1990, 43, 107–153. [Google Scholar]
- Mitra, J. Genetics and Genetic Improvement of Drought Resistance in Crop Plants. Curr. Sci. 2001, 80, 758–763. [Google Scholar]
- Hamwieh, A.; Imtiaz, M.; Hamwieh, A.; Imtiaz, M. Identifying Water-Responsive and Drought-Tolerant Chickpea Genotypes. Crop Pasture Sci. 2015, 66, 1003–1011. [Google Scholar] [CrossRef]
- Gaur, P.M.; Krishnamurthy, L.; Kashiwagi, J. Improving drought-avoidance root traits in chickpea (Cicer arietinum L.)-current status of research at ICRISAT. Plant Production Sci. 2008, 11, 3–11. [Google Scholar] [CrossRef]
- Turner, N.C.; Wright, G.C.; Siddique, K.H.M. Adaptation of Grain Legumes (Pulses) to Water-Limited Environments. In Advances in Agronomy; Academic Press: Cambridge, MA, USA, 2001; Volume 71, pp. 193–231. [Google Scholar]
- Berger, J.D. Ecogeographic and Evolutionary Approaches to Improving Adaptation of Autumn-Sown Chickpea (Cicer arietinum L.) to Terminal Drought: The Search for Reproductive Chilling Tolerance. Field Crops Res. 2007, 104, 112–122. [Google Scholar] [CrossRef]
- Kashiwagi, J.; Krishnamurthy, L.; Upadhyaya, H.D.; Krishna, H.; Chandra, S.; Vadez, V.; Serraj, R. Genetic Variability of Drought-Avoidance Root Traits in the Mini-Core Germplasm Collection of Chickpea (Cicer arietinum L.). Euphytica 2005, 146, 213–222. [Google Scholar] [CrossRef]
- Cattivelli, L.; Rizza, F.; Badeck, F.-W.; Mazzucotelli, E.; Mastrangelo, A.M.; Francia, E.; Marè, C.; Tondelli, A.; Stanca, A.M. Drought Tolerance Improvement in Crop Plants: An Integrated View from Breeding to Genomics. Field Crops Res. 2008, 105, 1–14. [Google Scholar] [CrossRef]
- Serraj, R.; Sinclair, T.R. Osmolyte Accumulation: Can It Really Help Increase Crop Yield under Drought Conditions? Plant Cell Environ. 2002, 25, 333–341. [Google Scholar] [CrossRef]
- Basu, P.S.; Singh, D.N. Physiology and Abiotic Stresses in Chickpea. In Chickpea Research in India; Indian Institute of Pulses Research: Kanpur, India, 2003; pp. 137–166. [Google Scholar]
- Moinuddin; Khanna-Chopra, R. Osmotic Adjustment in Chickpea in Relation to Seed Yield and Yield Parameters. Crop Sci. 2004, 44, 449–455. [Google Scholar] [CrossRef]
- Krishnamurthy, L.; Vadez, V.; Devi, M.J.; Serraj, R.; Nigam, S.N.; Sheshshayee, M.S.; Chandra, S.; Aruna, R. Variation in Transpiration Efficiency and Its Related Traits in a Groundnut (Arachis hypogaea L.) Mapping Population. Field Crops Res. 2007, 103, 189–197. [Google Scholar] [CrossRef]
- Nayyar, H.; Bains, T.; Kumar, S. Low Temperature Induced Floral Abortion in Chickpea: Relationship to Abscisic Acid and Cryoprotectants in Reproductive Organs. Environ. Exp. Bot. 2005, 53, 39–47. [Google Scholar] [CrossRef]
- Upadhyaya, H.D.; Kashiwagi, J.; Varshney, R.; Gaur, P.; Saxena, K.; Krishnamurthy, L.; Gowda, C.; Pundir, R.; Chaturvedi, S.; Basu, P.; et al. Phenotyping Chickpeas and Pigeonpeas for Adaptation to Drought. Front. Physiol. 2012, 3, 179. [Google Scholar] [CrossRef] [PubMed]
- Nayyar, H.; Chander, S. Protective Effects of Polyamines against Oxidative Stress Induced by Water and Cold Stress in Chickpea. J. Agron. Crop Sci. 2004, 190, 355–365. [Google Scholar] [CrossRef]
- Nandy, S.; Mandal, S.; Gupta, S.K.; Anand, U.; Ghorai, M.; Mundhra, A.; Rahman, M.H.; Ray, P.; Mitra, S.; Ray, D.; et al. Role of Polyamines in Molecular Regulation and Cross-Talks Against Drought Tolerance in Plants. J. Plant Growth Regul. 2022, 42, 4901–4917. [Google Scholar] [CrossRef]
- Esfahani, M.N.; Mostajeran, A. Rhizobial Strain Involvement in Symbiosis Efficiency of Chickpea–Rhizobia under Drought Stress: Plant Growth, Nitrogen Fixation and Antioxidant Enzyme Activities. Acta Physiol. Plant. 2011, 33, 1075–1083. [Google Scholar] [CrossRef]
- Wery, J.; Turc, O.; Lecoeur, J. Mechanism of Resistance to Cold, Heat and Drought in Cool-Season Legumes, with Special Reference to Chickpea and Pea. In Food Legumes; Wiley: Hoboken, NJ, USA, 1993; pp. 271–291. [Google Scholar]
- Kumar, S.; Kumari, P.; Kumar, U.; Grover, M.; Singh, A.K.; Singh, R.; Sengar, R.S. Molecular Approaches for Designing Heat Tolerant Wheat. J. Plant Biochem. Biotechnol. 2013, 22, 359–371. [Google Scholar] [CrossRef]
- Chakraborty, U.; Tongden, C. Evaluation of Heat Acclimation and Salicylic Acid Treatments as Potent Inducers of Thermotolerance in Cicer arietinum L. Curr. Sci. 2005, 89, 384–389. [Google Scholar]
- Kumar, S.; Kaushal, N.; Nayyar, H.; Gaur, P. Abscisic Acid Induces Heat Tolerance in Chickpea (Cicer arietinum L.) Seedlings by Facilitated Accumulation of Osmoprotectants. Acta Physiol. Plant. 2012, 34, 1651–1658. [Google Scholar] [CrossRef]
- Singh, K.B.; Malhotra, R.S.; Saxena, M.C. Additional Sources of Tolerance to Cold in Cultivated and Wild Cicer Species. Crop Sci. 1995, 35, 1491–1497. [Google Scholar] [CrossRef]
- Kumar, J.; van Rheenen, H. Brief Communication. A Major Gene for Time of Flowering in Chickpea. J. Hered. 2000, 91, 67–68. [Google Scholar] [CrossRef]
- Kashiwagi, J.; Krishnamurthy, L.; Purushothaman, R.; Upadhyaya, H.D.; Gaur, P.M.; Gowda, C.L.L.; Ito, O.; Varshney, R.K. Scope for Improvement of Yield under Drought through the Root Traits in Chickpea (Cicer arietinum L.). Field Crops Res. 2015, 170, 47–54. [Google Scholar] [CrossRef]
- Purushothaman, R.; Thudi, M.; Krishnamurthy, L.; Upadhyaya, H.D.; Kashiwagi, J.; Gowda, C.L.L.; Varshney, R.K. Association of Mid-Reproductive Stage Canopy Temperature Depression with the Molecular Markers and Grain Yields of Chickpea (Cicer arietinum L.) Germplasm under Terminal Drought. Field Crops Res. 2015, 174, 1–11. [Google Scholar] [CrossRef]
- Ramamoorthy, P.; Lakshmanan, K.; Upadhyaya, H.D.; Vadez, V.; Varshney, R.K. Shoot Traits and Their Relevance in Terminal Drought Tolerance of Chickpea (Cicer arietinum L.). Field Crops Res. 2016, 197, 10–27. [Google Scholar] [CrossRef] [PubMed]
- Ramamoorthy, P.; Lakshmanan, K.; Upadhyaya, H.D.; Vadez, V.; Varshney, R.K. Root Traits Confer Grain Yield Advantages under Terminal Drought in Chickpea (Cicer arietinum L.). Field Crops Res. 2017, 201, 146–161. [Google Scholar] [CrossRef]
- Chen, Y.; Ghanem, M.E.; Siddique, K.H. Characterising Root Trait Variability in Chickpea (Cicer arietinum L.) Germplasm. J. Exp. Bot. 2016, 68, 1987–1999. [Google Scholar] [CrossRef]
- Talebi, R.; Ensafi, M.H.; Baghebani, N.; Karami, E.; Mohammadi, K. Physiological Responses of Chickpea (Cicer arietinum) Genotypes to Drought Stress. Environ. Exp. Biol. 2013, 11, 9–15. [Google Scholar]
- Serraj, R.; Krishnamurthy, L.; Upadhyaya, H.D. Screening Chickpea Mini-Core Germplasm for Tolerance to Soil Salinity. Int. Chickpea Pigeonpea Newsl. 2004, 11, 29–32. [Google Scholar]
- Benjamin, J.G.; Nielsen, D.C. Water Deficit Effects on Root Distribution of Soybean, Field Pea and Chickpea. Field Crops Res. 2006, 97, 248–253. [Google Scholar] [CrossRef]
- Saradadevi, R.; Palta, J.A.; Siddique, K.H.M. ABA-Mediated Stomatal Response in Regulating Water Use during the Development of Terminal Drought in Wheat. Front. Plant Sci. 2017, 8, 1251. [Google Scholar] [CrossRef]
- Praba, M.L.; Cairns, J.E.; Babu, R.C.; Lafitte, H.R. Identification of Physiological Traits Underlying Cultivar Differences in Drought Tolerance in Rice and Wheat. J. Agron. Crop Sci. 2009, 195, 30–46. [Google Scholar] [CrossRef]
- Liu, F.; Jensen, C.R.; Andersen, M.N. Hydraulic and Chemical Signals in the Control of Leaf Expansion and Stomatal Conductance in Soybean Exposed to Drought Stress. Funct. Plant Biol. 2003, 30, 65–73. [Google Scholar] [CrossRef] [PubMed]
- Pang, J.; Turner, N.C.; Du, Y.-L.; Colmer, T.D.; Siddique, K.H.M. Pattern of Water Use and Seed Yield under Terminal Drought in Chickpea Genotypes. Front. Plant Sci. 2017, 8, 1375. [Google Scholar] [CrossRef] [PubMed]
- Ulemale, C.S.; Mate, S.N.; Deshmukh, D.V. Physiological Indices for Drought Tolerance in Chickpea (Cicer arietinum L.). World J. Agric. Sci. 2013, 9, 123–131. [Google Scholar]
- Farooq, M.; Ullah, A.; Lee, D.-J.; Alghamdi, S.S.; Siddique, K.H.M. Desi Chickpea Genotypes Tolerate Drought Stress Better than Kabuli Types by Modulating Germination Metabolism, Trehalose Accumulation, and Carbon Assimilation. Plant Physiol. Biochem. 2018, 126, 47–54. [Google Scholar] [CrossRef]
- Maqbool, M.A.; Aslam, M.; Ali, H. Breeding for Improved Drought Tolerance in Chickpea (Cicer arietinum L.). Plant Breed. 2017, 136, 300–318. [Google Scholar] [CrossRef]
- Nayak, S.N.; Zhu, H.; Varghese, N.; Datta, S.; Choi, H.-K.; Horres, R.; Jüngling, R.; Singh, J.; Kavi Kishor, P.B.; Sivaramakrishnan, S.; et al. Integration of Novel SSR and Gene-Based SNP Marker Loci in the Chickpea Genetic Map and Establishment of New Anchor Points with Medicago Truncatula Genome. Theor. Appl. Genet. 2010, 120, 1415–1441. [Google Scholar] [CrossRef]
- Sabaghpour, S.H.; Kumar, J.; Rao, T.N. Inheritance of Growth Vigour and Its Association with Other Characters in Chickpea. Plant Breed. 2003, 122, 542–544. [Google Scholar] [CrossRef][Green Version]
- Kanouni, H.; Kazemi, H.; Moghadam, M.; Neyshabori, M. Selection of Chickpea (Cicer arietinum L.) Entries for Drought Resistance. J. Agric. Sci. 2012, 12, 109–121. [Google Scholar]
- Jagadish, N.; Jayalakshmi, V. Combining Ability Studies for Drought Tolerance Attributes in Kabuli Chickpea (Cicer arietinum L.). Electron. J. Plant Breed. 2014, 5, 435–441. [Google Scholar]
- Arriagada, O.; Cacciuttolo, F.; Cabeza, R.A.; Carrasco, B.; Schwember, A.R. A Comprehensive Review on Chickpea (Cicer arietinum L.) Breeding for Abiotic Stress Tolerance and Climate Change Resilience. Int. J. Mol. Sci. 2022, 23, 6794. [Google Scholar] [CrossRef]
- Gangashetty, P.I.; Motagi, B.N.; Pavan, R.; Roodagi, M.B. Breeding Crop Plants for Improved Human Nutrition through Biofortification: Progress and Prospects. In Advances in Plant Breeding Strategies: Agronomic, Abiotic and Biotic Stress Traits; Al-Khayri, J.M., Jain, S.M., Johnson, D.V., Eds.; Springer International Publishing: Cham, Switzerland, 2016; pp. 35–76. ISBN 978-3-319-22518-0. [Google Scholar]
- Raina, A.; Laskar, R.A.; Tantray, Y.R.; Khursheed, S.; Wani, M.R.; Khan, S. Characterization of Induced High Yielding Cowpea Mutant Lines Using Physiological, Biochemical and Molecular Markers. Sci. Rep. 2020, 10, 3687. [Google Scholar] [CrossRef]
- Gaur, P.M.; Jukanti, A.K.; Varshney, R.K. Impact of Genomic Technologies on Chickpea Breeding Strategies. Agronomy 2012, 2, 199–221. [Google Scholar] [CrossRef]
- Devasirvatham, V.; Tan, D.K.Y. Impact of High Temperature and Drought Stresses on Chickpea Production. Agronomy 2018, 8, 145. [Google Scholar] [CrossRef]
- Rosielle, A.A.; Hamblin, J. Theoretical Aspects of Selection for Yield in Stress and Non-Stress Environment1. Crop Sci. 1981, 21, 943–946. [Google Scholar] [CrossRef]
- Fischer, R.A.; Maurer, R. Drought Resistance in Spring Wheat Cultivars. I. Grain Yield Responses. Aust. J. Agric. Res. 1978, 29, 897–912. [Google Scholar] [CrossRef]
- Fernandez, G.C. Effective Selection Criteria for Assessing Plant Stress Tolerance. In Proceedings of the International Symposium on Adaptation of Vegetables and Other Food Crops in Temperature and Water Stress, Shanhua, Taiwan, 13–16 August 1992; pp. 257–270. [Google Scholar]
- Gavuzzi, P.; Rizza, F.; Palumbo, M.; Campanile, R.G.; Ricciardi, G.L.; Borghi, B. Evaluation of Field and Laboratory Predictors of Drought and Heat Tolerance in Winter Cereals. Can. J. Plant Sci. 1997, 77, 523–531. [Google Scholar] [CrossRef]
- LIN, C.S.; BINNS, M.R. A Superiority Measure of Cultivar Performance for Cultivar × Location Data. Can. J. Plant Sci. 1988, 68, 193–198. [Google Scholar] [CrossRef]
- Kashiwagi, J.; Krishnamurthy, L.; Gaur, P.M.; Chandra, S.; Upadhyaya, H.D. Estimation of Gene Effects of the Drought Avoidance Root Characteristics in Chickpea (C. arietinum L.). Field Crops Res. 2008, 105, 64–69. [Google Scholar] [CrossRef]
- Kumar, J.; Rao, B.V. Super-Early Chickpea Developed at ICRISAT Asia Center. Int. Chickpea Newsl. 1996, 3, 17–18. [Google Scholar]
- Serraj, R.; Krishnamurthy, L.; Kashiwagi, J.; Kumar, J.; Chandra, S.; Crouch, J.H. Variation in Root Traits of Chickpea (Cicer arietinum L.) Grown under Terminal Drought. Field Crops Res. 2004, 88, 115–127. [Google Scholar] [CrossRef]
- Saxena, N.P. Management of Drought in Chickpea—A Holistic Approach. In Management of Agricultural Drought: Agronomic and Genetic Options; Cambridge University Press: Cambridge, MA, USA, 2003; pp. 103–122. [Google Scholar]
- Kashiwagi, J.; Krishnamurthy, L.; Gaur, P.M.; Upadhyaya, H.D.; Varshney, R.K.; Tobita, S. Traits of Relevance to Improve Yield under Terminal Drought Stress in Chickpea (C. arietinum L.). Field Crops Res. 2013, 145, 88–95. [Google Scholar] [CrossRef]
- Anonymous QRT Report 2010-11 to 2014-15; AICRP (SKNAU Jobner): Jaipur, India, 2016.
- Singh, K.B.; Omar, M.; Saxena, M.C.; Johansen, C. Screening for Drought Resistance in Spring Chickpea in the Mediterranean Region. J. Agron. Crop Sci. 1997, 178, 227–235. [Google Scholar] [CrossRef]
- Gupta, S.C.; Rathore, A.K.; Sharma, S.N.; Saini, R.S. Response of Chickpea Cultivars to Water Stress. Indian J. Plant Physiol. 2000, 5, 274–276. [Google Scholar]
- Farooq, M.; Hussain, M.; Siddique, K.H. Drought Stress in Wheat during Flowering and Grain-Filling Periods. Crit. Rev. Plant Sci. 2014, 33, 331–349. [Google Scholar] [CrossRef]
- Beebe, S.E.; Rao, I.M.; Cajiao, C.; Grajales, M. Selection for Drought Resistance in Common Bean Also Improves Yield in Phosphorus Limited and Favorable Environments. Crop Sci. 2008, 48, 582–592. [Google Scholar] [CrossRef]
- Torres, A.M.; Avila, C.M.; Gutierrez, N.; Palomino, C.; Moreno, M.T.; Cubero, J.I. Marker-Assisted Selection in Faba Bean (Vicia Faba L.). Field Crops Res. 2010, 115, 243–252. [Google Scholar] [CrossRef]
- Jha, U.C.; Nayyar, H.; Jha, R.; Nath, C.P.; Datta, D. Chickpea Breeding for Abiotic Stress: Breeding Tools and ‘Omics’ Approaches for Enhancing Genetic Gain. In Accelerated Plant Breeding, Volume 3: Food Legumes; Springer International Publishing: Cham, Switzerland, 2020; pp. 211–234. [Google Scholar]
- Venuprasad, R.; Lafitte, H.R.; Atlin, G.N. Response to Direct Selection for Grain Yield under Drought Stress in Rice. Crop Sci. 2007, 47, 285–293. [Google Scholar] [CrossRef]
- Fischer, R.A.; Byerlee, D.; Edmeades, G. Crop Yields and Global Food Security; ACIAR: Canberra, ACT, Australia, 2014; pp. 8–11. [Google Scholar]
- Varshney, R.K.; Thudi, M.; Nayak, S.N.; Gaur, P.M.; Kashiwagi, J.; Krishnamurthy, L.; Jaganathan, D.; Koppolu, J.; Bohra, A.; Tripathi, S. Genetic Dissection of Drought Tolerance in Chickpea (Cicer arietinum L.). Theor. Appl. Genet. 2014, 127, 445–462. [Google Scholar] [CrossRef]
- Krishnamurthy, L.; Kashiwagi, J.; Gaur, P.M.; Upadhyaya, H.D.; Vadez, V. Sources of Tolerance to Terminal Drought in the Chickpea (Cicer arietinum L.) Minicore Germplasm. Field Crops Res. 2010, 119, 322–330. [Google Scholar] [CrossRef]
- Acquaah, G. Conventional Plant Breeding Principles and Techniques. In Advances in Plant Breeding Strategies: Breeding, Biotechnology and Molecular Tools; Springer International Publishing: Cham, Switzerland, 2015; pp. 115–158. [Google Scholar]
- Watson, A.; Ghosh, S.; Williams, M.J.; Cuddy, W.S.; Simmonds, J.; Rey, M.-D.; Asyraf Md Hatta, M.; Hinchliffe, A.; Steed, A.; Reynolds, D.; et al. Speed Breeding Is a Powerful Tool to Accelerate Crop Research and Breeding. Nat. Plants 2018, 4, 23–29. [Google Scholar] [CrossRef] [PubMed]
- Krishnamurthy, L.; Gaur, P.M.; Basu, P.S.; Chaturvedi, S.K.; Tripathi, S.; Vadez, V.; Rathore, A.; Varshney, R.K.; Gowda, C.L.L. Large Genetic Variation for Heat Tolerance in the Reference Collection of Chickpea (Cicer arietinum L.) Germplasm. Plant Genet. Resour. 2011, 9, 59–69. [Google Scholar] [CrossRef]
- Varshney, R.K.; Thudi, M.; Roorkiwal, M.; He, W.; Upadhyaya, H.D.; Yang, W.; Bajaj, P.; Cubry, P.; Rathore, A.; Jian, J. Resequencing of 429 Chickpea Accessions from 45 Countries Provides Insights into Genome Diversity, Domestication and Agronomic Traits. Nat. Genet. 2019, 51, 857–864. [Google Scholar] [CrossRef]
- Toker, C. Mutagenesis for Resistance to Abiotic Stresses: Chickpea as Model Crop. In Mutagenesis: Exploring Novel Genes and Pathways; Wageningen Academic Publishers: Wageningen, The Netherlands, 2014; pp. 78–81. [Google Scholar]
- Rick, C.W. Plant Germplasm Resources. Handb. Plant Cell Cult. 1984, 2, 9–37. [Google Scholar]
- Maliro, M.F.A.; McNeil, D.; Redden, B.; Kollmorgen, J.F.; Pittock, C. Sampling Strategies and Screening of Chickpea (Cicer arietinum L.) Germplasm for Salt Tolerance. Genet. Resour. Crop Evol. 2008, 55, 53–63. [Google Scholar] [CrossRef]
- Warschefsky, E.; Penmetsa, R.V.; Cook, D.R.; Von Wettberg, E.J. Back to the Wilds: Tapping Evolutionary Adaptations for Resilient Crops through Systematic Hybridization with Crop Wild Relatives. Am. J. Bot. 2014, 101, 1791–1800. [Google Scholar] [CrossRef] [PubMed]
- Tekin, M.; Sari, D.; Catal, M.; Ikten, C.; Smykal, P.; Penmetsa, R.V.; Von Wettberg, E.J.; Toker, C. Eco-Geographic Distribution of Cicer Isauricum PH Davis and Threats to the Species. Genet. Resour. Crop Evol. 2018, 65, 67–77. [Google Scholar] [CrossRef]
- Sari, D.; Sari, H.; Eker, T.; Ikten, C.; Uzun, B.; Toker, C. Intraspecific versus Interspecific Crosses for Superior Progeny in Cicer Species. Crop Sci. 2022, 62, 2122–2137. [Google Scholar] [CrossRef]
- Gaur, P.M.; Chaturvedi, S.K.; Tripathi, S.; Gowda, C.L.; Krishnamurthy, L.; Vadez, V.; Mallikarjuna, N.; Varshney, R.K. Improving Heat Tolerance in Chickpea to Increase Its Resilience to Climate Change. In Proceedings of the 5th International Food Legumes Research Conference & 7th European Conference on Grain Legumes, Antalya, Turkey, 26–30 April 2010. [Google Scholar]
- Mallikarjuna, N.; Muehlbauer, F.J. Chickpea Hybridization Using In Vitro Techniques. In Plant Embryo Culture: Methods and Protocols; Springer International Publishing: Cham, Switzerland, 2011; pp. 93–105. [Google Scholar]
- Verma, M.M.; Sandhu, J.S.; Rraar, H.S.; Brar, J.S. Crossability Studies in Different Species of Cicer (L.). Crop Improv. 1990, 17, 179–181. [Google Scholar]
- Ahmad, F.; Slinkard, A.E. Genetic Relationships in the Genus Cicer L. as Revealed by Polyacrylamide Gel Electrophoresis of Seed Storage Proteins. Theor. Appl. Genet. 1992, 84, 688–692. [Google Scholar] [CrossRef]
- Badami, P.S.; Mallikarjuna, N.; Moss, J.P. Interspecific Hybridization between Cicer arietinum and C. pinnatifidum. Plant Breed. 1997, 116, 393–395. [Google Scholar] [CrossRef]
- Mallikarjuna, N.; Sharma, H.C.; Upadhyaya, H.D. Exploitation of Wild Relatives of Pigeonpea and Chickpea for Resistance to Helicoverpa armigera. J. SAT Agric. Res. 2007, 3, 4. [Google Scholar]
- Manickavelu, A.; Nadarajan, N.; Ganesh, S.K.; Gnanamalar, R.P.; Chandra Babu, R. Drought Tolerance in Rice: Morphological and Molecular Genetic Consideration. Plant Growth Regul. 2006, 50, 121–138. [Google Scholar] [CrossRef]
- Chamarthi, S.; Kumar, A.; Vuong, T.; Blair, M.; Gaur, P.; Nguyen, H.; Varshney, R. Trait Mapping and Molecular Breeding. In Biology and Breeding of Food Legumes; CABI: Wallingford, UK, 2011. [Google Scholar]
- Myles, S.; Peiffer, J.; Brown, P.J.; Ersoz, E.S.; Zhang, Z.; Costich, D.E.; Buckler, E.S. Association Mapping: Critical Considerations Shift from Genotyping to Experimental Design. Plant Cell 2009, 21, 2194–2202. [Google Scholar] [CrossRef] [PubMed]
- Rafalski, J.A. Association Genetics in Crop Improvement. Curr. Opin. Plant Biol. 2010, 13, 174–180. [Google Scholar] [CrossRef]
- Thudi, M.; Upadhyaya, H.D.; Rathore, A.; Gaur, P.M.; Krishnamurthy, L.; Roorkiwal, M.; Nayak, S.N.; Chaturvedi, S.K.; Basu, P.S.; Gangarao, N. Genetic Dissection of Drought and Heat Tolerance in Chickpea through Genome-Wide and Candidate Gene-Based Association Mapping Approaches. PLoS ONE 2014, 9, e96758. [Google Scholar] [CrossRef]
- Roorkiwal, M.; Bharadwaj, C.; Barmukh, R.; Dixit, G.P.; Thudi, M.; Gaur, P.M.; Chaturvedi, S.K.; Fikre, A.; Hamwieh, A.; Kumar, S. Integrating Genomics for Chickpea Improvement: Achievements and Opportunities. Theor. Appl. Genet. 2020, 133, 1703–1720. [Google Scholar] [CrossRef] [PubMed]
- Bharadwaj, C.; Tripathi, S.; Soren, K.R.; Thudi, M.; Singh, R.K.; Sheoran, S.; Roorkiwal, M.; Patil, B.S.; Chitikineni, A.; Palakurthi, R. Introgression of “QTL-Hotspot” Region Enhances Drought Tolerance and Grain Yield in Three Elite Chickpea Cultivars. Plant Genome 2021, 14, e20076. [Google Scholar] [CrossRef]
- Reynolds, M.; Chapman, S.; Crespo-Herrera, L.; Molero, G.; Mondal, S.; Pequeno, D.N.; Pinto, F.; Pinera-Chavez, F.J.; Poland, J.; Rivera-Amado, C. Breeder Friendly Phenotyping. Plant Sci. 2020, 295, 110396. [Google Scholar] [CrossRef] [PubMed]
- Choudhary, P.; Khanna, S.M.; Jain, P.K.; Bharadwaj, C.; Kumar, J.; Lakhera, P.C.; Srinivasan, R. Genetic Structure and Diversity Analysis of the Primary Gene pool of Chickpea Using SSR Markers. Genet. Mol. Res. 2012, 11, 891–905. [Google Scholar] [CrossRef]
- Hiremath, P.J.; Farmer, A.; Cannon, S.B.; Woodward, J.; Kudapa, H.; Tuteja, R.; Kumar, A.; BhanuPrakash, A.; Mulaosmanovic, B.; Gujaria, N. Large-Scale Transcriptome Analysis in Chickpea (Cicer arietinum L.), an Orphan Legume Crop of the Semi-Arid Tropics of Asia and Africa. Plant Biotechnol. J. 2011, 9, 922–931. [Google Scholar] [CrossRef]
- Vadez, V.; Krishnamurthy, L.; Thudi, M.; Anuradha, C.; Colmer, T.D.; Turner, N.C.; Siddique, K.H.; Gaur, P.M.; Varshney, R.K. Assessment of ICCV 2$\times$ JG 62 Chickpea Progenies Shows Sensitivity of Reproduction to Salt Stress and Reveals QTL for Seed Yield and Yield Components. Mol. Breed. 2012, 30, 9–21. [Google Scholar] [CrossRef]
- Paul, P.J.; Samineni, S.; Sajja, S.B.; Rathore, A.; Das, R.R.; Chaturvedi, S.K.; Lavanya, G.R.; Varshney, R.K.; Gaur, P.M. Capturing Genetic Variability and Selection of Traits for Heat Tolerance in a Chickpea Recombinant Inbred Line (RIL) Population under Field Conditions. Euphytica 2018, 214, 27. [Google Scholar] [CrossRef]
- Jha, U.C.; Nayyar, H.; Palakurthi, R.; Jha, R.; Valluri, V.; Bajaj, P.; Chitikineni, A.; Singh, N.P.; Varshney, R.K.; Thudi, M. Major QTLs and Potential Candidate Genes for Heat Stress Tolerance Identified in Chickpea (Cicer arietinum L.). Front. Plant Sci. 2021, 12, 655103. [Google Scholar] [CrossRef]
- Jha, U.C.; Jha, R.; Singh, N.P.; Shil, S.; Kole, P.C. Heat Tolerance Indices and Their Role in Selection of Heat Stress Tolerant Chickpea (Cicer arietinum) Genotypes. Indian J. Agric. Sci. 2018, 88, 260–263. [Google Scholar] [CrossRef]
- Reddy, D.S.; Bhatnagar-Mathur, P.; Vadez, V.; Sharma, K.K. Grain Legumes (Soybean, Chickpea, and Peanut): Omics Approaches to Enhance Abiotic Stress Tolerance. In Improving Crop Resistance to Abiotic Stress; Wiley: Hoboken, NJ, USA, 2012; pp. 995–1032. [Google Scholar]
- Jaganathan, D.; Thudi, M.; Kale, S.; Azam, S.; Roorkiwal, M.; Gaur, P.M.; Kishor, P.K.; Nguyen, H.; Sutton, T.; Varshney, R.K. Genotyping-by-Sequencing Based Intra-Specific Genetic Map Refines a ‘‘QTL-Hotspot” Region for Drought Tolerance in Chickpea. Mol. Genet. Genom. 2015, 290, 559–571. [Google Scholar] [CrossRef] [PubMed]
- Garg, R.; Shankar, R.; Thakkar, B.; Kudapa, H.; Krishnamurthy, L.; Mantri, N.; Varshney, R.K.; Bhatia, S.; Jain, M. Transcriptome Analyses Reveal Genotype-and Developmental Stage-Specific Molecular Responses to Drought and Salinity Stresses in Chickpea. Sci. Rep. 2016, 6, 19228. [Google Scholar] [CrossRef]
- Badhan, S.; Kole, P.; Ball, A.; Mantri, N. RNA Sequencing of Leaf Tissues from Two Contrasting Chickpea Genotypes Reveals Mechanisms for Drought Tolerance. Plant Physiol. Biochem. 2018, 129, 295–304. [Google Scholar] [CrossRef] [PubMed]
- Thudi, M.; Bohra, A.; Nayak, S.N.; Varghese, N.; Shah, T.M.; Penmetsa, R.V.; Thirunavukkarasu, N.; Gudipati, S.; Gaur, P.M.; Kulwal, P.L. Novel SSR Markers from BAC-End Sequences, DArT Arrays and a Comprehensive Genetic Map with 1,291 Marker Loci for Chickpea (Cicer arietinum L.). PLoS ONE 2011, 6, e27275. [Google Scholar] [CrossRef]
- Molina, C.; Rotter, B.; Horres, R.; Udupa, S.M.; Besser, B.; Bellarmino, L.; Baum, M.; Matsumura, H.; Terauchi, R.; Kahl, G. SuperSAGE: The Drought Stress-Responsive Transcriptome of Chickpea Roots. BMC Genom. 2008, 9, 553. [Google Scholar] [CrossRef] [PubMed]
- Deokar, A.A.; Kondawar, V.; Jain, P.K.; Karuppayil, S.M.; Raju, N.L.; Vadez, V.; Varshney, R.K.; Srinivasan, R. Comparative Analysis of Expressed Sequence Tags (ESTs) between Drought-Tolerant and-Susceptible Genotypes of Chickpea under Terminal Drought Stress. BMC Plant Biol. 2011, 11, 70. [Google Scholar] [CrossRef]
- Mantri, N.L.; Ford, R.; Coram, T.E.; Pang, E.C. Transcriptional Profiling of Chickpea Genes Differentially Regulated in Response to High-Salinity, Cold and Drought. BMC Genom. 2007, 8, 303. [Google Scholar] [CrossRef] [PubMed]
- Molina, C.; Zaman-Allah, M.; Khan, F.; Fatnassi, N.; Horres, R.; Rotter, B.; Steinhauer, D.; Amenc, L.; Drevon, J.-J.; Winter, P. The Salt-Responsive Transcriptome of Chickpea Roots and Nodules via DeepSuperSAGE. BMC Plant Biol. 2011, 11, 31. [Google Scholar] [CrossRef]
- Varshney, R.K.; Hiremath, P.J.; Lekha, P.; Kashiwagi, J.; Balaji, J.; Deokar, A.A.; Vadez, V.; Xiao, Y.; Srinivasan, R.; Gaur, P.M. A Comprehensive Resource of Drought-and Salinity-Responsive ESTs for Gene Discovery and Marker Development in Chickpea (Cicer arietinum L.). BMC Genom. 2009, 10, 523. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Liu, Y.; Jia, Y.; Gu, H.; Ma, H.; Yu, T.; Zhang, H.; Chen, Q.; Ma, L.; Gu, A. Transcriptional Responses to Drought Stress in Root and Leaf of Chickpea Seedling. Mol. Biol. Rep. 2012, 39, 8147–8158. [Google Scholar] [CrossRef] [PubMed]
- Mahdavi Mashaki, K.; Garg, V.; Nasrollahnezhad Ghomi, A.A.; Kudapa, H.; Chitikineni, A.; Zaynali Nezhad, K.; Thudi, M. RNA-Seq analysis revealed genes associated with drought stress response in kabuli chickpea (Cicer arietinum L.). PLoS ONE 2018, 13, e0199774. [Google Scholar] [CrossRef]
- Kumar, M.; Chauhan, A.S.; Kumar, M.; Yusuf, M.A.; Sanyal, I.; Chauhan, P.S. Transcriptome Sequencing of Chickpea (Cicer arietinum L.) Genotypes for Identification of Drought-Responsive Genes under Drought Stress Condition. Plant Mol. Biol. Rep. 2019, 37, 186–203. [Google Scholar] [CrossRef]
- Kudapa, H.; Agarwal, G.; Chitikineni, A.; Gaur, P.M.; Krishnamurthy, L.; Varshney, R.K. Mining for Heat Stress Responsive Genes by RNA-Seq Based Comprehensive Gene Expression Analyses in Chickpea (Cicer arietinum L.). In Proceedings of the InterDrought-V 2017, Hyderabad, India, 21–25 February 2017. [Google Scholar]
- Jones-Rhoades, M.W.; Bartel, D.P.; Bartel, B. MicroRNAs and Their Regulatory Roles in Plants. Annu. Rev. Plant Biol. 2006, 57, 19–53. [Google Scholar] [CrossRef]
- Hajyzadeh, M.; Turktas, M.; Khawar, K.M.; Unver, T. MiR408 Overexpression Causes Increased Drought Tolerance in Chickpea. Gene 2015, 555, 186–193. [Google Scholar] [CrossRef]
- Dasmandal, T.; Rao, A.R.; Sahu, S. Identification and Characterization of Circular RNAs Regulating Genes Responsible for Drought Stress Tolerance in Chickpea and Soybean. Indian J. Genet. Plant Breed. 2020, 80, 1–8. [Google Scholar] [CrossRef]
- Pawłowski, T.A. Proteome Analysis of Norway Maple (Acer platanoides L.) Seeds Dormancy Breaking and Germination: Influence of Abscisic and Gibberellic Acids. BMC Plant Biol. 2009, 9, 48. [Google Scholar] [CrossRef]
- Jaiswal, D.K.; Mishra, P.; Subba, P.; Rathi, D.; Chakraborty, S.; Chakraborty, N. Membrane-Associated Proteomics of Chickpea Identifies Sad1/UNC-84 Protein (CaSUN1), a Novel Component of Dehydration Signaling. Sci. Rep. 2014, 4, 4177. [Google Scholar] [CrossRef]
- Faghani, E.; Gharechahi, J.; Komatsu, S.; Mirzaei, M.; Khavarinejad, R.A.; Najafi, F.; Farsad, L.K.; Salekdeh, G.H. Comparative Physiology and Proteomic Analysis of Two Wheat Genotypes Contrasting in Drought Tolerance. J. Proteom. 2015, 114, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Pandey, A.; Chakraborty, S.; Datta, A.; Chakraborty, N. Proteomics Approach to Identify Dehydration Responsive Nuclear Proteins from Chickpea (Cicer arietinum L.). Mol. Cell. Proteom. 2008, 7, 88–107. [Google Scholar] [CrossRef]
- Kudapa, H.; Garg, V.; Chitikineni, A.; Varshney, R.K. The RNA-Seq-based high resolution gene expression atlas of chickpea (Cicer arietinum L.) reveals dynamic spatio-temporal changes associated with growth and development. Plant Cell Environ. 2018, 41, 2209–2225. [Google Scholar] [CrossRef] [PubMed]
- Çevik, S.; Akpinar, G.; Yildizli, A.; Kasap, M.; Karaosmanoğlu, K.; Ünyayar, S. Comparative Physiological and Leaf Proteome Analysis between Drought-Tolerant Chickpea Cicer reticulatum and Drought-Sensitive Chickpea C. arietinum. J. Biosci. 2019, 44, 1–13. [Google Scholar] [CrossRef]
- Gayen, D.; Gayali, S.; Barua, P.; Lande, N.V.; Varshney, S.; Sengupta, S.; Chakraborty, S.; Chakraborty, N. Dehydration-Induced Proteomic Landscape of Mitochondria in Chickpea Reveals Large-Scale Coordination of Key Biological Processes. J. Proteom. 2019, 192, 267–279. [Google Scholar] [CrossRef]
- Pinheiro, C.; Chaves, M.M. Photosynthesis and Drought: Can We Make Metabolic Connections from Available Data? J. Exp. Bot. 2011, 62, 869–882. [Google Scholar] [CrossRef] [PubMed]
- Garg, R.; Bhattacharjee, A.; Jain, M. Genome-Scale Transcriptomic Insights into Molecular Aspects of Abiotic Stress Responses in Chickpea. Plant Mol. Biol. Rep. 2015, 33, 388–400. [Google Scholar] [CrossRef]
- Khan, N.; Bano, A.; Rahman, M.A.; Rathinasabapathi, B.; Babar, M.A. UPLC-HRMS-Based Untargeted Metabolic Profiling Reveals Changes in Chickpea (Cicer arietinum) Metabolome Following Long-Term Drought Stress. Plant Cell Environ. 2019, 42, 115–132. [Google Scholar] [CrossRef]
- Kesari, R.; Lasky, J.R.; Villamor, J.G.; Des Marais, D.L.; Chen, Y.-J.C.; Liu, T.-W.; Lin, W.; Juenger, T.E.; Verslues, P.E. Intron-Mediated Alternative Splicing of Arabidopsis P5CS1 and Its Association with Natural Variation in Proline and Climate Adaptation. Proc. Natl. Acad. Sci. USA 2012, 109, 9197–9202. [Google Scholar] [CrossRef] [PubMed]
- Muscolo, A.; Junker, A.; Klukas, C.; Weigelt-Fischer, K.; Riewe, D.; Altmann, T. Phenotypic and Metabolic Responses to Drought and Salinity of Four Contrasting Lentil Accessions. J. Exp. Bot. 2015, 66, 5467–5480. [Google Scholar] [CrossRef] [PubMed]
- Ullah, N.; Yüce, M.; Neslihan Öztürk Gökçe, Z.; Budak, H. Comparative Metabolite Profiling of Drought Stress in Roots and Leaves of Seven Triticeae Species. BMC Genom. 2017, 18, 969. [Google Scholar] [CrossRef] [PubMed]
- Kotak, S.; Larkindale, J.; Lee, U.; von Koskull-Döring, P.; Vierling, E.; Scharf, K.-D. Complexity of the Heat Stress Response in Plants. Curr. Opin. Plant Biol. 2007, 10, 310–316. [Google Scholar] [CrossRef] [PubMed]
- Yadav, R.; Juneja, S.; Kumar, S. Cross Priming with Drought Improves Heat-Tolerance in Chickpea (Cicer arietinum L.) by Stimulating Small Heat Shock Proteins and Antioxidative Defense. Environ. Sustain. 2021, 4, 171–182. [Google Scholar] [CrossRef]
- Bhogireddy, S.; Kudapa, H.; Bajaj, P.; Garg, V.; Chitikineni, A.; Nayak, S.; Varshney, R.K. Transcriptome Analysis of Chickpea during Heat Stress Unveils the Signatures of Long Intergenic Non-Coding RNAs (LincRNAs) and MRNAs in the Heat-QTL Region. Crop Des. 2023, 2, 100026. [Google Scholar] [CrossRef]
- Kudapa, H.; Barmukh, R.; Garg, V.; Chitikineni, A.; Samineni, S.; Agarwal, G.; Varshney, R.K. Comprehensive Transcriptome Profiling Uncovers Molecular Mechanisms and Potential Candidate Genes Associated with Heat Stress Response in Chickpea. Int. J. Mol. Sci. 2023, 24, 1369. [Google Scholar] [CrossRef]
- Parankusam, S.; Bhatnagar-Mathur, P.; Sharma, K.K. Heat Responsive Proteome Changes Reveal Molecular Mechanisms Underlying Heat Tolerance in Chickpea. Environ. Exp. Bot. 2017, 141, 132–144. [Google Scholar] [CrossRef]
- Makonya, G.M.; Ogola, J.B.; Gabier, H.; Rafudeen, M.S.; Muasya, A.M.; Crespo, O.; Maseko, S.; Valentine, A.J.; Ottosen, C.-O.; Rosenqvist, E. Proteome Changes and Associated Physiological Roles in Chickpea (Cicer arietinum) Tolerance to Heat Stress under Field Conditions. Funct. Plant Biol. 2021, 49, 13–24. [Google Scholar] [CrossRef]
- Kishore, B.; Hong, Z.; Miao, G.; Hu, C.A.A.A.; Verma, D. Over Expression of [Delta]-Pyrroline-5-Carboxylate Synthetase Increases Proline Production and Confers Osmotolerance in Transgenic Plants. Plant Physiol 1995, 108, 1387–1394. [Google Scholar] [CrossRef]
- Xu, D.; Duan, X.; Wang, B.; Hong, B.; Ho, T.-H.D.; Wu, R. Expression of a Late Embryogenesis Abundant Protein Gene, HVA1, from Barley Confers Tolerance to Water Deficit and Salt Stress in Transgenic Rice. Plant Physiol. 1996, 110, 249–257. [Google Scholar] [CrossRef]
- McKersie, B.D.; Bowley, S.R.; Harjanto, E.; Leprince, O. Water-Deficit Tolerance and Field Performance of Transgenic Alfalfa Overexpressing Superoxide Dismutase. Plant Physiol. 1996, 111, 1177–1181. [Google Scholar] [CrossRef] [PubMed]
- Ishizaki-Nishizawa, O.; Fujii, T.; Azuma, M.; Sekiguchi, K.; Murata, N.; Ohtani, T.; Toguri, T. Low-Temperature Resistance of Higher Plants Is Significantly Enhanced by a Nonspecific Cyanobacterial Desaturase. Nat. Biotechnol. 1996, 14, 1003–1006. [Google Scholar] [CrossRef]
- Nakashima, K.; Tran, L.-S.P.; Van Nguyen, D.; Fujita, M.; Maruyama, K.; Todaka, D.; Ito, Y.; Hayashi, N.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Functional Analysis of a NAC-Type Transcription Factor OsNAC6 Involved in Abiotic and Biotic Stress-Responsive Gene Expression in Rice. Plant J. 2007, 51, 617–630. [Google Scholar] [CrossRef]
- Hu, H.; You, J.; Fang, Y.; Zhu, X.; Qi, Z.; Xiong, L. Characterization of Transcription Factor Gene SNAC2 Conferring Cold and Salt Tolerance in Rice. Plant Mol. Biol. 2008, 67, 169–181. [Google Scholar] [CrossRef]
- Sharmila, P.; Phanindra, M.L.V.; Anwar, F.; Singh, K.; Gupta, S.; Saradhi, P.P. Targeting Prokaryotic Choline Oxidase into Chloroplasts Enhance the Potential of Photosynthetic Machinery of Plants to Withstand Oxidative Damage. Plant Physiol. Biochem. 2009, 47, 391–396. [Google Scholar] [CrossRef]
- Liu, Q.; Kasuga, M.; Sakuma, Y.; Abe, H.; Miura, S.; Yamaguchi-Shinozaki, K.; Shinozaki, K. Two transcription factors, DREB1 and DREB2, with anEREBP/AP2 DNA binding domain separate two cellular signal transduction pathways in drought- and lowtemperature-responsive gene expression, respectively, in Arabidopsis. Plant Cell. 1998, 10, 1391–1406. [Google Scholar] [CrossRef] [PubMed]
- Azeem, F.; Ahmad, B.; Atif, R.M.; Ali, M.A.; Nadeem, H.; Hussain, S.; Manzoor, H.; Azeem, M.; Afzal, M. Genome-Wide Analysis of Potassium Transport-Related Genes in Chickpea (Cicer arietinum L.) and Their Role in Abiotic Stress Responses. Plant Mol. Biol. Rep. 2018, 36, 451–468. [Google Scholar] [CrossRef]
- Cho, S.; Kumar, J.; Shultz, J.L.; Anupama, K.; Tefera, F.; Muehlbauer, F.J. Mapping Genes for Double Podding and Other Morphological Traits in Chickpea. Euphytica 2002, 128, 285–292. [Google Scholar] [CrossRef]
- Bhatnagar-Mathur, P.; Vadez, V.; Jyostna Devi, M.; Lavanya, M.; Vani, G.; Sharma, K.K. Genetic Engineering of Chickpea (Cicer arietinum L.) with the P5CSF129A Gene for Osmoregulation with Implications on Drought Tolerance. Mol. Breed. 2009, 23, 591–606. [Google Scholar] [CrossRef]
- Peng, H.; Cheng, H.-Y.; Chen, C.; Yu, X.-W.; Yang, J.-N.; Gao, W.-R.; Shi, Q.-H.; Zhang, H.; Li, J.-G.; Ma, H. A NAC Transcription Factor Gene of Chickpea (Cicer arietinum), CarNAC3, Is Involved in Drought Stress Response and Various Developmental Processes. J. Plant Physiol. 2009, 166, 1934–1945. [Google Scholar] [CrossRef]
- Movahedi, A.; Zhang, J.; Gao, P.; Yang, Y.; Wang, L.; Yin, T.; Kadkhodaei, S.; Ebrahimi, M.; Zhuge, Q. Expression of the Chickpea CarNAC3 Gene Enhances Salinity and Drought Tolerance in Transgenic Poplars. Plant Cell Tissue Organ Cult. PCTOC 2015, 120, 141–154. [Google Scholar] [CrossRef]
- Nguyen, K.H.; Ha, C.V.; Watanabe, Y.; Tran, U.T.; Nasr Esfahani, M.; Nguyen, D.V.; Tran, L.-S.P. Correlation between Differential Drought Tolerability of Two Contrasting Drought-Responsive Chickpea Cultivars and Differential Expression of a Subset of CaNAC Genes under Normal and Dehydration Conditions. Front. Plant Sci. 2015, 6, 449. [Google Scholar] [CrossRef]
- Wardhan, V.; Jahan, K.; Gupta, S.; Chennareddy, S.; Datta, A.; Chakraborty, S.; Chakraborty, N. Overexpression of CaTLP1, a Putative Transcription Factor in Chickpea (Cicer arietinum L.), Promotes Stress Tolerance. Plant Mol. Biol. 2012, 79, 479–493. [Google Scholar] [CrossRef] [PubMed]
- Anbazhagan, K.; Bhatnagar-Mathur, P.; Vadez, V.; Dumbala, S.R.; Kishor, P.K.; Sharma, K.K. DREB1A Overexpression in Transgenic Chickpea Alters Key Traits Influencing Plant Water Budget across Water Regimes. Plant Cell Rep. 2015, 34, 199–210. [Google Scholar] [CrossRef]
- Joshi-Saha, A.; Reddy, K.S. Repeat Length Variation in the 5ʹUTR of Myo-Inositol Monophosphatase Gene Is Related to Phytic Acid Content and Contributes to Drought Tolerance in Chickpea (Cicer arietinum L.). J. Exp. Bot. 2015, 66, 5683–5690. [Google Scholar] [CrossRef]
- Gupta, S.; Garg, V.; Kant, C.; Bhatia, S. Genome-Wide Survey and Expression Analysis of F-Box Genes in Chickpea. BMC Genom. 2015, 16, 67. [Google Scholar] [CrossRef] [PubMed]
- Meena, M.K.; Ghawana, S.; Dwivedi, V.; Roy, A.; Chattopadhyay, D. Expression of Chickpea CIPK25 Enhances Root Growth and Tolerance to Dehydration and Salt Stress in Transgenic Tobacco. Front. Plant Sci. 2015, 6, 683. [Google Scholar] [CrossRef]
- Ramalingam, A.; Kudapa, H.; Pazhamala, L.T.; Garg, V.; Varshney, R.K. Gene Expression and Yeast Two-Hybrid Studies of 1R-MYB Transcription Factor Mediating Drought Stress Response in Chickpea (Cicer arietinum L.). Front. Plant Sci. 2015, 6, 1117. [Google Scholar] [CrossRef]
- Deokar, A.A.; Kondawar, V.; Kohli, D.; Aslam, M.; Jain, P.K.; Karuppayil, S.M.; Varshney, R.K.; Srinivasan, R. The CarERF Genes in Chickpea (Cicer arietinum L.) and the Identification of CarERF116 as Abiotic Stress Responsive Transcription Factor. Funct. Integr. Genom. 2015, 15, 27–46. [Google Scholar] [CrossRef]
- Sachdeva, S.; Bharadwaj, C.; Singh, R.K.; Jain, P.K.; Patil, B.S.; Roorkiwal, M.; Varshney, R. Characterization of ASR Gene and Its Role in Drought Tolerance in Chickpea (Cicer arietinum L.). PLoS ONE 2020, 15, e0234550. [Google Scholar] [CrossRef]
- Khandal, H.; Gupta, S.K.; Dwivedi, V.; Mandal, D.; Sharma, N.K.; Vishwakarma, N.K.; Pal, L.; Choudhary, M.; Francis, A.; Malakar, P. Root-Specific Expression of Chickpea Cytokinin Oxidase/Dehydrogenase 6 Leads to Enhanced Root Growth, Drought Tolerance and Yield without Compromising Nodulation. Plant Biotechnol. J. 2020, 18, 2225–2240. [Google Scholar] [CrossRef]
- Badhan, S.; Ball, A.S.; Mantri, N. First Report of CRISPR/Cas9 Mediated DNA-Free Editing of 4CL and RVE7 Genes in Chickpea Protoplasts. Int. J. Mol. Sci. 2021, 22, 396. [Google Scholar] [CrossRef]
- Meenakshi; Kumar, A.; Kumar, V.; Dubey, A.K.; Narayan, S.; Sawant, S.V.; Pande, V.; Shirke, P.A.; Sanyal, I. CAMTA Transcription Factor Enhances Salinity and Drought Tolerance in Chickpea (Cicer arietinum L.). Plant Cell Tissue Organ Cult. PCTOC 2022, 148, 319–330. [Google Scholar] [CrossRef]
- Varshney, R.K.; Roorkiwal, M.; Sun, S.; Bajaj, P.; Chitikineni, A.; Thudi, M.; Singh, N.P.; Du, X.; Upadhyaya, H.D.; Khan, A.W.; et al. A Chickpea Genetic Variation Map Based on the Sequencing of 3,366 Genomes. Nature 2021, 599, 622–627. [Google Scholar] [CrossRef]
- Gowda, S.J.M.; Radhika, P.; Mhase, L.B.; Jamadagni, B.M.; Gupta, V.S.; Kadoo, N.Y. Mapping of QTLs Governing Agronomic and Yield Traits in Chickpea. J. Appl. Genet. 2011, 52, 9–21. [Google Scholar] [CrossRef] [PubMed]
- Rehman, A.U.; Malhotra, R.S.; Bett, K.; Tar’An, B.; Bueckert, R.; Warkentin, T.D. Mapping QTL Associated with Traits Affecting Grain Yield in Chickpea (Cicer arietinum L.) under Terminal Drought Stress. Crop Sci. 2011, 51, 450–463. [Google Scholar] [CrossRef]
- Varshney, R.K.; Gaur, P.M.; Chamarthi, S.K.; Krishnamurthy, L.; Tripathi, S.; Kashiwagi, J.; Samineni, S.; Singh, V.K.; Thudi, M.; Jaganathan, D. Fast-Track Introgression of “QTL-Hotspot” for Root Traits and Other Drought Tolerance Traits in JG 11, an Elite and Leading Variety of Chickpea. Plant Genome 2013, 6, plantgenome2013.07.0022. [Google Scholar] [CrossRef]
- Hamwieh, A.; Imtiaz, M.; Malhotra, R.S. Multi-Environment QTL Analyses for Drought-Related Traits in a Recombinant Inbred Population of Chickpea (Cicer arientinum L.). Theor. Appl. Genet. 2013, 126, 1025–1038. [Google Scholar] [CrossRef] [PubMed]
- Kale, S.M.; Jaganathan, D.; Ruperao, P.; Chen, C.; Punna, R.; Kudapa, H.; Thudi, M.; Roorkiwal, M.; Katta, M.A.; Doddamani, D. Prioritization of Candidate Genes in “QTL-Hotspot” Region for Drought Tolerance in Chickpea (Cicer arietinum L.). Sci. Rep. 2015, 5, 15296. [Google Scholar] [CrossRef]
- Srivastava, R.; Bajaj, D.; Malik, A.; Singh, M.; Parida, S.K. Transcriptome Landscape of Perennial Wild Cicer Microphyllum Uncovers Functionally Relevant Molecular Tags Regulating Agronomic Traits in Chickpea. Sci. Rep. 2016, 6, 33616. [Google Scholar] [CrossRef]
- Sivasakthi, K.; Thudi, M.; Tharanya, M.; Kale, S.M.; Kholová, J.; Halime, M.H.; Jaganathan, D.; Baddam, R.; Thirunalasundari, T.; Gaur, P.M. Plant Vigour QTLs Co-Map with an Earlier Reported QTL Hotspot for Drought Tolerance while Water Saving QTLs Map in Other Regions of the Chickpea Genome. BMC Plant Biol. 2018, 18, 29. [Google Scholar] [CrossRef] [PubMed]
- Lei, L.; Goltsman, E.; Goodstein, D.; Wu, G.A.; Rokhsar, D.S.; Vogel, J.P. Plant Pan-Genomics Comes of Age. Annu. Rev. Plant Biol. 2021, 72, 411–435. [Google Scholar] [CrossRef] [PubMed]
- Tay Fernandez, C.G.; Nestor, B.J.; Danilevicz, M.F.; Gill, M.; Petereit, J.; Bayer, P.E.; Finnegan, P.M.; Batley, J.; Edwards, D. Pangenomes as a Resource to Accelerate Breeding of Under-Utilised Crop Species. Int. J. Mol. Sci. 2022, 23, 2671. [Google Scholar] [CrossRef] [PubMed]
- Das, A.; Basu, P.S.; Kumar, M.; Ansari, J.; Shukla, A.; Thakur, S.; Singh, P.; Datta, S.; Chaturvedi, S.K.; Sheshshayee, M.S. Transgenic Chickpea (Cicer arietinum L.) Harbouring AtDREB1a Are Physiologically Better Adapted to Water Deficit. BMC Plant Biol. 2021, 21, 39. [Google Scholar] [CrossRef] [PubMed]
- Bhowmik, P.; Konkin, D.; Polowick, P.; Hodgins, C.L.; Subedi, M.; Xiang, D.; Yu, B.; Patterson, N.; Rajagopalan, N.; Babic, V. CRISPR/Cas9 Gene Editing in Legume Crops: Opportunities and Challenges. Legume Sci. 2021, 3, e96. [Google Scholar] [CrossRef]
- Razzaq, M.K.; Akhter, M.; Ahmad, R.M.; Cheema, K.L.; Hina, A.; Karikari, B.; Raza, G.; Xing, G.; Gai, J.; Khurshid, M. CRISPR-Cas9 Based Stress Tolerance: New Hope for Abiotic Stress Tolerance in Chickpea (Cicer arietinum). Mol. Biol. Rep. 2022, 49, 8977–8985. [Google Scholar] [CrossRef]

| Marker Trait | Mechanism | Ref. |
|---|---|---|
| Early flowering and maturity | Escape | [7,90,109,110] |
| Root traits like a prolific root system, length, density, dry weight, diameter, surface area, and volume | Avoidance | [111,112,113,114,115] |
| Carotenoid concentrations and ion accumulation (Na+ and K+) | Tolerance | [116] |
| Water use efficiency | Tolerance | [99,117,118,119] |
| Osmotic adjustment | Tolerance | [118,120] |
| Relative water content | Tolerance | [116] |
| Chlorophyll content | Tolerance | [57,116] |
| Shoot biomass, canopy temperature depression, and leaf area index | Avoidance | [113] |
| Stomatal conductance | Tolerance | [121] |
| ABA regulation | Tolerance | [122] |
| Antioxidant scavenging enzymes, proline, and molecular chaperones | Tolerance | [57,123,124] |
| Tolerant Sources (Donors) | Basis for Tolerance | Ref. |
|---|---|---|
| ICC 4958 | Length, dry weight, and density of root | [140] |
| ICCC 37, ICCV 2, ICCV 10, and ICCV 90629 | Early phenology traits | [48,141] |
| ICC series: 4958, 1356, 3512, 8261, 4872, 15697, and 13523 | Deep root systems and root length density | [142] |
| ICC 10480 and ICC 5680 | Narrow fewer pinnules and smaller leaf areas | [143] |
| H208, RS11, H355, RS10, S26, and G24 Azerbaijan 583 | Deep root, root mass, and root volume | [43] |
| ICC 4958 and ICC 8261 | Deep root, rapid rates of root development and water extraction, and drought adaptive root traits | [91,143] |
| ICC 4958 | Root volume and root length | [43] |
| ACC317 and ACC316 | Early phenological trait | [7] |
| ICC 7571 | Harvest index and DRI | [144] |
| RSG 143-1, RSG 973, and CSJ 73 | Drought susceptibility index, WUE, plant dry matter, and low membrane injury | [145] |
| ICCV 10 and ICC 14778 | Better partitioning and superior yield | [114] |
| ICC 16374B, ICC 9586, ICC 15510, and ICC 867 | Deep rooting | [115] |
| FLIP 87-59 C | Stress yield | [146] |
| RSG 991, RSG 888, and RSG 973 | Root penetration depth, WUE, and plant dry matter | [145] |
| ICC 4958 | Osmotic regulation | [147] |
| IG5844, IG5856, IG5883, IG5867, IG5884, IG5887, IG5894, IG5896, IG5906, IG5908, ILWC 118, ICC 17207, and ILWC 21 | Relative water content (RWC), membrane stability index (MSI), and drought tolerance index | [49] |
| IG5844a, 5856, 5867, 5884, 5887, 5894, 5896, 5906, and 5908 | (MSI) and (RWC) | [7,41] |
| ICC 8261 and ICC 4958 | Root traits | [91] |
| Sources | Genotype | Ref. |
|---|---|---|
| Landraces | ICC 637, ICC 762, ICC 1180, ICC 1205, ICC 2065, ICC 4567, ICC 4958, ICC 8950, ICC 10393, ICC 15618, and ICC 16524 | [114,159] |
| Breeding lines | ICCV 07104, ICCV 07105, ICCV 07108, ICCV 07109, ICCV 07110, ICCV 07115, ICCV 07117, ICCV 07118, and ICCV 98902 | [112,115,159] |
| Elite cultivars | GG 2, ICCV 92069, JG 6, PhuleG 12, Vaibhav, ICCV 89314, Rajas, ICCL 83149, ICCC 37, DigVijay, ICCL 87207, KPG 59, ICCL 83110, ICCL 82108, Pusa 547, and Pusa 391 | [116,159] |
| Wild Cicer relatives | C. anatolicum, C. microphyllum, C. montbretii, C. oxydon, C. reticulatum, and C. songaricum | [7] |
| Stress | Trait | LG | Markers/Locus | Cross/Genotypes | Ref. |
|---|---|---|---|---|---|
| Heat | SY | CaLG05 | Ca5_44667768-Ca5_46955940 | ICC 4567 × ICC 15614 | [183] |
| CC | CaLG06 | CPGR206-H3G031 | DCP 92-3 × ICCV 92944 | [184] | |
| MSI | CaLG05 | NCPGR267 | Desi genotypes | [185] | |
| MSI | CaLG06 | H2L102 | Desi genotypes | [185] | |
| MSI | CaLG07 | TS 53 | Desi genotypes | [185] |
| S.N. | Gene Name | Promoter | Functional Remarks | Reference |
|---|---|---|---|---|
| 1 | P5CSF129A | CaMV 35S | Osmoregulatory gene encoding the mutagenized D1-pyrroline-5-carboxylate synthetase (P5CS) for the over production of proline. | [232] |
| 2 | CarNAC | - | A potential regulatory gene contributing to the differential tissue-specific drought tolerability. | [233,234,235] |
| 3 | CaTLP1 | CaMV 35S | CaTLP1 is upregulated by dehydration, and its stress-responsive function is associated with an ABA-dependent network. | [236] |
| 4 | Dehydration-responsive element-binding protein 1A (DREB1A) and 2A (DREB2A) genes | Atrd29A | rd29A influences DREB1A on mechanisms underlying water uptake, stomata response, transpiration efficiency, and rooting architecture in water-stressed plants. | [201,237] |
| 5 | Rd17, Rd29a, and Rd29b genes | miR408 | DREB1A and DREB2A transcription factors act on Rd17, Rd29a, and Rd29b genes andregulate their expression levels under drought conditions. | [201] |
| 6 | CaIMP | - | Regulating phytic acid levels to confer drought tolerance in natural populations of chickpeas. | [238] |
| 7 | CaRRP1 | - | Secretome analysis reveals dynamic extracellular remodeling that was used to maintain cell structure and biogenesis, in addition to acting in signaling events crucial for cellular homeostasis during stress adaptation. | [239] |
| 8 | CaCIPK25 | - | Gene expression in chickpeas increases upon salt, dehydration, and different hormonal treatments and is involved in root development and abiotic stress tolerance. | [240] |
| 9 | 1R-MYB | - | The 1R-MYB transcription factors play an important role in co-regulating drought tolerance in chickpea roots. | [241] |
| 10 | CarERF116 gene | CaMV 35S | Transcriptional factor CarERF116 differentially expressed and upregulates several stress-related genes involved in resistance to osmotic stress and reduced sensitivity to ABA during seed germination. | [242] |
| 11 | Probable mannitol dehydrogenase, serine hydroxymethyltransferase 4-like, 17.5 kDa class I heat shock protein-like, cytochrome P450 81E8-like, galactinol-sucrose galactosyltransferase-like, xyloglucan endotransglucosylase/hydrolase protein 23, abscisic acid 8′-hydroxylase 1-like, calmodulin-like protein 11, and proline dehydrogenase 2 mitochondrial-like genes | - | The C. arietinum drought-responsive genes (CaDRGs) modulate the expression of transcription factors (TFs) AP2-EREBP, bHLH, bZIP, C3H, MYB, NAC, WRKY, and MADS under simulated drought conditions. | [198] |
| 12 | Abscissic acid and the stress-ripening (ASR) gene | - | Play a role in drought tolerance in chickpeas. | [243] |
| 13 | CaCKX6 | CaWRKY31 | Increased cytokinin oxidases/dehydrogenases (CKX) activity in root and advanced chickpea transgenic lines exhibited a higher root-to-shoot biomass ratio and enhanced long-term drought tolerance. | [244] |
| 14 | 4CL (4-coumarate ligase) and RVE7 (Reveille 7) | - | First report of CRISPR/Cas9-mediated DNA-free editing of 4CL and RVE7 genes for drought tolerance. | [245] |
| 15 | CAMTA (calmodulin-binding transcription activator) gene | - | The CAMTA gene overexpression in response to drought and salinity stress has shown enhanced activities of various antioxidant enzymes (ascorbate peroxidase (APX), catalase (CAT), glutathione S-transferase (GST), superoxide dismutase (SOD), and monodehydroascorbate reductase (MDHAR)). | [246] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jain, S.K.; Wettberg, E.J.v.; Punia, S.S.; Parihar, A.K.; Lamichaney, A.; Kumar, J.; Gupta, D.S.; Ahmad, S.; Pant, N.C.; Dixit, G.P.; et al. Genomic-Mediated Breeding Strategies for Global Warming in Chickpeas (Cicer arietinum L.). Agriculture 2023, 13, 1721. https://doi.org/10.3390/agriculture13091721
Jain SK, Wettberg EJv, Punia SS, Parihar AK, Lamichaney A, Kumar J, Gupta DS, Ahmad S, Pant NC, Dixit GP, et al. Genomic-Mediated Breeding Strategies for Global Warming in Chickpeas (Cicer arietinum L.). Agriculture. 2023; 13(9):1721. https://doi.org/10.3390/agriculture13091721
Chicago/Turabian StyleJain, Shailesh Kumar, Eric J. von Wettberg, Sumer Singh Punia, Ashok Kumar Parihar, Amrit Lamichaney, Jitendra Kumar, Debjyoti Sen Gupta, Sarfraz Ahmad, Naveen Chandra Pant, Girish Prasad Dixit, and et al. 2023. "Genomic-Mediated Breeding Strategies for Global Warming in Chickpeas (Cicer arietinum L.)" Agriculture 13, no. 9: 1721. https://doi.org/10.3390/agriculture13091721
APA StyleJain, S. K., Wettberg, E. J. v., Punia, S. S., Parihar, A. K., Lamichaney, A., Kumar, J., Gupta, D. S., Ahmad, S., Pant, N. C., Dixit, G. P., Sari, H., Sari, D., Ma’ruf, A., Toker, P., & Toker, C. (2023). Genomic-Mediated Breeding Strategies for Global Warming in Chickpeas (Cicer arietinum L.). Agriculture, 13(9), 1721. https://doi.org/10.3390/agriculture13091721










