Development of a High Yielded Chlorsulfuron-Resistant Soybean (Glycine max L.) Variety through Mutation Breeding
Abstract
1. Introduction
2. Materials and Methods
2.1. Development of EMS-Mutated Populations
2.2. Determination of Herbicide Effective Dose
2.3. Detection of SU-Resistant Mutants
2.4. The AHAS Gene Analysis of the Mutant Progeny
2.5. Comparison of Agronomic Characters between the Mutant and the Wild Type
3. Results
3.1. Screening for the Resistance under Field Condition
3.2. Confirmation Tests under Greenhouse Conditions
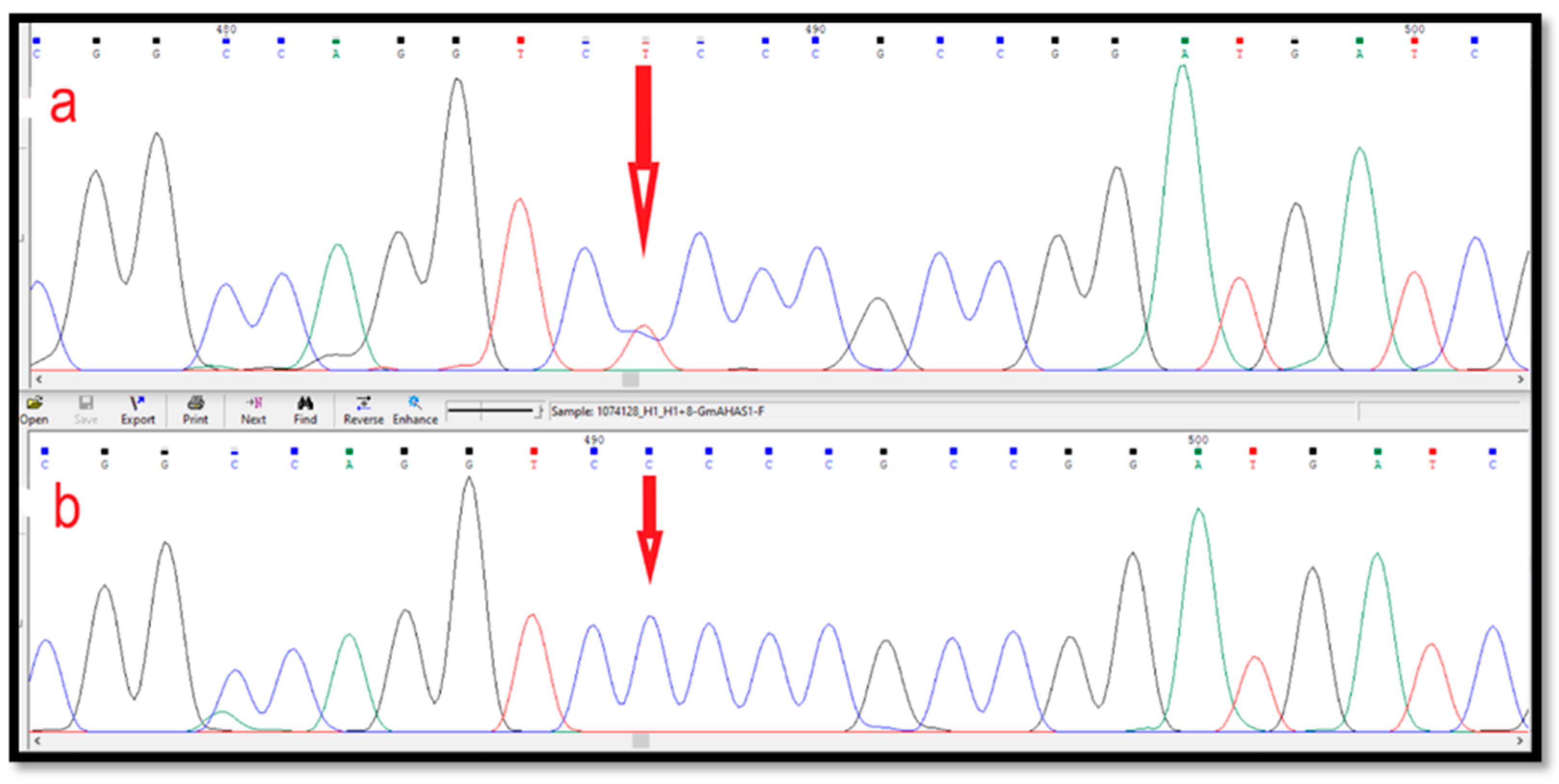
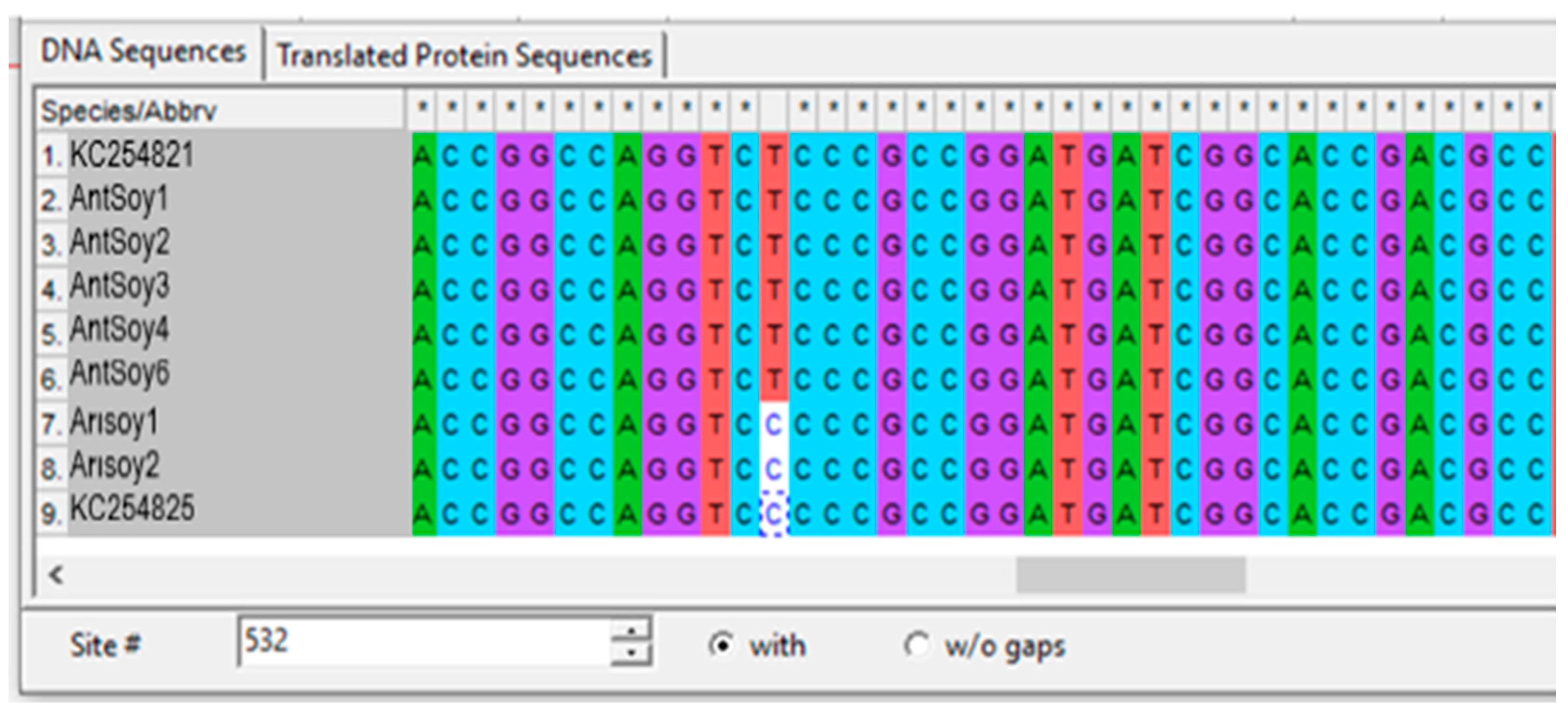
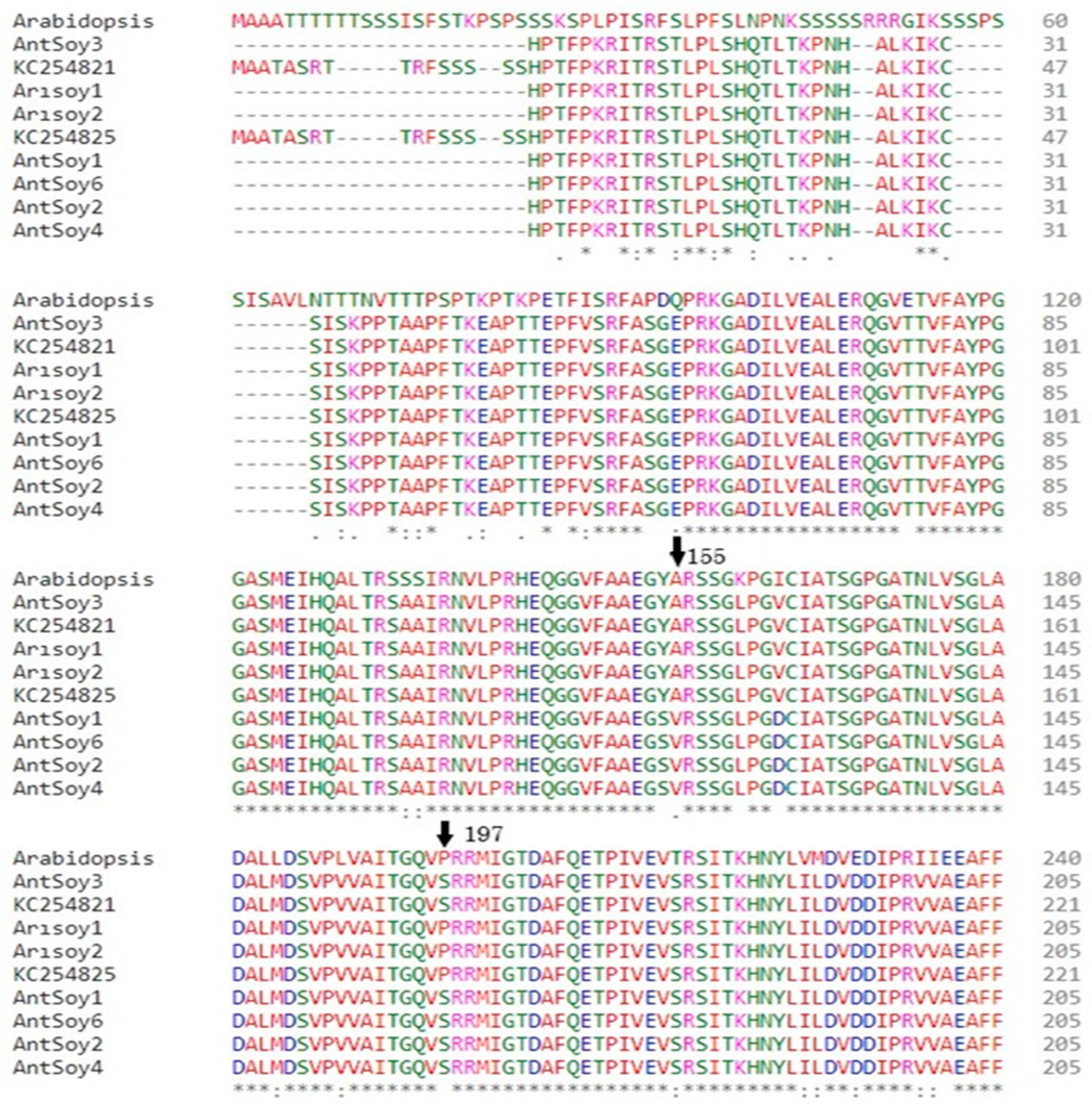
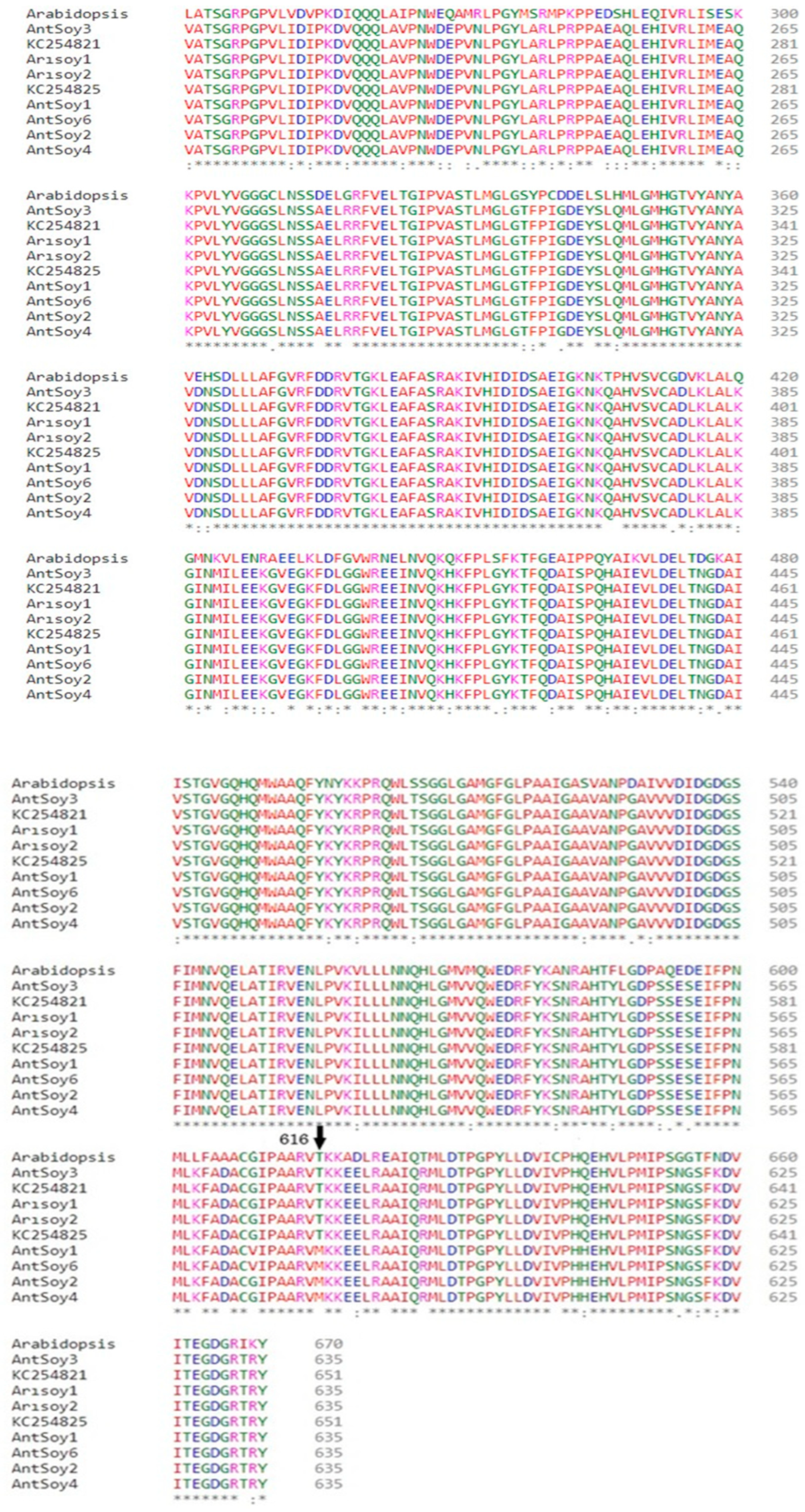
3.3. Identification of SNPs in the AHAS Gene
3.4. Comparison of Agronomic Characters between the Mutant and the Wild Type
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Balta, I.; Stef, L.; Pet, I.; Iancu, T.; Stef, D.; Corcionivoschi, N. Essential Fatty Acids as Biomedicines in Cardiac Health. Biomedicines 2021, 9, 1466. [Google Scholar] [CrossRef]
- Vedavanam, K.; Srijayanta, S.; O’Reilly, J.; Raman, A.; Wiseman, H. Antioxidant action and potential antidiabetic properties of an isoflavonoid-containing soyabean phytochemical extract (SPE). Phytother. Res. 1999, 13, 601–608. [Google Scholar] [CrossRef]
- Elhady, A.; Hallmann, J.; Heuer, H. Symbiosis of soybean with nitrogen fixing bacteria affected by root lesion nematodes in a density-dependent manner. Sci. Rep. 2020, 10, 1619. [Google Scholar] [CrossRef]
- Oerke, E.C. Crop losses to pests. J. Agric. Sci. 2006, 144, 31–43. [Google Scholar] [CrossRef]
- Soltani, N.; Dille, J.A.; Burke, I.C.; Everman, W.J.; VanGessel, M.J.; Davis, V.M.; Sikkema, P.H. Perspectives on Potential Soybean Yield Losses from Weeds in North America. Weed Technol. 2017, 31, 148–154. [Google Scholar] [CrossRef]
- Busi, R.; Neve, P.; Powles, S. Evolved polygenic herbicide resistance in Lolium rigidum by low-dose herbicide selection within standing genetic variation. Evol. Appl. 2013, 6, 231–242. [Google Scholar] [CrossRef] [PubMed]
- Ellis, A.T.; Steckel, L.E.; Main, C.L.; de Melo, M.S.C.; West, D.R.; Mueller, T.C. A Survey for Diclofop-Methyl Resistance in Italian Ryegrass from Tennessee and How to Manage Resistance in Wheat. Weed Technol. 2010, 24, 303–309. [Google Scholar] [CrossRef]
- Grey, T.L.; Bridges, D.C. Alternatives to diclofop for the control of Italian ryegrass (Lolium multiflorum) in winter wheat (Triticum aestivum). Weed Technol. 2003, 17, 219–223. [Google Scholar] [CrossRef]
- Grey, T.L.; Braxton, L.B.; Richburg, J.S. Effect of Wheat Herbicide Carryover on Double-Crop Cotton and Soybean. Weed Technol. 2012, 26, 207–212. [Google Scholar] [CrossRef]
- Green, J.M. Review of glyphosate and ALS-inhibiting herbicide crop resistance and resistant weed management. Weed Technol. 2007, 21, 547–558. [Google Scholar] [CrossRef]
- Duggleby, R.G.; McCourt, J.A.; Guddat, L.W. Structure and mechanism of inhibition of plant acetohydroxyacid synthase. Plant Physiol. Biochem. 2008, 46, 309–324. [Google Scholar] [CrossRef]
- Duggleby, R.G.; Pang, S.S. Acetohydroxyacid synthase. J. Biochem. Mol. Biol. 2000, 33, 1–36. [Google Scholar]
- HRAC. Herbicide Classification. Available online: https://hracglobal.com/tools/classification-lookup (accessed on 10 July 2020).
- Yu, Q.; Powles, S.B. Resistance to AHAS inhibitor herbicides: Current understanding. Pest Manag. Sci. 2014, 70, 1340–1350. [Google Scholar] [CrossRef]
- Delye, C. Unravelling the genetic bases of non-target-site-based resistance (NTSR) to herbicides: A major challenge for weed science in the forthcoming decade. Pest Manag. Sci. 2013, 69, 176–187. [Google Scholar] [CrossRef]
- Beckie, H.J.; Tardif, F.J. Herbicide cross resistance in weeds. Crop Prot. 2012, 35, 15–28. [Google Scholar] [CrossRef]
- Shaner, D.L. Role of Translocation as a Mechanism of Resistance to Glyphosate. Weed Sci. 2009, 57, 118–123. [Google Scholar] [CrossRef]
- Bai, S.; Zhang, F.W.; Li, Z.R.; Wang, H.Z.; Wang, Q.; Wang, J.X.; Liu, W.T.; Bai, L.Y. Target-site and non-target-site-based resistance to tribenuron-methyl in multiply-resistant Myosoton aquaticum L. Pestic. Biochem. Physiol. 2019, 155, 8–14. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.Y.; Chu, Z.Z.; Han, H.P.; Goggin, D.E.; Yu, Q.; Sayer, C.; Powles, S.B. A Val-202-Phe alpha-tubulin mutation and enhanced metabolism confer dinitroaniline resistance in a single Lolium rigidum population. Pest Manag. Sci. 2020, 76, 645–652. [Google Scholar] [CrossRef] [PubMed]
- Heap. The International Herbicide-Resistant Weed Database. 2020. Available online: https://www.weedscience.org/Home.aspx (accessed on 5 July 2020).
- Tranel, P.J.; Wright, T.R.; Heap, I.M. Mutations in Herbicide-Resistant Weeds to ALS Inhibitors. Available online: https://www.weedscience.com (accessed on 12 January 2020).
- Tan, S.; Evans, R.; Singh, B. Herbicidal inhibitors of amino acid biosynthesis and herbicide-tolerant crops. Amino Acids 2006, 30, 195–204. [Google Scholar] [CrossRef]
- Anderson, P.C.; Georgeson, M. Herbicide-Tolerant Mutants of Corn. Genome 1989, 31, 994–999. [Google Scholar] [CrossRef]
- Miller, J.F.; Al-Khatib, K. Registration of imidazolinone herbicide-resistant sunflower maintainer (HA 425) and fertility restorer (RHA 426 and RHA 427) germplasms. Crop Sci. 2002, 42, 988–989. [Google Scholar] [CrossRef]
- Pozniak, C.J.; Hucl, P.J. Genetic analysis of imidazolinone resistance in mutation-derived lines of common wheat. Crop Sci. 2004, 44, 23–30. [Google Scholar]
- Sebastian, S.A.; Fader, G.M.; Ulrich, J.F.; Forney, D.R.; Chaleff, R.S. Semidominant Soybean Mutation for Resistance to Sulfonylurea Herbicides. Crop Sci. 1989, 29, 1403–1408. [Google Scholar] [CrossRef]
- Swanson, E.B.; Herrgesell, M.J.; Arnoldo, M.; Sippell, D.W.; Wong, R.S.C. Microspore Mutagenesis and Selection—Canola Plants with Field Tolerance to the Imidazolinones. Theor. Appl. Genet. 1989, 78, 525–530. [Google Scholar] [CrossRef]
- Ghio, C.; Ramos, M.L.; Altieri, E.; Bulos, M.; Sala, C.A. Molecular characterization of Als1, an acetohydroxyacid synthase mutation conferring resistance to sulfonylurea herbicides in soybean. Theor. Appl. Genet. 2013, 126, 2957–2968. [Google Scholar] [CrossRef]
- Walter, K.L.; Strachan, S.D.; Ferry, N.M.; Albert, H.H.; Castle, L.A.; Sebastian, S.A. Molecular and phenotypic characterization of Als1 and Als2 mutations conferring tolerance to acetolactate synthase herbicides in soybean. Pest Manag. Sci. 2014, 70, 1831–1839. [Google Scholar] [CrossRef] [PubMed]
- Greene, E.A.; Codomo, C.A.; Taylor, N.E.; Henikoff, J.G.; Till, B.J.; Reynolds, S.H.; Enns, L.C.; Burtner, C.; Johnson, J.E.; Odden, A.R.; et al. Spectrum of chemically induced mutations from a large-scale reverse-genetic screen in arabidopsis. Genetics 2003, 164, 731–740. [Google Scholar] [CrossRef] [PubMed]
- Vila-Aiub, M.M.; Neve, P.; Powles, S.B. Fitness costs associated with evolved herbicide resistance alleles in plants. New Phytol. 2009, 184, 751–767. [Google Scholar] [CrossRef]
- Holt, J.; Thill, D. Growth and productivity of resistant plants. In Herbicid Resistance in Plants; CRC Press: Boca Raton, FL, USA, 2018; pp. 299–316. [Google Scholar]
- Tranel, P.J.; Wright, T.R. Resistance of weeds to ALS-inhibiting herbicides: What have we learned? Weed Sci. 2002, 50, 700–712. [Google Scholar] [CrossRef]
- Gressel, J. Molecular biology of weed control. Transgenic Res. 2000, 9, 355–382. [Google Scholar] [CrossRef]
- Doyle, J.J.; Doyle, J.L. A rapid total DNA preparation procedure for fresh plant tissue. Focus 1990, 12, 13–15. [Google Scholar]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547. [Google Scholar] [CrossRef] [PubMed]
- Powles, S.B.; Yu, Q. Evolution in Action: Plants Resistant to Herbicides. Annu. Rev. Plant Biol. 2010, 61, 317–347. [Google Scholar] [CrossRef] [PubMed]
- Li, H.T.; Li, J.J.; Zhao, B.; Wang, J.; Yi, L.C.; Liu, C.; Wu, J.S.; King, G.J.; Liu, K.D. Generation and characterization of tribenuron-methyl herbicide-resistant rapeseed (Brasscia napus) for hybrid seed production using chemically induced male sterility. Theor. Appl. Genet. 2015, 128, 107–118. [Google Scholar] [CrossRef] [PubMed]

| Primers | Sequence | Amplicon Size (bp) | Chromosome |
|---|---|---|---|
| GmAHAS1-F | ATGGCGGCCACCGCTTC | 2041 | 4 |
| GmAHAS1-R | ACAGGCCAAATCCTGCAACTAGGAC | 2041 | 4 |
| GmAHAS2-F | ATGGCGGCCACAGCTTCCAG | 1936 | 6 |
| GmAHAS2-R | TCAGTACCTCGTTCTACCGTCTCCCTCC | 1936 | 6 |
| GmAHAS3-F | TTTAGATTATTGTGGTATTGGAAGATG | 2105 | 13 |
| GmAHAS3-R | GAATATTTAGTACTAAAAGAAACCAACATC | 2015 | 13 |
| GmAHAS4-F | ACCTTTTGGTGCTATTTGAAAATG | 2122 | 15 |
| GmAHAS4-R | ACATATAATTAACAAAAATAACCAACATTG | 2122 | 15 |
| Characters/Years | Wild-Type | AntSoy | ||||
|---|---|---|---|---|---|---|
| 2020 | 2021 | Average | 2021 | 2022 | Average | |
| Yield (kg da−1) | 340.87 | 353.97 | 347.43 | 341.93 | 351.21 | 346.57 |
| 1000 seeds weight (g) | 109.1 | 110.43 | 109.76 | 96.7 | 99.4 | 98.07 |
| Number of pods per plant | 47.33 | 53.0 | 50.16 | 48.47 | 54.6 | 51.53 |
| First pod height (cm) | 19.07 | 21.4 | 20.24 | 20.73 | 22.06 | 21.40 |
| Plant height (cm) | 92.4 | 98.73 | 95.56 | 97.07 | 99.33 | 98.2 |
| Physiological maturity (day) | 120.67 | 117 | 118.56 | 121.33 | 118.66 | 120.0 |
| Genotypes | Sum of Squares | Mean Square | F Ratio | Prob > F | |
|---|---|---|---|---|---|
| Yield | 2.20 | 2.20 | <0.001 | 0.99 | NS |
| 1000 seeds weight | 410.67 | 410.67 | 3.20 | 0.10 | NS |
| Number of pods per plant | 5.60 | 5.60 | 0.06 | 0.81 | NS |
| First pod height | 4.08 | 4.08 | 1.66 | 0.24 | NS |
| Plant height | 3.85 | 3.85 | 0.13 | 0.73 | NS |
| Physiological maturity | 6.16 | 6.16 | 1.40 | 0.26 | NS |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ustun, R.; Uzun, B. Development of a High Yielded Chlorsulfuron-Resistant Soybean (Glycine max L.) Variety through Mutation Breeding. Agriculture 2023, 13, 559. https://doi.org/10.3390/agriculture13030559
Ustun R, Uzun B. Development of a High Yielded Chlorsulfuron-Resistant Soybean (Glycine max L.) Variety through Mutation Breeding. Agriculture. 2023; 13(3):559. https://doi.org/10.3390/agriculture13030559
Chicago/Turabian StyleUstun, Rustem, and Bulent Uzun. 2023. "Development of a High Yielded Chlorsulfuron-Resistant Soybean (Glycine max L.) Variety through Mutation Breeding" Agriculture 13, no. 3: 559. https://doi.org/10.3390/agriculture13030559
APA StyleUstun, R., & Uzun, B. (2023). Development of a High Yielded Chlorsulfuron-Resistant Soybean (Glycine max L.) Variety through Mutation Breeding. Agriculture, 13(3), 559. https://doi.org/10.3390/agriculture13030559







