Characterizing Agronomic and Shoot Morphological Diversity across 263 Wild Emmer Wheat Accessions
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials
2.2. Glasshouse and Field Experiment
2.3. Data Collection
2.4. Statistical Analysis
3. Results
3.1. Diversity in Shoot Morphology
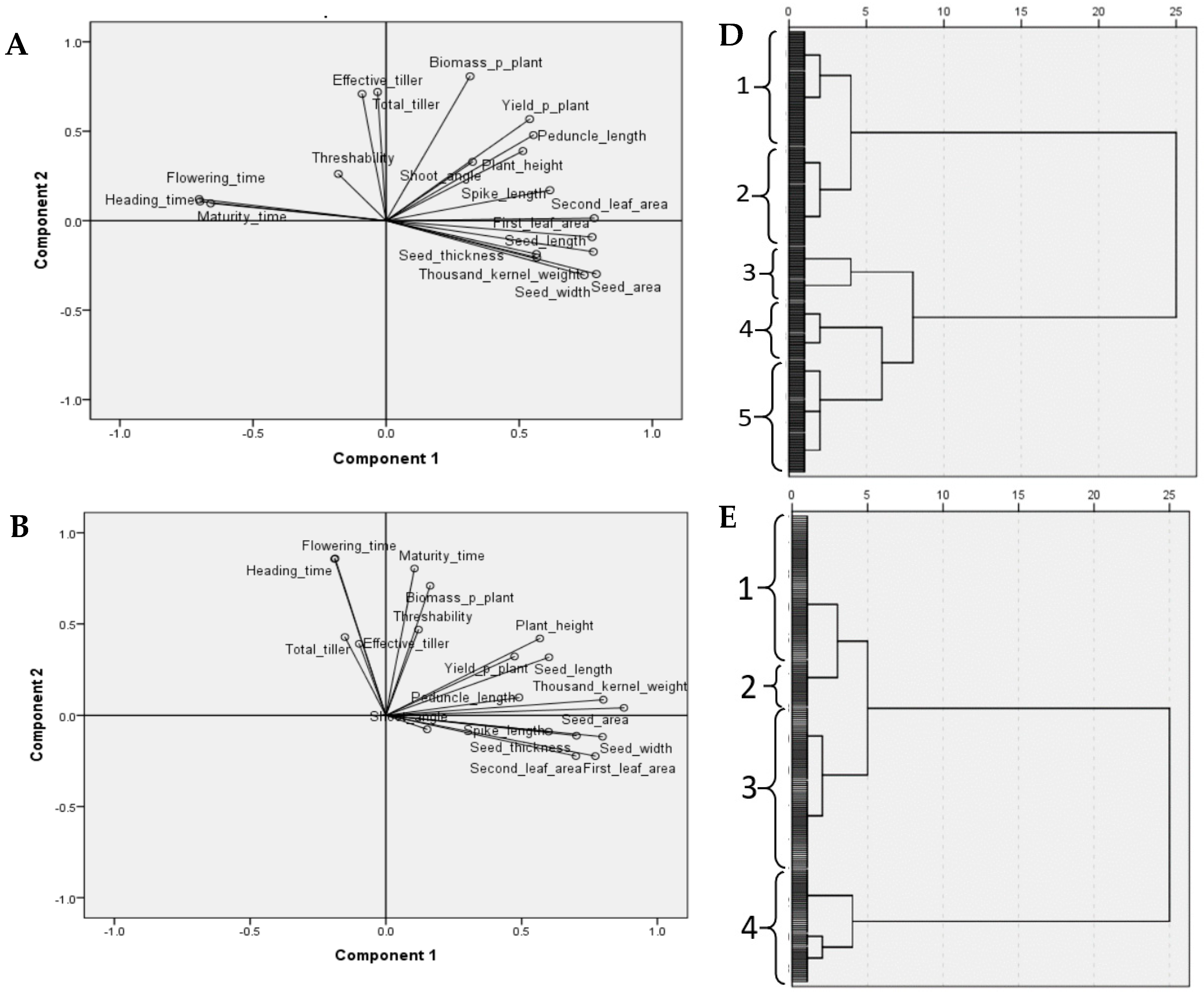
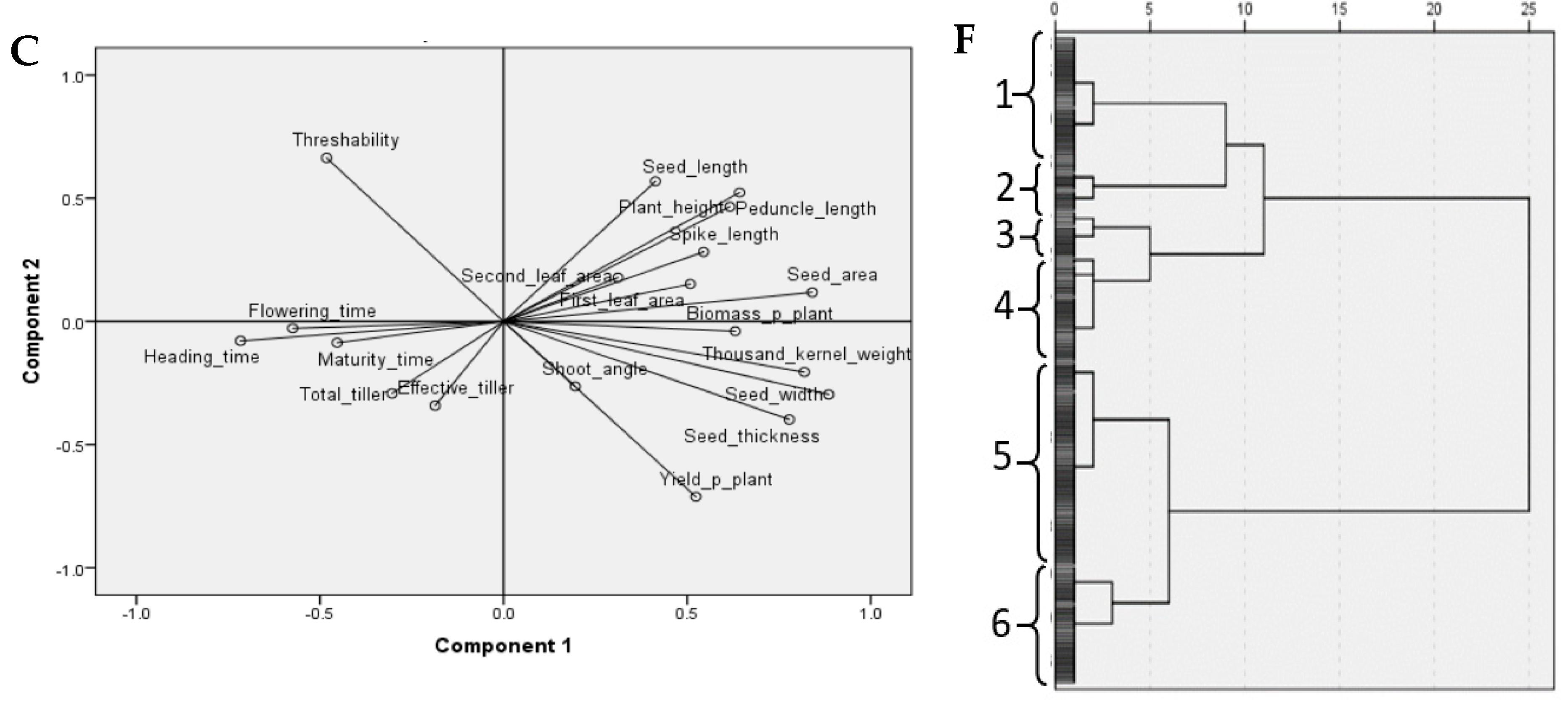
3.2. Multivariate Analysis
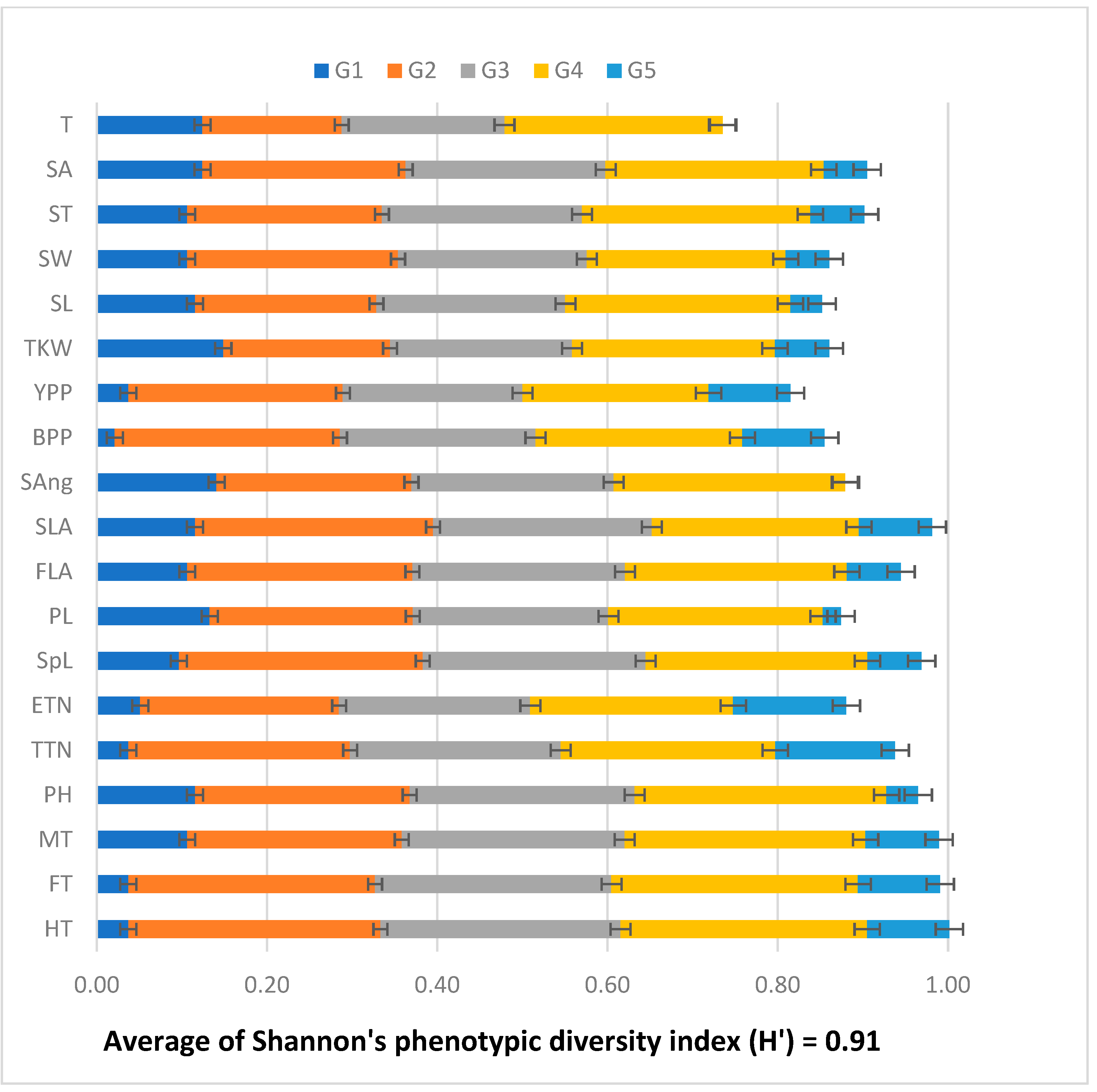
3.3. Shannon’s Phenotypic Diversity Index
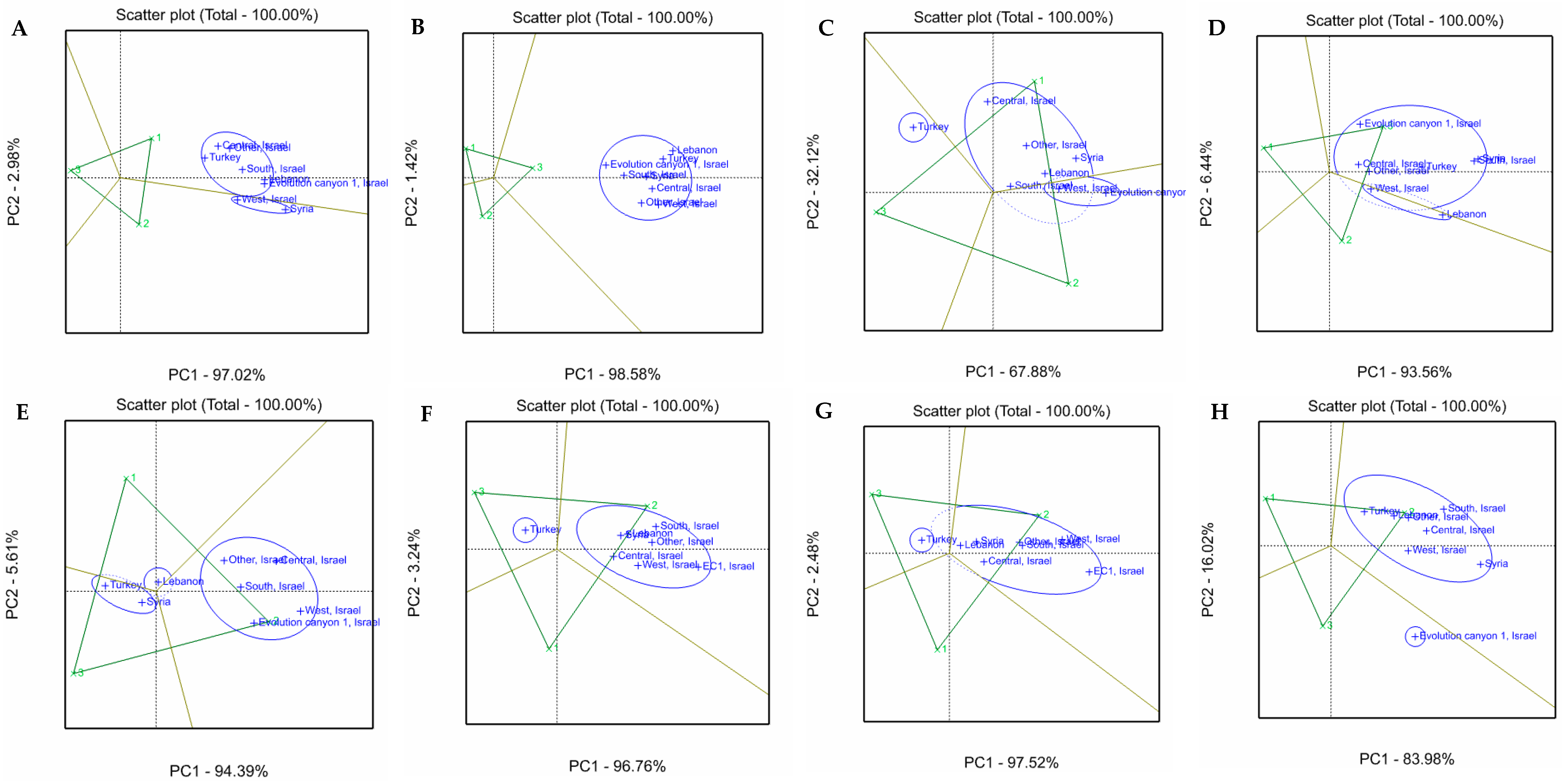
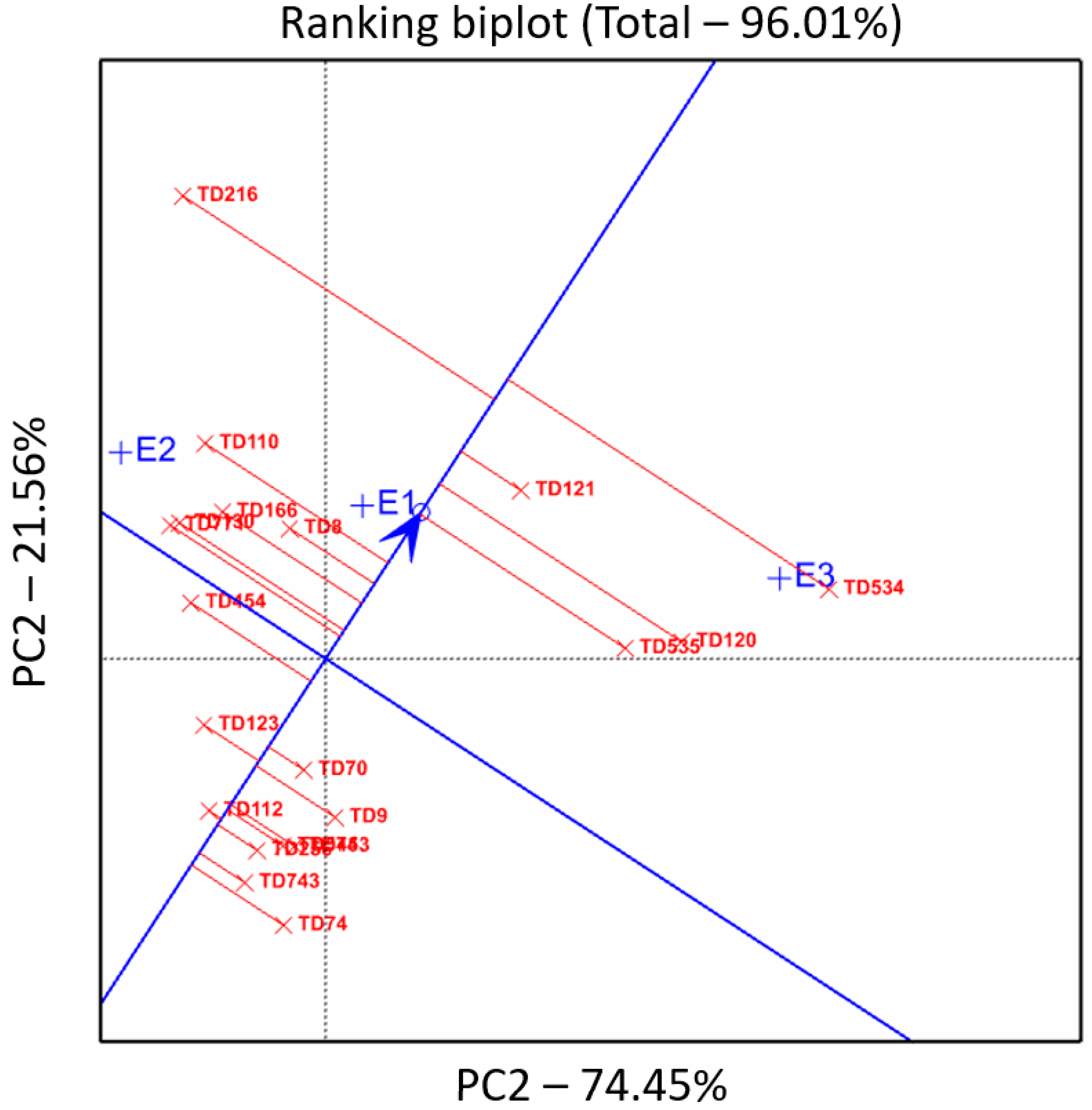
3.4. Phenotypic Variability Explained by Accession Origin
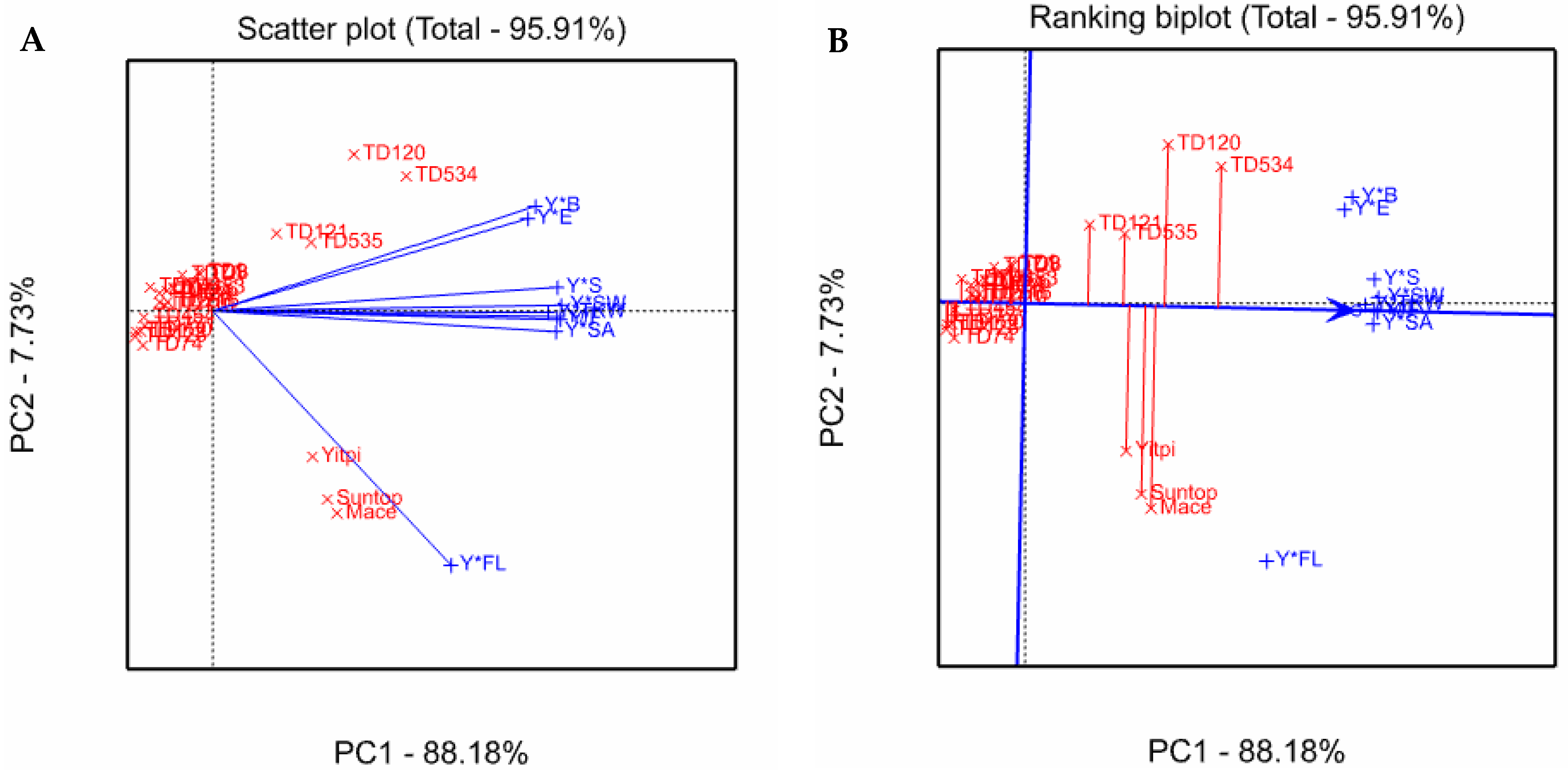
3.5. Ranking Genotype Superiority
4. Discussion
4.1. Wild Emmer Possesses Wide Variation in Shoot Morphology
4.2. Some Traits of Wild Emmer with Potential Breeding Value
4.3. Performances of the Wild Emmer Populations in the Australian Environment Is Trait-Specific
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- FAO. World Food and Agriculture—Statistical Yearbook 2020; FAO: Rome, Italy, 2020. [Google Scholar]
- Enghiad, A.; Ufer, D.; Countryman, A.M.; Thilmany, D.D. An Overview of Global Wheat Market Fundamentals in an Era of Climate Concerns. Int. J. Agron. 2017, 2017, 3931897. [Google Scholar] [CrossRef]
- Avni, R.; Nave, M.; Barad, O.; Baruch, K.; Twardziok, S.O.; Gundlach, H.; Hale, I.; Mascher, M.; Spannagl, M.; Wiebe, K. Wild emmer genome architecture and diversity elucidate wheat evolution and domestication. Science 2017, 357, 93–97. [Google Scholar] [CrossRef] [PubMed]
- Rahman, S.; Islam, S.; Yu, Z.; She, M.; Nevo, E.; Ma, W. Current progress in understanding and recovering the wheat genes lost in evolution and domestication. Int. J. Mol. Sci. 2020, 21, 5836. [Google Scholar] [CrossRef]
- Peng, J.; Sun, D.; Peng, Y.; Nevo, E. Gene discovery in Triticum dicoccoides, the direct progenitor of cultivated wheats. Cereal Res. Commun. 2013, 41, 1–22. [Google Scholar] [CrossRef]
- Jorgensen, C.; Luo, M.C.; Ramasamy, R.; Dawson, M.; Gill, B.S.; Korol, A.B.; Distelfeld, A.; Dvorak, J. A High-Density Genetic Map of Wild Emmer Wheat from the Karaca Dag Region Provides New Evidence on the Structure and Evolution of Wheat Chromosomes. Front Plant Sci 2017, 8, 1798. [Google Scholar] [CrossRef]
- Matsuoka, Y. Evolution of polyploid Triticum wheats under cultivation: The role of domestication, natural hybridization and allopolyploid speciation in their diversification. Plant Cell Physiol. 2011, 52, 750–764. [Google Scholar] [CrossRef]
- Gornicki, P.; Zhu, H.; Wang, J.; Challa, G.S.; Zhang, Z.; Gill, B.S.; Li, W. The chloroplast view of the evolution of polyploid wheat. New Phytol. 2014, 204, 704–714. [Google Scholar] [CrossRef]
- Kato, K.; Mori, Y.; Beiles, A.; Nevo, E. Geographical variation in heading traits in wild emmer wheat, Triticum dicoccoides. I. Variation in vernalization response and ecological differentiation. Theor. Appl. Genet. 1997, 95, 546–552. [Google Scholar]
- Plekhanova, E.; Vishnyakova, M.A.; Bulyntsev, S.; Chang, P.L.; Carrasquilla-Garcia, N.; Negash, K.; von Wettberg, E.; Noujdina, N.; Cook, D.R.; Samsonova, M.G. Genomic and phenotypic analysis of Vavilov’s historic landraces reveals the impact of environment and genomic islands of agronomic traits. Sci. Rep. 2017, 7, 4816. [Google Scholar] [CrossRef]
- Khodadadi, M.; Fotokian, M.H.; Miransari, M. Genetic diversity of wheat (Triticum aestivum L.) genotypes based on cluster and principal component analyses for breeding strategies. Aust. J. Crop Sci. 2011, 5, 17–24. [Google Scholar]
- Mengistu, D.K.; Kiros, A.Y.; Pè, M.E. Phenotypic diversity in Ethiopian durum wheat (Triticum turgidum var. durum) landraces. Crop J. 2015, 3, 190–199. [Google Scholar] [CrossRef]
- Talini, R.F.; Brandolini, A.; Miculan, M.; Brunazzi, A.; Vaccino, P.; Pè, M.E.; Dell’Acqua, M. Genome-wide association study of agronomic and quality traits in a world collection of the wild wheat relative Triticum urartu. Plant J. 2020, 102, 555–568. [Google Scholar] [CrossRef]
- Bohra, A.; Kilian, B.; Sivasankar, S.; Caccamo, M.; Mba, C.; McCouch, S.R.; Varshney, R.K. Reap the crop wild relatives for breeding future crops. Trends Biotechnol. 2021, 40, 412–431. [Google Scholar] [CrossRef]
- Peng, J.; Ronin, Y.; Fahima, T.; Röder, M.S.; Li, Y.; Nevo, E.; Korol, A. Domestication quantitative trait loci in Triticum dicoccoides, the progenitor of wheat. Proc. Natl. Acad. Sci. USA 2003, 100, 2489–2494. [Google Scholar] [CrossRef]
- Zaharieva, M.; Ayana, N.G.; Hakimi, A.A.; Misra, S.C.; Monneveux, P. Cultivated emmer wheat (Triticum dicoccon Schrank), an old crop with promising future: A review. Genet. Resour. Crop Evol. 2010, 57, 937–962. [Google Scholar] [CrossRef]
- Chatzav, M.; Peleg, Z.; Ozturk, L.; Yazici, A.; Fahima, T.; Cakmak, I.; Saranga, Y. Genetic diversity for grain nutrients in wild emmer wheat: Potential for wheat improvement. Ann. Bot. 2010, 105, 1211–1220. [Google Scholar] [CrossRef]
- Law, C.; Sutka, J.; Worland, A. A genetic study of day-length response in wheat. Heredity 1978, 41, 185–191. [Google Scholar] [CrossRef]
- Simkin, A.J.; López-Calcagno, P.E.; Raines, C.A. Feeding the world: Improving photosynthetic efficiency for sustainable crop production. J. Exp. Bot. 2019, 70, 1119–1140. [Google Scholar] [CrossRef]
- Rawashdeh, N.K.; Haddad, N.I.; Al-Ajlouni, M.M.; Turk, M.A. Phenotypic diversity of durum wheat (Triticum durum Desf.) from Jordan. Genet. Resour. Crop Evol. 2007, 54, 129–138. [Google Scholar] [CrossRef]
- Stanley, J.D.; Mehring, G.H.; Wiersma, J.J.; Ransom, J.K. A Standardized Method for Determining Tillering Capacity of Wheat Cultivars. Am. J. Plant Sci. 2020, 11, 604. [Google Scholar] [CrossRef]
- Huang, L.; Feng, L.; He, Y.; Tang, Z.; He, J.; Sela, H.; Krugman, T.; Fahima, T.; Liu, D.; Wu, B. Variation in stripe rust resistance and morphological traits in wild emmer wheat populations. Agronomy 2019, 9, 44. [Google Scholar] [CrossRef]
- Tan, L.; Li, X.; Liu, F.; Sun, X.; Li, C.; Zhu, Z.; Fu, Y.; Cai, H.; Wang, X.; Xie, D. Control of a key transition from prostrate to erect growth in rice domestication. Nat. Genet. 2008, 40, 1360–1364. [Google Scholar] [CrossRef]
- Dong, H.; Zhao, H.; Xie, W.; Han, Z.; Li, G.; Yao, W.; Bai, X.; Hu, Y.; Guo, Z.; Lu, K. A novel tiller angle gene, TAC3, together with TAC1 and D2 largely determine the natural variation of tiller angle in rice cultivars. PLoS Genet. 2016, 12, e1006412. [Google Scholar] [CrossRef]
- Mohammadi, R.; Amri, A. Phenotypic diversity and relationships among a worldwide durum wheat (Triticum turgidum L. var. durum) germplasm collection under rainfed conditions of Iran. Crop Pasture Sci. 2013, 64, 87–99. [Google Scholar] [CrossRef]
- Hailu, F.; Merker, A.; Belay, G.; Johansson, E. Multivariate analysis of diversity of tetraploid wheat germplasm from Ethiopia. Genet. Resour. Crop Evol. 2006, 53, 1089–1098. [Google Scholar] [CrossRef]
- Chakravorty, A.; Ghosh, P.; Sahu, P. Multivariate analysis of phenotypic diversity of landraces of rice of West Bengal. J. Exp. Agric. Int. 2013, 3, 110–123. [Google Scholar] [CrossRef]
- Yan, W.; Frégeau-Reid, J. Genotype by Yield* Trait (GYT) Biplot: A novel approach for genotype selection based on multiple traits. Sci. Rep. 2018, 8, 8242. [Google Scholar] [CrossRef]
- Ghaedrahmati, M. Assessment of relationships among traits and selection of superior bread wheat genotypes using genotype by yield× trait biplot method. Cereal Res. 2020, 10, 61–72. [Google Scholar]
- Mohammadi, R. Genotype by yield* trait biplot for genotype evaluation and trait profiles in durum wheat. Cereal Res. Commun. 2019, 47, 541–551. [Google Scholar] [CrossRef]
- Woyann, L.G.; Meira, D.; Zdziarski, A.D.; Matei, G.; Milioli, A.S.; Rosa, A.C.; Madella, L.A.; Benin, G. Multiple-trait selection of soybean for biodiesel production in Brazil. Ind. Crops Prod. 2019, 140, 111721. [Google Scholar] [CrossRef]
- Boureima, S.; Abdoua, Y. Genotype by Yield* Trait Combination Biplot Approach to Evaluate Sesame Genotypes on Multiple Traits Basis. Turk. J. Field Crops 2019, 24, 237–244. [Google Scholar] [CrossRef]
- de Oliveira, T.R.A.; de Amaral Gravina, G.; de Moura Rocha, M.; de Alcântara Neto, F.; da Cruz, D.P.; de Oliveira, G.H.F.; da Costa Jaeggi, M.E.P.; Rocha, R.S. GYT biplot analysis: A new approach for cowpea line selection. J. Exp. Agric. Int. 2019, 41, 1–9. [Google Scholar] [CrossRef]
- Nevo, E. Genetic resources of wild emmer, Triticum dicoccoides, for wheat improvement in the third millennium. Isr. J. Plant Sci. 2001, 49, 77–92. [Google Scholar] [CrossRef]
- Faris, J.D.; Zhang, Z.; Chao, S. Map-based analysis of the tenacious glume gene Tg-B1 of wild emmer and its role in wheat domestication. Gene 2014, 542, 198–208. [Google Scholar] [CrossRef]
- Johnson, H.W.; Robinson, H.; Comstock, R. Estimates of genetic and environmental variability in soybeans 1. Agron. J. 1955, 47, 314–318. [Google Scholar] [CrossRef]
- Burton, G.W. Quantitative inheritance in grasses. Pro VI Int. Grassl Cong. 1952, 1952, 277–283. [Google Scholar]
- Comstock, R.; Robinson, H. Genetic parameters, their estimation and significance. In Proceedings of the 6th International Grassland Congress, State College, PA, USA, 17–23 August 1952; pp. 248–291. [Google Scholar]
- Liu, Y.; Lin, Y.; Gao, S.; Li, Z.; Ma, J.; Deng, M.; Chen, G.; Wei, Y.; Zheng, Y. A genome-wide association study of 23 agronomic traits in Chinese wheat landraces. Plant J. 2017, 91, 861–873. [Google Scholar] [CrossRef]
- Ormoli, L.; Costa, C.; Negri, S.; Perenzin, M.; Vaccino, P. Diversity trends in bread wheat in Italy during the 20th century assessed by traditional and multivariate approaches. Sci. Rep. 2015, 5, 8574. [Google Scholar] [CrossRef]
- Gurcan, K.; Demirel, F.; Tekin, M.; Demirel, S.; Akar, T. Molecular and agro-morphological characterization of ancient wheat landraces of turkey. BMC Plant Biol. 2017, 17, 171. [Google Scholar] [CrossRef]
- Zarkti, H.; Ouabbou, H.; Udupa, S.M.; Gaboun, F.; Hilali, A. Agro-morphological variability in durum wheat landraces of Morocco. Aust. J. Crop Sci. 2012, 6, 1172–1178. [Google Scholar]
- Al-Habbar, Z. Improve Nitrogen Use Efficiency of Wheat Cultivars under Western Australian Conditions. Ph.D. Thesis, Murdoch University, Murdoch, Australia, 2018. [Google Scholar]
- Sultana, N. Characterization of TaNAC-S Gene in Australian Wheat Cultivars in Relation to Senescence and Nitrogen Stress Response. Ph.D. Thesis, Murdoch University, Murdoch, Australia, 2020. [Google Scholar]
- Ali, M.; Hussain, M.; Khan, M.; Ali, Z.; Zulkiffal, M.; Anwar, J.; Sabir, W.; Zeeshan, M. Source-sink relationship between photosynthetic organs and grain yield attributes during grain filling stage in spring wheat (Triticum aestivum). Int. J. Agric. Biol 2010, 12, 509–515. [Google Scholar]
- Chen, X.; Luo, Y.; Xia, X.; Xia, L.; Chen, X.; Ren, Z.; He, Z.; Jia, J. Chromosomal location of powdery mildew resistance gene Pm16 in wheat using SSR marker analysis. Plant Breed. 2005, 124, 225–228. [Google Scholar] [CrossRef]
- Hovmøller, M. Sources of seedling and adult plant resistance to Puccinia striiformis f. sp. tritici in European wheats. Plant Breed. 2007, 126, 225–233. [Google Scholar] [CrossRef]
- DePauw, R.; Townley-Smith, T.; Humphreys, G.; Knox, R.; Clarke, F.; Clarke, J. Lillian hard red spring wheat. Can. J. Plant Sci. 2005, 85, 397–401. [Google Scholar] [CrossRef]
- Desheva, G.; Kyosev, B. Genetic diversity assessment of common winter wheat (Triticum aestivum L.) genotypes. Emir. J. Food Agric. 2015, 27, 283–290. [Google Scholar]
- Shukla, S.; Bhargava, A.; Chatterjee, A.; Srivastava, A.; Singh, S. Genotypic variability in vegetable amaranth (Amaranthus tricolor L. for foliage yield and its contributing traits over successive cuttings and years. Euphytica 2006, 151, 103–110. [Google Scholar] [CrossRef]
- Shashikala, S. Analysis of Genetic Diversity in Wheat. Ph.D. Thesis, University of Agricultural Sciences GKVK, Banglore, Imdia, 2006. [Google Scholar]
- Robinson, H.; Comstock, R.E.; Harvey, P. Estimates of heritability and the degree of dominance in corn. Agron. J. 1949, 41, 353–359. [Google Scholar] [CrossRef]
- Anshuman, V.; Dixit, N.; Sharma, S.; Marker, S. Studies on heritability and genetic advance estimates in maize genotypes. Biosci. Discov. 2013, 4, 165–168. [Google Scholar]
- Özkan, H.; Willcox, G.; Graner, A.; Salamini, F.; Kilian, B. Geographic distribution and domestication of wild emmer wheat (Triticum dicoccoides). Genet. Resour. Crop Evol. 2011, 58, 11–53. [Google Scholar] [CrossRef]



| Populations | Collection Location | Total Number of Accessions Studied | ||
|---|---|---|---|---|
| Central area | Qazrin | 3 | 129 |
| Yehudiyya | 77 | |||
| Gamla | 4 | |||
| Rosh-Pinha | 4 | |||
| Tabigha | 41 | |||
| South area | Mt. Gilboa | 1 | 17 | |
| Mt. Gerizim | 5 | |||
| Gitit | 2 | |||
| Kokhav-Hashahar | 1 | |||
| Taiyiba | 1 | |||
| Sanhedriyya | 2 | |||
| Bet-Meir | 2 | |||
| J’aba | 3 | |||
| West area | Amirim | 2 | 11 | |
| Nesher | 1 | |||
| Beit-Oren | 3 | |||
| Daliyya | 3 | |||
| Bat-Shelomo | 2 | |||
| North area | Mt. Hermon | 2 | ||
| Evolutionary canyon 1 | 13 | |||
| Non-specified | 41 | |||
| 14 | |||
| 16 | |||
| 19 | |||
| 1 | |||
| Total | 263 | |||
| Traits | Genotypic Variance | Phenotypic Variance | GCV | PCV | Hb | GA | GA% |
|---|---|---|---|---|---|---|---|
| HT | 1214.35 | 1240.63 | 21.83 | 22.06 | 97.88 | 97.94 | 61.35 |
| FT | 1161.73 | 1180.59 | 20.6 | 20.77 | 98.4 | 97.75 | 59.08 |
| MT | 629.09 | 683.83 | 11.04 | 11.51 | 92 | 103.9 | 45.74 |
| PH | 347.55 | 417.62 | 14.87 | 16.3 | 83.22 | 39.09 | 31.19 |
| TTN | 2.05 | 3.87 | 21.68 | 29.81 | 52.91 | 2.48 | 37.62 |
| ETN | 0.99 | 1.7 | 25.15 | 32.94 | 58.31 | 1.92 | 48.45 |
| SpL | 0.89 | 1.48 | 12.54 | 16.15 | 60.31 | 2.18 | 28.95 |
| PL | 88.16 | 131.92 | 18.66 | 22.83 | 66.83 | 19.68 | 39.12 |
| FLA | 75.69 | 108 | 34.45 | 41.15 | 70.08 | 17.42 | 69 |
| SLA | 77.74 | 105.4 | 27.56 | 32.09 | 73.76 | 18.21 | 56.91 |
| Sang | 190.52 | 233.51 | 24.3 | 26.9 | 81.59 | 27.86 | 49.04 |
| BPP | 10.7 | 11.81 | 33.89 | 35.60 | 90.63 | 8.72 | 90.29 |
| YPP | 0.21 | 0.24 | 40.78 | 43.98 | 85.98 | 1.07 | 96.19 |
| TKW | 26.08 | 34.34 | 18.44 | 21.17 | 75.93 | 10.61 | 38.31 |
| SL | 0.35 | 0.66 | 6.72 | 9.17 | 53.65 | 0.86 | 9.7 |
| SW | 0.05 | 0.1 | 8.31 | 12.04 | 47.61 | 0.3 | 11.62 |
| ST | 0.04 | 0.1 | 8.44 | 13.21 | 40.86 | 0.28 | 11.84 |
| SA | 2.93 | 4.26 | 11.36 | 13.71 | 68.75 | 3.04 | 20.17 |
| T | 0.3 | 0.45 | 18.23 | 22.26 | 67.01 | 0.88 | 29.36 |
| Genotypes | Y/F | Y*E | Y*S | Y*FL | Y*SA | Y*B | Y*TKW | Y*SW | Y/T | Mean (Superior-ity Index) |
|---|---|---|---|---|---|---|---|---|---|---|
| TD534 | 2.43 | 1.87 | 2.27 | 0.35 | 1.89 | 3.02 | 2.91 | 2.49 | 1.97 | 2.13 |
| TD120 | 1.46 | 2.34 | 1.89 | −0.09 | 1.92 | 2.17 | 0.84 | 1.53 | 1.89 | 1.55 |
| Mace | 1.61 | 1.18 | 1.10 | 2.91 | 1.51 | 0.59 | 1.32 | 1.40 | 1.43 | 1.45 |
| Suntop | 1.50 | 0.58 | 1.11 | 2.53 | 1.46 | 0.58 | 1.47 | 1.36 | 1.38 | 1.33 |
| Yitpi | 0.96 | 0.38 | 1.42 | 2.07 | 1.50 | 0.72 | 0.89 | 1.11 | 1.32 | 1.15 |
| TD535 | 1.34 | 1.30 | 1.21 | 0.03 | 0.82 | 0.86 | 1.62 | 1.35 | 1.18 | 1.08 |
| TD121 | 0.63 | 1.30 | 1.01 | −0.23 | 0.84 | 0.48 | 0.33 | 0.69 | 1.18 | 0.69 |
| TD8 | −0.17 | −0.54 | 0.01 | −0.42 | 0.12 | 0.48 | −0.24 | −0.12 | −0.50 | −0.15 |
| TD9 | −0.24 | 0.18 | −0.30 | −0.42 | −0.12 | −0.10 | −0.04 | −0.11 | −0.32 | −0.16 |
| TD70 | −0.43 | −0.26 | −0.16 | −0.39 | −0.54 | 0.16 | −0.38 | −0.42 | −0.66 | −0.34 |
| TD453 | −0.48 | 0.08 | −0.64 | −0.50 | −0.38 | −0.61 | −0.55 | −0.42 | −0.47 | −0.44 |
| TD216 | −0.45 | −0.22 | −0.64 | −0.43 | −0.63 | −0.61 | −0.44 | −0.57 | −0.57 | −0.51 |
| TD166 | −0.56 | −0.10 | −0.73 | −0.51 | −0.53 | −0.57 | −0.73 | −0.55 | −0.37 | −0.52 |
| TD110 | −0.50 | −0.91 | −0.44 | −0.44 | −0.66 | −0.20 | −0.51 | −0.50 | −0.68 | −0.54 |
| TD574 | −0.70 | −0.09 | −0.65 | −0.50 | −0.55 | −0.66 | −0.78 | −0.72 | −0.71 | −0.59 |
| TD256 | −0.59 | −0.37 | −0.83 | −0.49 | −0.62 | −0.62 | −0.48 | −0.64 | −0.72 | −0.60 |
| TD743 | −0.80 | −0.08 | −0.89 | −0.55 | −1.12 | −0.69 | −0.54 | −0.82 | −0.78 | −0.70 |
| TD771 | −0.79 | −1.01 | −0.83 | −0.47 | −0.83 | −0.71 | −0.65 | −0.77 | −0.77 | −0.76 |
| TD454 | −0.89 | −1.06 | −0.63 | −0.54 | −0.66 | −0.62 | −0.99 | −0.91 | −0.65 | −0.77 |
| TD74 | −0.76 | −1.22 | −0.76 | −0.44 | −0.66 | −0.97 | −0.75 | −0.73 | −0.76 | −0.78 |
| TD130 | −0.82 | −1.05 | −0.83 | −0.50 | −0.83 | −0.90 | −0.72 | −0.84 | −0.79 | −0.81 |
| TD112 | −0.87 | −1.21 | −0.84 | −0.49 | −1.06 | −0.79 | −0.77 | −0.87 | −0.80 | −0.85 |
| TD123 | −0.89 | −1.08 | −0.84 | −0.50 | −0.87 | −1.03 | −0.83 | −0.96 | −0.81 | −0.87 |
| Mean | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | - |
| SD | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | - |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rahman, S.; Islam, S.; Nevo, E.; Saieed, M.A.U.; Liu, Q.; Varshney, R.K.; Ma, W. Characterizing Agronomic and Shoot Morphological Diversity across 263 Wild Emmer Wheat Accessions. Agriculture 2023, 13, 759. https://doi.org/10.3390/agriculture13040759
Rahman S, Islam S, Nevo E, Saieed MAU, Liu Q, Varshney RK, Ma W. Characterizing Agronomic and Shoot Morphological Diversity across 263 Wild Emmer Wheat Accessions. Agriculture. 2023; 13(4):759. https://doi.org/10.3390/agriculture13040759
Chicago/Turabian StyleRahman, Shanjida, Shahidul Islam, Eviatar Nevo, Md Atik Us Saieed, Qier Liu, Rajeev Kumar Varshney, and Wujun Ma. 2023. "Characterizing Agronomic and Shoot Morphological Diversity across 263 Wild Emmer Wheat Accessions" Agriculture 13, no. 4: 759. https://doi.org/10.3390/agriculture13040759
APA StyleRahman, S., Islam, S., Nevo, E., Saieed, M. A. U., Liu, Q., Varshney, R. K., & Ma, W. (2023). Characterizing Agronomic and Shoot Morphological Diversity across 263 Wild Emmer Wheat Accessions. Agriculture, 13(4), 759. https://doi.org/10.3390/agriculture13040759









