Abstract
With the rapid population growth, increasing agricultural productivity is an extreme requirement to meet demands. Early identification of crop diseases is essential to prevent yield loss. Nevertheless, it is a tedious task to manually monitor leaf diseases, as it demands in-depth knowledge of plant pathogens as well as a lot of work, and excessive processing time. For these purposes, various methods based on image processing, deep learning, and machine learning are developed and examined by researchers for crop leaf disease identification and often have obtained significant results. Motivated by this existing work, we conducted an extensive comparative study between traditional machine learning (SVM, LDA, KNN, CART, RF, and NB) and deep transfer learning (VGG16, VGG19, InceptionV3, ResNet50, and CNN) models in terms of precision, accuracy, f1-score, and recall on a dataset taken from the PlantVillage Dataset composed of diseased and healthy crop leaves for binary classification. Moreover, we applied several activation functions and deep learning optimizers to further enhance these CNN architectures’ performance. The classification accuracy (CA) of leaf diseases that we obtained by experimentation is quite impressive for all models. Our findings reveal that NB gives the least CA at 60.09%, while the InceptionV3 model yields the best CA, reaching an accuracy of 98.01%.
1. Introduction
Crop diseases have always been one of the main threats to agricultural production as they drastically reduce the productivity and quality of agricultural products [1]. Therefore, early detection of diseases is a major challenge for crop production [2]. Identifying diseases in crops as early as possible is essential to curb the spread of disease in fields and foremost to optimize the usage of pesticides to mitigate the environmental and health risks associated with their application. For these purposes, many techniques in phytopathology are exploited to detect diseases, namely methods related to chemical analysis of the affected area of a plant and indirect methods such as the use of physical techniques such as spectroscopy [3]. Such techniques demand the intervention of highly qualified personnel with expertise in the field and relatively expensive rates [4]. Therefore, the search for an accurate, fast, automatic, and less expensive disease detection method has become an absolute necessity for researchers and farmers. In this line, researchers are employing image processing [4,5], machine learning [6,7], and deep learning techniques [8,9] to aid in the task of identifying diseases at a very early stage. In addition, crops are subjected to a variety of diseases caused by pathogens such as bacteria, fungi, and viruses [10] that appear in different parts such as leaves, stems, and roots. In our paper, we focus on leaves as they are the most common disease detection feature.
Our motivation is to enhance the overall detection model performance. Notably, we aim to strike a compromise between high accuracy and a low misclassification rate. Thus, we compared different competitive machine learning and deep learning algorithms, using several computer vision techniques and various performance evaluation metrics to evaluate our results to find out the most efficient model that can be exploited for real-time leaf disease identification and classification. So, from the findings, we were able to pin down the advantages and inconveniences of each model.
This present comparative study stands out from other studies in that it is conducted on a massive dataset containing several types of crops and the use of both ML/DL techniques, resulting in weeks of training for the models to be completed. The novelty of this study lies in obtaining the most appropriate combination of the DL optimizer and the CNN model, which delivered greatly better results compared to prior research. The remainder of this article is arranged as follows: the next section provides a review of the existing research on this topic. Then, the third section presents the materials and methodology needed to carry out this study. Afterward, the fourth section reports the experimental results along with their extensive discussion, and finally, the article concludes with the fifth section, which tackles possible future directions for the present work.
Related Work
In previous research, various ML and DL algorithms have already been proposed for crop leaf disease detection [11], and some of them are explored in the following section. Among these studies, Orchi et al. [12] established a brief survey of methods of identifying crop diseases using ML and DL techniques, in particular SVM, ANN, CNN, and RNN, to help researchers recognize the disadvantages and advantages of the different techniques used for plant disease detection.
Today, a growing trend to replace machine learning algorithms with those based on deep learning methods for leaf image classification is gaining momentum. In fact, Argüeso et al. [13] deployed an approach to the distance metric within few-short learning that relied on triplet loss as well as a shallow classifier at class limits that proved efficient for classifying plant leaf diseases with little training images annotated using deep learning. Their approach showed greater than 90% accuracy using only 80 images per class for six diseases. In contrast, Pantazi et al. [14] suggested a technique for identifying diseases (downy mildew, powdery mildew, healthy, and black rot) on grape leaves by applying class classification and the local binary pattern to extract features and the algorithm Grab cut to segment the images. The novelty in their model is the high generalization capacity, which has been proven and tested on different leaf samples from diverse plant species, reaching an accuracy of 95%. Specifically, 44 combinations of the plant diseases tested out of 46 were classified perfectly, resulting in a 95% total success rate. The resolution of conflicts between classifiers into one class is crucial. This enables correct identification when ambulatory data belong to single or multiple conditions, i.e., more than 50% of the cases have reached 100% identification capacity.
Arora et al. [15] applied a Deep Forest algorithm for maize leaf disease classification from an image set containing three classes of diseased leaves and one class of healthy leaves. Their approach has surpassed deep neural networks in terms of accuracy, as it combines both the robustness of assembled decision trees and the architecture of neural networks. Ubiquitous, automatic detection via mobile devices has become a trend in the area of disease diagnosis. Tang et al. [16] introduce a hybrid lightweight CNN-based approach for identifying grape diseases, such as black measles, and black rot, by leveraging channel-wise attention (CA) mechanisms. Thus, they enhanced the ShuflleNet architectures by using the compression and excitation blocks as the channel-wise attention mechanism as well as the ShuffleNet v1 and v2 architectures as the backbone. Their model was compressed from 227.5 MB to 4.2 MB and tested on a dataset of 4062 grape leaf images that gave a high accuracy of 99.14% in real-time.
Another special type of SNN is the ELM algorithm. In this perspective, Bhatia et al. [17] implemented the Extreme Learning Machine (ELM) based on a TPMD dataset (Tomato Powdery Mildew Disease) collected in real-time for leaf disease prediction, stating that this TPMD dataset was very unbalanced, so they used many resampling techniques such as Random Under Sampling (RUS), Random Over Sampling (ROS) [18], Synthetic Minority Over-sampling (SMOTE) [19], and Importance Sampling (IMPS) to balance the dataset before feeding it into the selected predictive model. Subsequently, ELM algorithms were developed for the unbalanced dataset as well as for the balanced dataset acquired using resampling techniques. The performance analysis of these ELM algorithms is performed using two predictive accuracy measures, namely Area Under the Curve (AUC) and Classification Accuracy (CA). Therefore, the findings indicated that the ELM algorithm works much better for the dataset obtained from resampling techniques. In particular, the better outcome was attained using the IMPS technique, with the maximum values for AUC and CA, or 88.57% and 89.19%, respectively. The power of their Deep Forest model is justified by achieving an accuracy of 96.25%.
2. Materials and Methods
We have conducted a thorough comparative study between cutting-edge DL and ML models for the case study of binary crop disease classification on Python using many libraries such as Numpy, Pandas, Scikit-learn, Keras, and TensorFlow. Moreover, a performance enhancement of the CNN architectures has been undertaken by training the most performing model (obtained in the former step) by using different activation and optimization functions of DL.
2.1. Dataset
Our dataset was drawn from the PlantVillage Dataset [20]. Crop leaf images were taken under different weather conditions with a standard digital camera. They were gathered from many sources, which makes the dataset more diversified and tailored to the application of machine learning algorithms, notably deep learning ones. In total, there are 42854 images of 8 different crops, divided into 23 distinct classes, given as a pair: healthy leaf and diseased leaf.
The crop leaf images are sized at 256 × 256 pixels in RGB color, as shown in Figure 1, which displays a few randomly selected leaf images from the dataset to give a glimpse of the raw image form. Hence, color change, background suppression, and contrast tuning are required for removing any potential bias. The species in this set of images are grape, apple, peach, cherry, strawberry, corn, bell pepper, potato, and tomato. These crop leaves are affected by viral, fungal, and bacterial diseases. The dataset in detail is presented in Table 1. Note that the image distribution is not uniform, presenting a problem of data imbalance.

Figure 1.
An overview of some crop leaf images selected randomly from the PlantVillage dataset.

Table 1.
The detailed dataset was extracted from the PlantVillage dataset.
2.2. Experimental Setup
The experimental study was performed on a Google Compute Engine instance called Google Collaboratory along with a local HP Pavilion workstation having a RAM capacity of 16 GB. The Collaboratory notebook [21] is powered by Jupyter, which runs like a Google Docs object. Moreover, the notebooks are preconfigured with leading machine learning libraries, such as Keras, TensorFlow, as well as Matplotlib. The Google Collaboratory provides access to fast TPUs and operates on Ubuntu 17.10 64-bit. This one is made up of an Intel Xeon processor with 13 GB of RAM and powered by an NVIDIA Tesla K80 processor, 12 GB of RAM, and 2496 CUDA. Our experiments are developed and run in Python, by employing the PyTorch library, taking advantage of automated differentiation on graphs of varying computations. Moreover, we experimented with the PyTorch Zoo model, including different pre-trained models upon the ImageNet dataset. We also used the TPU Collaboratory accelerated runtime, which is suitable not only for accelerating operations such as deep learning but also for handling other GPU-centric applications. TPUs are also known to be faster than GPUs, and each TPU provides up to 180 teraflops of floating-point performance and 64 GB of memory with high bandwidth on a single board.
2.3. Machine Learning Approach
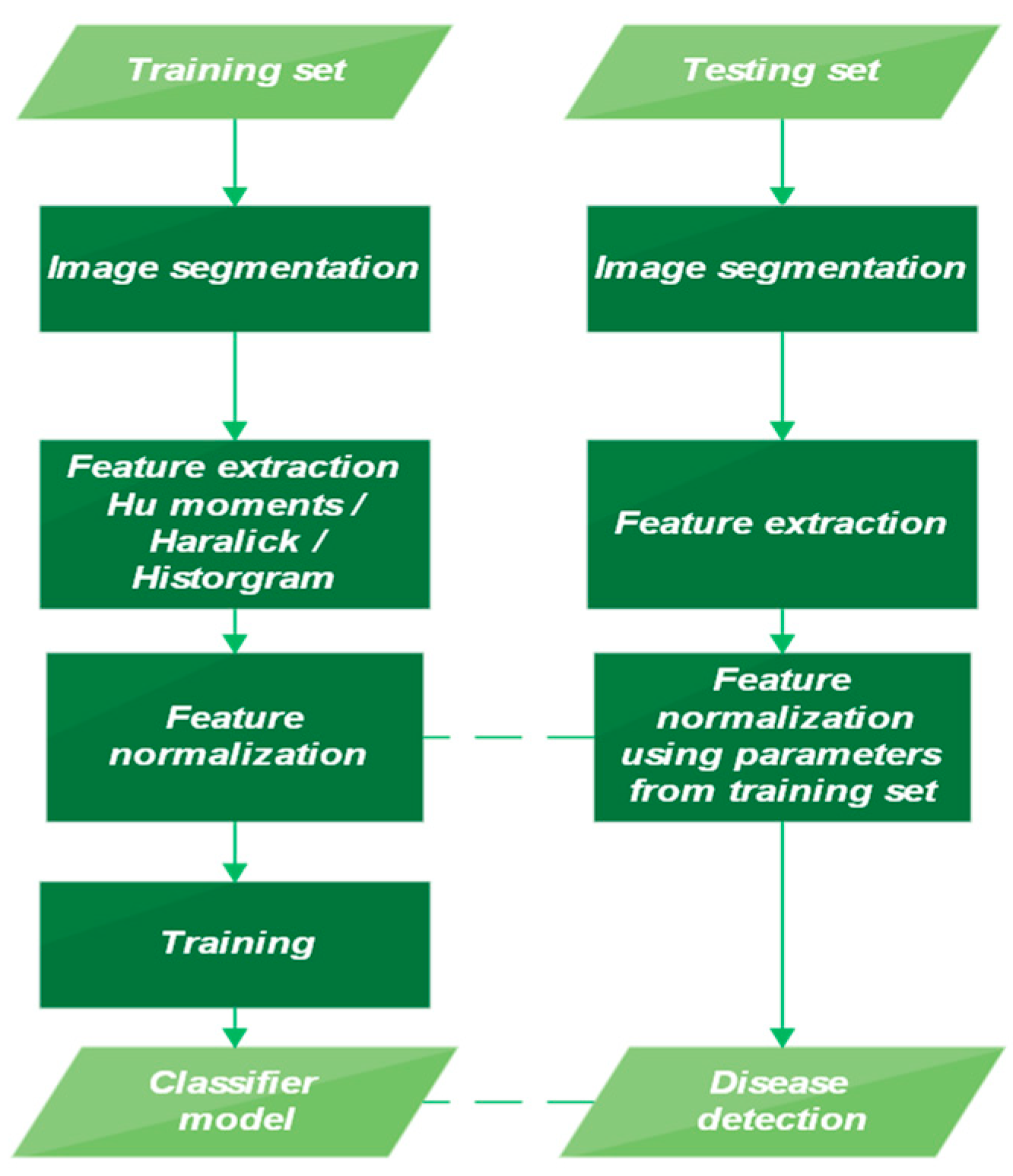
When working with conventional ML algorithms, some basic pre-processing steps must be followed. These core steps are illustrated below in Figure 2.
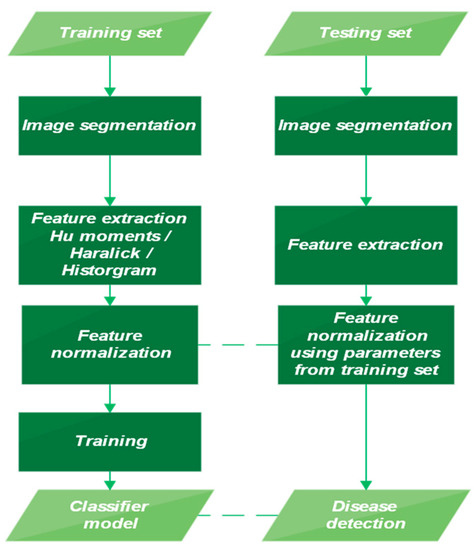
Figure 2.
Diagram illustrating the training and testing phases.
2.3.1. Image Preprocessing
First, the RGB images are resized to a smaller pixel size to accelerate the computations, followed by using Gaussian blur for noise removal. Since the RGB format cannot separate the image’s intensity, it has shadows and lights that make it less suitable for removing the background. So, the RGB crop leaf images are converted to another color space called HSV (Hue, Saturation, Value), which can separate the color from the intensity.
2.3.2. Background Removal and Segmentation of the Diseased Region
Afterward, the habitual preprocessing steps, including removing background and artifacts, are carried out as shown in Figure 3. Indeed, the background suppression step is pivotal in the process because it can bring down the quality of the features retrieved from the crop leaf images. Thus, background elimination is performed to preclude any potential skew in the extracted features. We opted for a mask generation-based technique for segmentation purposes using the color information, color intensity, and brightness of the HSV color space (which is the most straightforward technique for image segmentation as it reduces memory usage and computation time). We thresholded the HSV image for the green and brown color range, separating the images’ areas of interest. Green means that the image samples are healthy, and brown means they are diseased. Thresholding of the HSV images results in a mask for a healthy and diseased leaf image in RGB color space. Then, these segmented images are transferred to feature descriptors as shown in the flowchart below.

Figure 3.
Sample input image (left) and background removal from the leaf (right).
2.3.3. Feature Extraction
Selecting the appropriate features is arguably the most difficult and important part of implementing ML algorithms, which demands a profound analysis as well as proficiency in the concerned domain. In our program, we have deployed three feature descriptors, namely:
Hu Moment’s descriptor [22]: we used it to describe and quantify the shape of objects, which normally stands for the object outlines. Then we converted the color images into grayscale images and calculated the seven invariant moments that are dedicated to the rotation, translation, and object scale change, to recognize the object independently of these factors. These seven moments [23] are given by the following Formula (1).
where represents the relative moments that are centered on the centroid, and which can be computed as stated beneath:
I(x, y) stands for the pixel intensity value at the coordinates (x, y), and p, q are non-negative integers. As well, the centroid is the center of the coordinates (x, y), which we have set as and , respectively.
where is a shape’s regular moment in a binary image defined by:
These seven moments form the feature vector V = [,,,,,,].
Haralick Texture descriptor [24]: We used the Haralick texture descriptor to quantify the texture of the images. The color images must be converted to grayscale for the Haralick descriptor to extract texture features. Moreover, the fundamental concept involved in the calculation of these Haralick texture characteristics is the GLCM gray-level co-occurrence matrix [25] which comprises 13 features. From this GLCM matrix , which is defined on an n * m image I with the (∆x, ∆y) offset as stated below:
Note that the co-occurrence matrix can be seen as a frequency matrix of neighboring pixels in an image I with a lag (∆x, ∆y), where there are two pixels, one with grey level i and the other with grey level j. It is noteworthy that this matrix is symmetric. The textural characteristics are computed based on a statistical theory. Hence, the Haralick texture features are calculated as listed in Table 2 [26]:

Table 2.
Calculation of Haralick texture features.
Table 2.
Calculation of Haralick texture features.
| Angular Second Moment | It denotes the summation of the squares in the grey-level co-occurrence matrix. |
| Contrast | It represents the local intensity difference sum, where i ≠ j. |
| Correlation | It denotes the gray-level linear dependence of adjacent pixels. |
| Squares Sum | Variance |
| Sum Mean (μ) | |
| Inverse Different Moment | |
| Sum Variance | |
| Sum Entropy | |
| Entropy | It indicates the required amount of information on the image for compression. |
| Difference Variance | |
| Difference Entropy | |
| Correlation information measures | |
| where: HXY = − | |
| Furthermore, HX and HY are the (is the marginal likelihood matrix entry obtained by adding the rows) and (stands for the marginal likelihood matrix entry obtained by adding the columns) entropies. | |
| Maximal Correlation Coefficient |
Color Histogram descriptor [27]: We have computed a histogram of 26 bins for each channel and applied the number of pixels per bin as characteristics, then multiplied by three channels, which yields 78 characteristics.
So, after extracting the features from images as shown in Figure 4, Figure 5, Figure 6 and Figure 7, they are stacked together using ‘np. stack’ Numpy function. Then, according to the images located in the file, the labels are encoded in digital format for better understandability by the machine. The dataset is partitioned into training and testing sets with a proportion of 80/20. Following this, we have employed Min–Max scaling. This scaling puts the value between 0 and 1. Feature scalability is a method of standardizing independent characteristics within data to a specified range. It is conducted during data preprocessing to deal with highly variable magnitudes, units, or values. If scaling of features is not carried out, a machine learning algorithm will tend to emphasize large values and regard small values as inferior, regardless of the unit of the values. Hence, once the characteristics are pulled from the images, they are saved into an HDF5 file. Note that the Hierarchical Data Format Version 5 [28] is an open-source file format for handling heterogeneous, enormous, and intricate data. The HDF5 employs a “file folder” framework that enables you to arrange the file data in several structured ways, like how you might arrange directories on your computer. Eventually, the model is trained on 6 machine learning models, and it is vetted through the 10 k-fold cross-validation method.
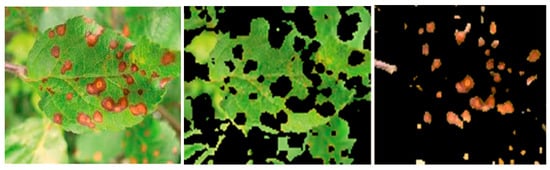
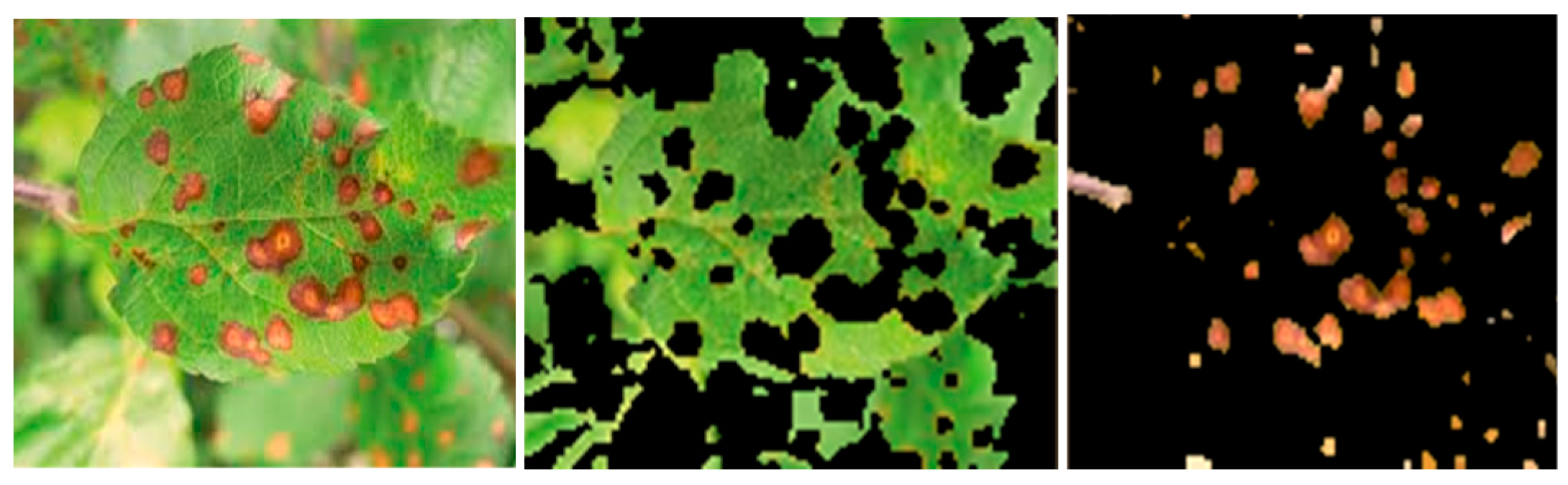
Figure 4.
Sample input image of a Cercospora-affected leaf (left) with its segmented healthy area (middle) and extracted disease spot (right).
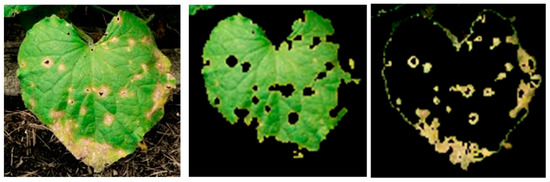
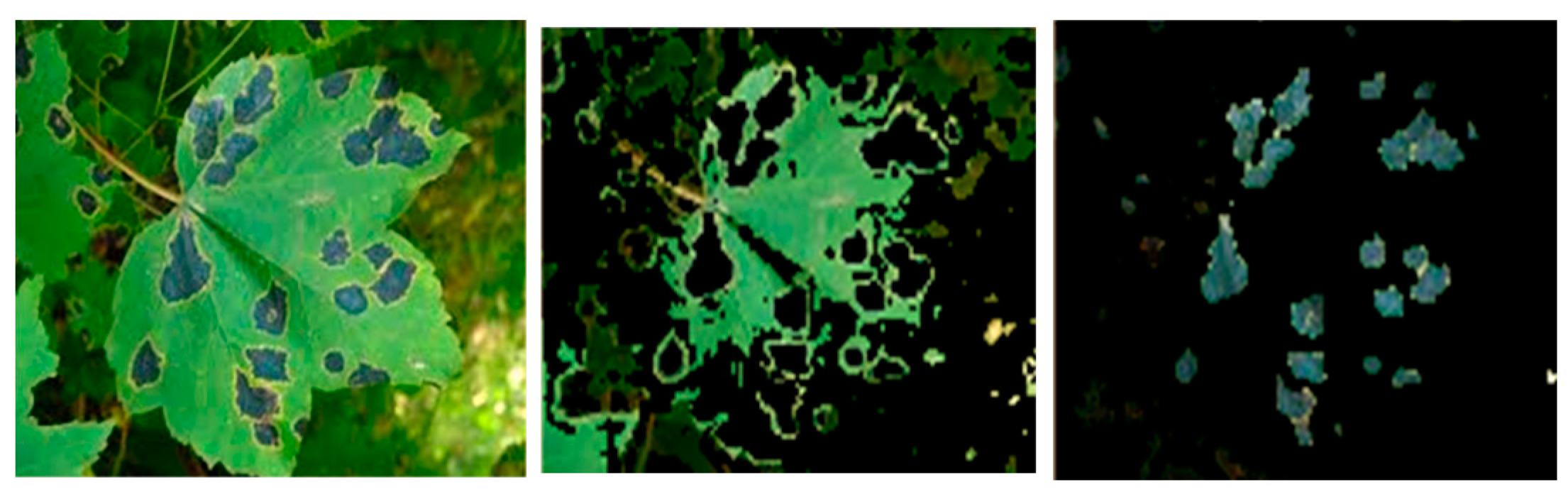
Figure 5.
Sample input image of an Anthracnose-affected leaf (left) with its segmented healthy area (middle) and extracted disease spot (right).

Figure 6.
Sample input image of a fire blight-affected leaf (left) with its segmented healthy area (middle) and extracted disease spot (right).
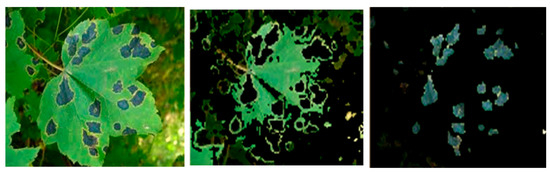
Figure 7.
Sample input image of an early blight-affected leaf (left) with. its segmented healthy area (middle) and extracted disease spot (right).
2.3.4. Classification
Support Vector Machine (SVM) [29] is a supervised learning algorithm that is used to solve regression and classification problems. Classification is performed by setting a separation hyperplane in the function space. In its basic form, it performs a linear classification on two clusters. Thanks to the kernels, nonlinear classification can also be carried out. They are employed to transform the underlying feature space into an infinite or high-dimensional feature space through a kernel function and then to construct an OSH (Optimal Separating Hyper Plane) between the two classes in the transformed space, thus achieving highly nonlinear hyperplanes and gaining expert knowledge of the problem through kernel engineering. Among these great advantages is also the regularization parameter, which encourages the user to avoid overfitting. To build a non-linear support vector classifier, we substitute the inner product (x, y) for the kernel function K (x, y), as shown in (24).
where f(x) sets the belongingness of x. At this point, we suppose that abnormal subjects were tagged as +1 and others as −1. The SVM consists of two parts. Throughout the learning phase, the first phase chooses the basis K (xi, x) = 1, 2…N from the specified kernel set, whereas the second phase builds a linear function in the space. Doing so is analogous to locating the optimum hyperplane in the resulting characteristic space.
Three principal kinds of kernel functions presently exist, whose formula is given in Table 3:

Table 3.
Principal kernel functions and their formulas.
Table 3.
Principal kernel functions and their formulas.
| Polynomial kernel function | |
| The outcome is a polynomial classifier of the rank q | |
| Radial Basis Kernel (RBF) function | |
| Sigmoid kernel function |
Note that polynomial kernel and RBF functions are broadly used owing to their powerful classification capabilities [30]. As a result, we performed numerous setup experiments and discovered that the Radial Basis Function with C = 100 as the normalization parameter produced the best outcomes. The accuracy obtained on the test set is 84.10%, as shown in Figure 8.
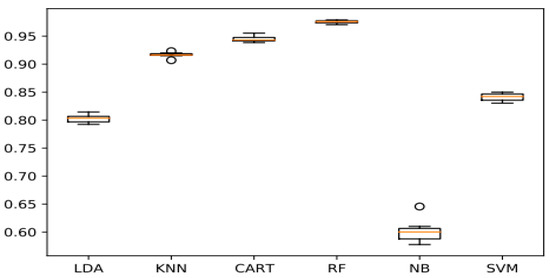
Figure 8.
Comparison of machine learning algorithms.
K-Nearest Neighbors (KNN) [31] is a fairly straightforward algorithm that is frequently used to address classification issues. It does not have a training phase and is non-parametric. When selecting a sample class, KNN looks at its k nearest neighbors and decides which class it pertains to by plain majority rule, assuming that many of the samples in the same class are near to each other in the functionality space. Note that lower values of k enable greater non-linearity but are very susceptible to aberrations. In contrast, higher values of k are not suitable for complex boundaries, but they allow good generalization.
The KNN classification steps are as follows:
: learning data
: class labels
: the unknown sample
Step 1: for i = 1 to n, perform the above steps
Step 2: calculate the distance d (xi, x)
Step 3: Finish for
Step 4: Repeat Step 1 while keeping the indices for the K shortest distances d (, x) in mind
Step 5: Turn the majority label over.
We found that small values of k gave better results, so ranging k from 1 to 9 does not influence the accuracy significantly. The highest outcome is 91.67%, much higher than the one obtained by the SVM.
Random Forest (RF) [32] relies on a heuristic partitioning of the description space. The decision structure of a tree is built on the recursive division of this space, making the ultimate decisions highly dependent on the upstream divisions. The search space of possible decision tree structures is then strongly constrained by these dependencies. A random forest consists of a basic classifier set such as a decision tree presented in:
Random forests consist of a collection of binary decision trees into which randomness has been introduced. RF was defined by Breiman et al. [33] with the following broad definition.
Consider () a set of tree predictors, with independent random variables of . The random forest predictor is acquired by aggregating this set of random trees in the following way:
The average prediction of the individual trees in the regression.
A majority vote among the classification’s prediction trees.
The term random forest arises from considering that individual predictors here are explicitly per-tree predictors as well as each tree relies on a supplementary random variable. Using RF, we achieved better and greater accuracy over other machine learning algorithms 97.54%.
Naive Bayes (NB) [34]: The Naive Bayes algorithm represents one of the most straightforward and efficient ways of classifying. This classifier relies on the Bayesian network concept, which stands for a possible graphical model featuring a random set of variables along with their conditional autonomy. There are many efficient algorithms in Bayesian networks that perform assimilation and learning. The only prerequisite is that the features of the data operate independently. Data features are interdependent owing to their genetic roots. However, the dependence does not seem to be stout. Consequently, classification algorithms may assume that the features are independent and use the Naive Bayes approach. It first generates probabilities for each sequence in the data, then calculates the probable sequence number for each sample and compares it to the number of occurrences in the database. The sample is then ranked based on the number of probable sequences. The Naive Bayes algorithm is a simple probabilistic classifier that uses the Bayes theory and assumes a high level of independence among the data features. The data storage possibilities for X with a category label are:
By applying this classifier to our dataset, we obtained the least precision of 60.09% compared to the other classifiers.
Latent Dirichlet Allocation (LDA) [35] is part of the ML toolkit used to explain sets of observations through the identification of unobserved groups defined by similarities in the data. Using this model, we obtained an accuracy of 80.28%.
Classification And Regression Trees (CART) [36] can be applied to solve classification and regression predictive modelling problems. It serves as the foundation for essential algorithms including random forest, bagged, and boosted decision trees. CART classifier uses a binary tree representation and employs recursive binary splitting to divide the input space. Therefore, an expensive approach is employed to split the space, named recursive binary splitting. The accuracy obtained is 94.45%.
2.4. Deep Learning Approach
DL models represent a subset of machine learning algorithms that use many layers to make feature learning more hierarchical and have been consistently proven to successfully learn extremely intricate models with sufficient data. The learning process has two primary phases [37]:
The input is transformed nonlinearly in the first phase, and the output is built using a statistical model. The second phase aims at fine-tuning the model by using a derivative technique. The network then repeats these two phases several times till it reaches a satisfactory accuracy level. With DL models, no feature engineering is required as these networks are driven by raw data and can train those features as appropriate. Convolutional neural networks (CNN) are frequently employed to solve image recognition tasks. Hence, we compared this model to four other pre-trained models, namely InceptionV3, VGG19, VGG16, and ResNet50.
2.4.1. Image Preprocessing
Data preprocessing is the fundamental milestone in any deep learning approach. Moreover, data augmentation must be performed, so that the system can work properly in all out-of-sample images as well as prevent the overfitting problem from arising. The image size is 256 × 256. Thus, it is essential to resize the input images and subsequently normalize the image pixel values by dividing them by 255 and then proceed to the implementation of different data augmentation techniques, such as rotating the images by using transformations such as affine transformation, turning the data on the horizontal axis, photo brightness, as well as changing randomly the hue and saturation values.
2.4.2. Convolutional Neural Networks CNN
CNN represent a leading category for image recognition and classification [38]. CNN can strongly model nonlinear functions. Unlike SVM and KNN, this one does not converge toward the global optimum. It can also be used directly on raw data without the need for handcrafted features. The artificial neurons in a CNN are inspired by the structure of biological neurons, consisting of a summation term, connection weights, and a nonlinear activation function. The number of hidden layers and neurons in a CNN is often determined through trial and error. Each neuron X is characterized by certain of its bias, weights, and activation function. Images are introduced into the deep learning model through the input layer, where the neuron carries out a transformation as depicted in Equation (31).
X = (weights ∗ input) + bias
An artificial neural network without an activation function is weaker and less suitable for tasks involving complex patterns. Without the activation function, it is equivalent to a linear regression model. So, the proper choice of activation function enables the DL to effortlessly learn very challenging patterns and the following are among the prominent purposes of the activation function:
- − To hold the output restricted to the desired range.
- − To encompass a non-linear functionality in the data.
Different Kinds of Activation Functions
Several activation functions are available in the literature [39,40,41] and some of them are outlined in detail below:
-Sigmoid: It stands for a non-linear function producing output values ranging from 0 to 1, and the resulting output will be non-linear with an identical sign. The function has no symmetry around zero:
-Tanh: It is akin to a sigmoid function but is symmetrical around 0 and produces output values in the range [−1, 1], and the output sign can vary.
The generated output for the sigmoid and tanh functions features upper and lower bounds.
-ReLu: It denotes a rectified linear unit and is a non-linear function. The ReLu function possesses an edge over other functions because it rarely activates all neurons at once. If the output value is inferior to 0, the neuron is disabled. It is substantially more efficient computationally than the tanh and sigmoid functions:
f(x) = max (0, x)
-Softmax: It is built up of several sigmoids. It yields values ranging from 0 to 1 and processes data probabilities related to the class employed in the multiclass task:
-Softplus: It is a sort of conventional function released in 2001. It is differentiable and simple to prove thanks to its derivative.
This is an optional replacement for the dead ReLu. The outcome ranges from [0 to ]:
-Softsign: It resembles the tanh much more; except that tanh is exponentially converging while Softsign is polynomially converging. The resultant outcome lies between [−1, 1]:
-Swish function: It is one of the earliest hybrid combination AFs proposed, combining the input function and the sigmoid AF to produce a hybrid AF. It was introduced by Ramachandran et al. [42] and uses an autonomous search technique based on reinforcement learning to compute the function. The Swish function has properties of non-monotonicity, and smoothness being bounded below and unbounded above. Its smoothness makes it effective for training deep learning (DL) architectures and improving generalization and optimization outcomes.
-Leaky ReLU: It was introduced to address the issue of “dead neurons” in the rectified linear unit (ReLU) by adding a small negative slope to keep weight updates active throughout the entire propagation process [43]. The parameter was then introduced to ensure that gradients never reach zero during training. LReLU calculates the gradient with a low fixed value for the negative gradient within a range of 0.01, so the LReLU function is calculated as below.
-Exponential Linear Units (ELUs): ELU is a variant of the rectified linear unit (ReLU) proposed by Clevert et al. [44]. It is used to speed up deep neural network (DNN) training and has the advantage of overcoming the vanishing gradient problem by using the identity function for positive values and enhancing learning features. ELUs also have negative values that push the average unit activation closer to zero, reducing computational complexity and improving training speed. ELUs are a good alternative to ReLUs because they can reduce bias shifts by driving the average activation toward zero during training.
The ELU is expressed as:
The gradient or derivative of the ELU equation is provided as follows:
where hyperparameter α = ELU monitors the point of saturation for the negative net inputs and is typically set to 1.0. There is a well-defined saturation plateau in the negative mode of the ELU, which allows learning robust representations and provides faster learning and higher generalization than LReLU and ReLU with a particular network structure, especially over five layers, and ensures state-of-the-art results regarding the ReLU variants. However, a critical restriction of the ELU is it fails to center the values at zero as well as the parametric ELU was suggested to solve this problem [45].
Table 4 provides a detailed comparison of the different activation functions used in this study, including their advantages and disadvantages.

Table 4.
The pros and cons of the applied activation functions.
Table 4.
The pros and cons of the applied activation functions.
| Activation Function | Advantages | Disadvantages | When Might It Be Used? |
|---|---|---|---|
| Sigmoid [41] | -Smooth gradient, avoiding “skips” in the output values -Consistently differentiable | -Decreasing gradient: It refuses to learn more and is quite slow to reach an accurate prediction. -Computationally demanding | If the output lies between (0,1) then the sigmoid may be employed |
| Tanh [41] | -Centered on zero -Continuously differentiable at any point | -Vanishing gradient issue -Since the function is nonlinear, it can effortlessly backpropagate errors/faults | If the output lies between (0,1) or (−1, 1), then the tanh can be used |
| ReLU [41] | -It is computationally efficient and allows the net to get together quickly -Backpropagation is allowable -Non-linear: Although it possesses a derivative function | -Once the inputs contact zero/negative, the gradient becomes zero, making the network unable to perform backpropagation and failing at learning -Unlimited and undifferentiable at zero | ReLU is widely used, when we want to predict output values higher than 1 because tanh or sigmoid are not appropriate for this purpose |
| Softmax [39] | -Proficient in managing different classes, single class among other functions -Provides the probability of the input value to be in a particular class. | -Does not consider the rejection of null values -This is unlikely to work if data are non-linearly separable | when predicting a probability for a multi-class task, the Softmax function must be implemented in the last layer |
| Softplus [41] | -Soft derivative employed in backpropagation and it is equivalent to a sigmoid function | -Operation not as affordable as ReLU | Rather never |
| Softsign [41] | -Better and faster learning due to the absence of difficulties related to the vanishing gradient -Softsign prevents neuron saturation, which enables much more efficient learning. | -Frequently, the gradient yields either an extremely high or low value -More costly to calculate than tanh | Rather never |
| Swish [42] | -It yields an efficient propagation of information during training and has an improved accuracy as it is devoid of leakage gradient problems | -It is very costly in terms of calculation. | It is used in very deep networks, when the number of hidden layers is high (nearly 30) |
| LeakyReLU [42] | -Seeks to overcome the “dead neuron” -Straightforward implementation and low-cost operation | -Large gradients can change the weights so that neural units are permanently disabled (never activated). | To be used only if you expect a “dying ReLU” problem, so it should be applied in hidden layers. |
| ELU [44] | -It can deliver negative outputs -Resolves both the vanishing gradient and the dying ReLU problem | -Computationally demanding -For x 0, it can jump the activation along with the output range of [0, ] | When the risk of overfitting is high, and it should be used in hidden layers. |
Convolutional neural networks (CNNs) classify incoming images into specific categories (e.g., healthy or affected) by processing them as pixel arrays with a resolution represented as h × w × d (h = height, w = width, d = dimension). For example, an image with dimensions 8 × 8 × 3 is a matrix of RGB values, while an image with dimensions 4 × 4 × 1 is a matrix of grayscale values. The CNN applies a series of convolutional layers with kernels to extract features from the input image using the convolution mathematical function, defined as the continuous function of two functions f and g:
The analogous convolution function in the discrete state is stated as such:
The above 1D convolution for the numerical image can be expanded to a 2D convolution, such as:
In this application, a kernel or filter (called the “g function”) is applied to an input image (called “f”). The process of applying the kernel to the image is known as 2D convolution, which involves sliding the kernel over the image and performing a convolution operation at each point. This produces a 2D array called a feature map. The feature map is then processed by a nonlinear activation layer, which determines whether a neuron should be activated based on the weighted summation and bias. Activation functions that can be used in this layer include ReLU, Leaky ReLU, ELU, and Softmax. The purpose of determining the activation function is to illustrate non-linearity in the neuron’s output. Spatial pooling also termed down-sampling or subsampling, decreases each feature map’s dimensionality while retaining the relevant information. It also helps to prevent overfitting by reducing the number of computations and parameters. There are various types of pooling, such as average, sum, and maximum pooling. The final layer in a convolutional neural network (CNN) is a dense or fully connected layer, where every neuron receives input from all the neurons in the previous layer.
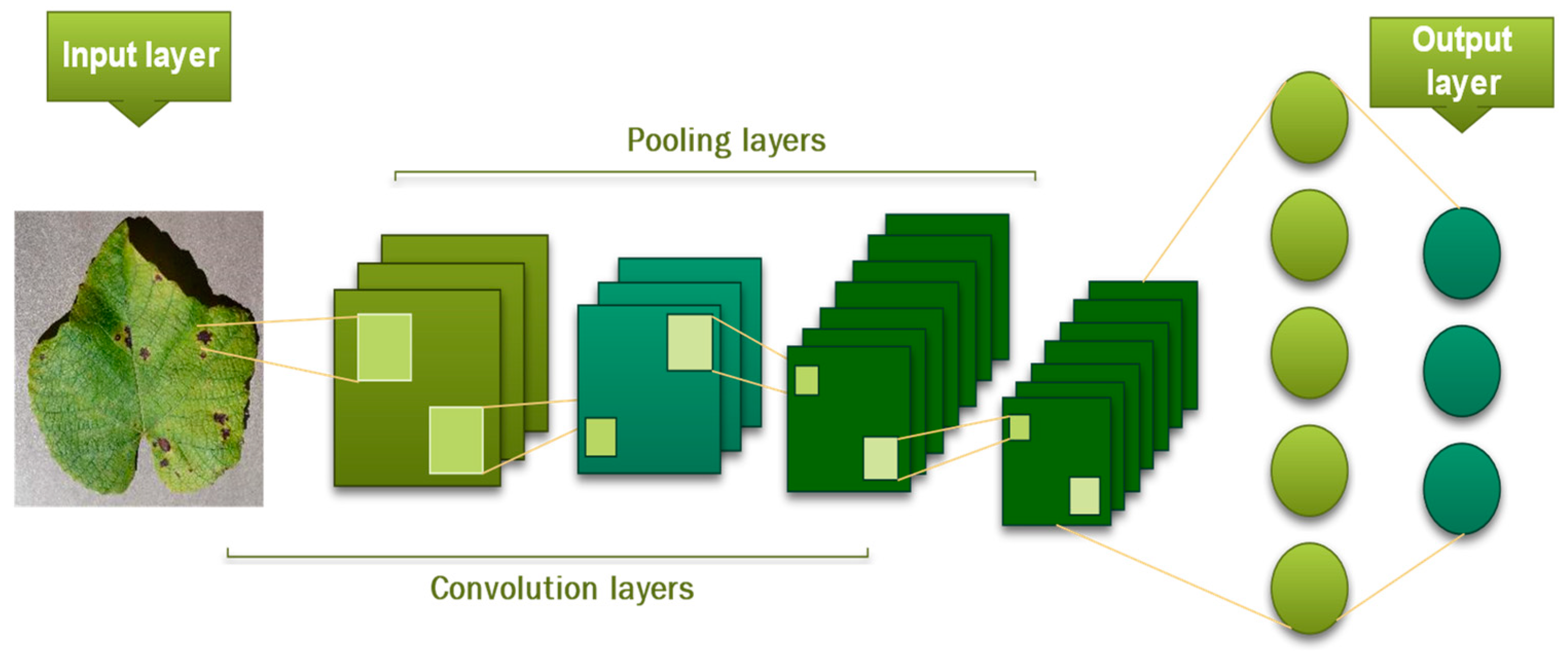
This layer produces the model’s output with probabilities between 0 and 1 [46]. The model depicted in Figure 9 represents a complete CNN process for input image classification according to assigned values.
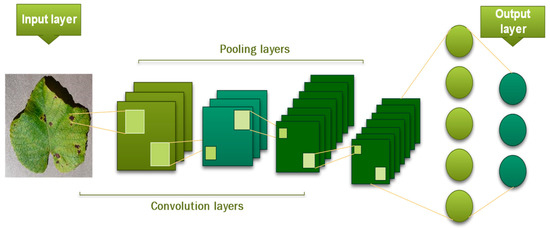
Figure 9.
The architecture of CNN.
The key configuration settings for a convolutional neural network (CNN) include: The number of concealed layers, the activation function used in each layer, the method of regularization, the optimization technique, the technique of regularization, and method of optimization.
In this work, we not only compared several activation functions that exist in the DL literature and applied them to leaf disease detection tasks, but also implemented several DL optimizers and compared them to our target problem of classifying leaf diseases.
Optimization Method
SGD optimizer was employed to train all CNN architectures in the first step. After obtaining the best DL model, an attempt was also made to upgrade the plant disease classification results. To this end, we applied five cutting-edge DL optimizers to train the InceptionV3 architecture that achieved both the best f1-score as well as validation accuracy over 10 epochs at the first stage of the parsing process. Some specifications of these optimizers are presented below:
-SGD: It is one of the most straightforward deep learning optimizers. It requires the rate of static learning for all parameters throughout the learning process and has a fast convergence capability [47].
-Adagrad: This employs various learning rates for each model parameter. It adjusts the learning rate in accordance with the update frequency of each parameter [48].
-RMSProp: To shorten the learning time seen in Adagrad, the RMSProp optimization functions have been suggested and its learning rate decreases exponentially [49].
-Adadelta: It represents an expanded version of the Adagrad optimizer that stacks up against prior gradients over a frozen time frame, ensuring continual learning even after numerous repetitions. It leverages the Hessian approximation to provide the current direction inside the negative gradient and suppress the update rule’s learning rate [50].
-Adam: The Adaptive Moment Estimation optimizer estimates first- and second-moment adaptive learning rates of gradients for several parameters [51]. It combines the benefits of two expanded forms of the SGD optimizer, namely Adagrad as well as RMSProp. Unlike RMSProp, it averages the second gradient moment and also employs the preceding gradients to accelerate learning [51].
Optimizer Training Specifications
The PlantVillage dataset was used to train all the CNN architecture-based models from scratch. The random search approach was used to tweak the deep learning optimizers’ hyperparameters [52]. The problem of internal covariate shift arises in the neural network due to the variation of the input data distribution owing to a modification in the previous layer’s number of parameters. This issue was solved by using batch normalization, which is a highly efficient and successful technique for a high learning rate [53].
In the first step, for training all CNN architectures, the ReLU was used due to its computational efficiency [54,55] and its ability to diminish the likelihood of gradient vanishing. Table 5 lists the parameters of all DL optimizers.

Table 5.
Hyperparameters of DL optimizers.
Table 5.
Hyperparameters of DL optimizers.
| Optimizers | Specifications |
|---|---|
| SGD [47] | -weight decay = 0.0005, momentum = 0.9, nesterov = False, lr = 0.001 |
| Adagrad [48] | -epsilon = 1 × 10−7, lr = 0.001 |
| RMSProp [49,56] | -epsilon = 1 × 10−7, rho = 0.9, lr = 0.001 |
| Adadelta [50] | -epsilon = 1 × , lr = 0.001, beta1 = 0.9, beta2 = 0.999, amsgrad = False |
| Adam [51] | -epsilon = 1 × , beta1 = 0.9, beta2 = 0.999, lr = 0.002 |
Hyperparameter Tunning
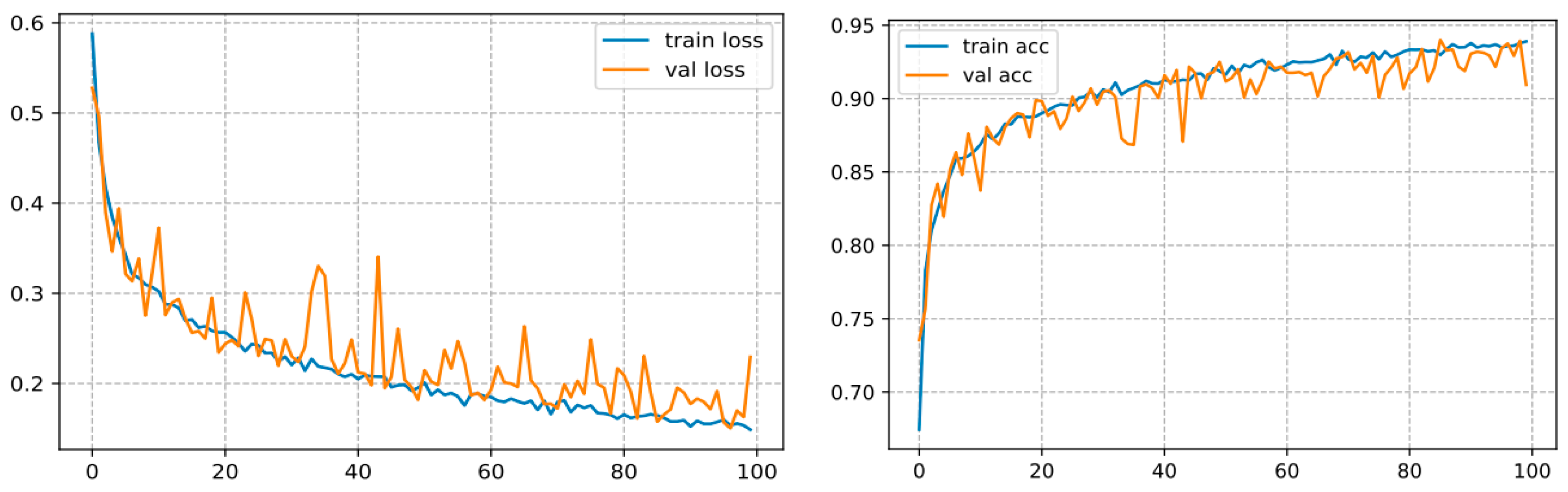
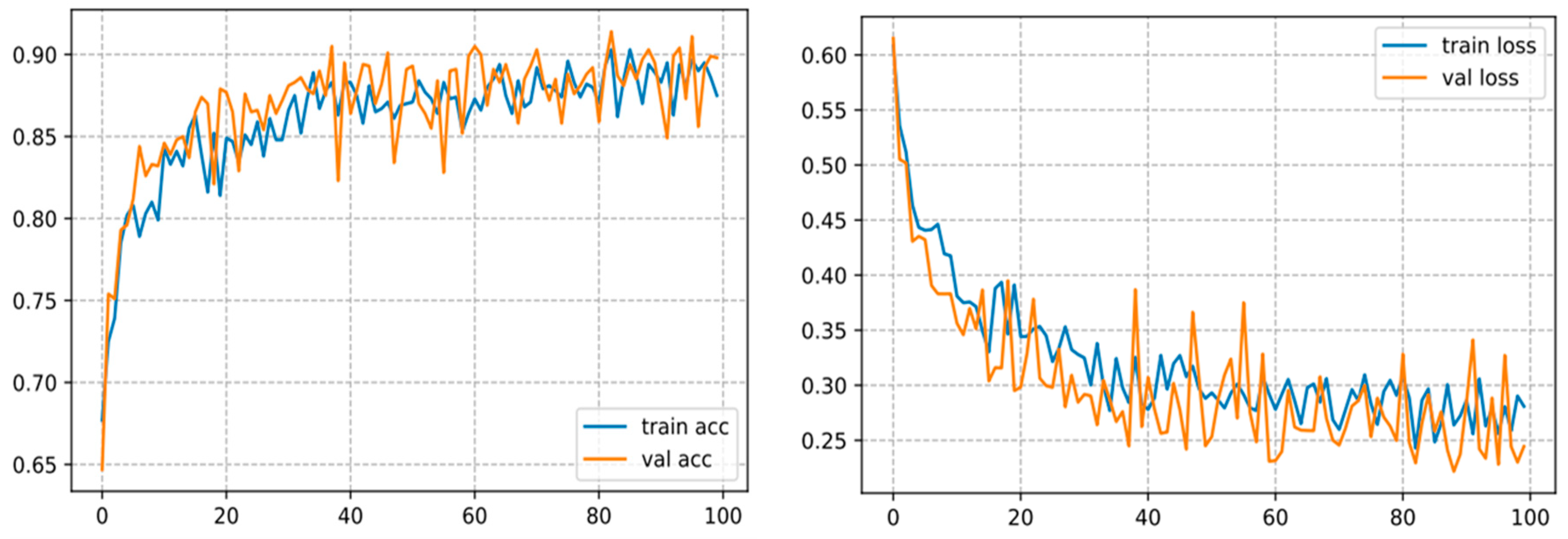
The backbone of any deep learning model is the tuning of the hyperparameters. Finding the best parameters is an incredibly laborious task, requiring numerous experiments to be undertaken when building the model. The hyperparameters comprise the batch size, loss function, epoch number, and learning rate, as well as the optimizer, which are generally regarded to tune the model. For the sake of building the classification model for three classes, a specific range of each hyper-parameter is considered. Thereby, several experiments were carried out to build an efficient model. After running some experiments with varying the given hyperparameters, it was inferred that the accuracy of the model strongly depends on the learning rate, batch size, epoch number, and the dataset size. In the present research, the following hyperparameters were used: batch size = 32, optimizer = RMSProp, loss function = binary cross-entropy, decay learning = 0.0001, initial learning rate = 0.01, epochs = 100. By employing these parameters, the model yielded satisfactory classification accuracy as shown in Figure 10 on our test set. In our setup, we used a CNN with ten hidden layers, we firstly inserted a 2D convolutional layer with 32 filters, a 3 × 3 kernel, the input size as the dimensions of our image, which is 256 × 256 × 3, and the ReLU (Linear Rectified Unit) as the activation functions in the hidden layers, then we added Leaky ReLUs because this one solves the problem of the dying linear rectified units. Next, we added a maximum pooling layer with MaxPooling2D () that halves the image size, and so forth. We stacked up 10 of these layers together, with each subsequent CNN adding more filters (32, 64,128, etc.). Lastly, we flattened the output of the CNN layers, fed it into a fully connected layer, and then into the final dense layer, which is a sigmoid activation function with 2 units, which is necessary for addressing a binary classification task. For the specification setup when training the model. We trained our CNN model with the binary cross-entropy loss, which assesses the model performance and whose output is a likelihood value ranging from 0 to 1. Moreover, we used the RMSProp optimizer. Indeed, the RMSProp is a judicious optimization algorithm as it automates the learning rate tuning for us. We appended the accuracy to the metrics so that the model monitors the accuracy during training. This configuration gave us an accuracy of 93.89% as illustrated in Figure 10 on our test set.
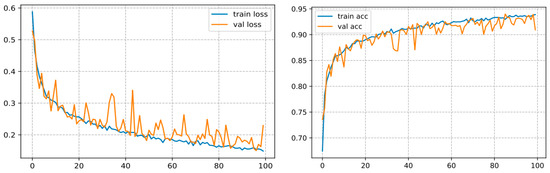
Figure 10.
Curve represents the loss and accuracy value for the CNN model.
2.4.3. ResNet50
The ResNet50 model stands for Residual Networks, which was a baseline in the ILSVRC and COCO classification competitions carried out in 2015 and was presented by He et al. [57]. Their architecture is the winner, having an error rate of 3.57%. The multiple nonlinear layers’ inability to make identity mappings learnable and the concern of degradation prompted the framework of deep residual learning. It is a robust deep learning architecture relying on numerous piled-up residual units. The latter constitute the building set of blocks employed to build the network. These residual units are composed of convolution and pooling layers. The architecture is quite like the VGG network made up of 3 × 3 filters, except that ResNet is around 8 times more profound than the VGG architecture. This is due to the use of global pooling layers instead of fully connected layers. ResNet was updated again [58] to achieve higher accuracy by upgrading the residual module to employ identity mappings. Note that the number beside ResNet indicates the number of network layers, and we took ResNet50, implying that it has 50 layers for processing. The ResNet50 receives inputs in 256 dimensions, and the prime benefit of this architecture is the embedding of skip connections, that also aids to address the issue of vanishing gradient descent. In this paper, we deployed the ResNet50 architecture with pre-trained weights on the ImageNet dataset.
The following are the parameters used:
- DL Optimizer: Stochastic Gradient Descent (SGD).
- Shear range: 0.2.
- Activation function: ReLu/Sigmoid.
- Loss function: Binary-cross-entropy.
- The number of epochs: 100.
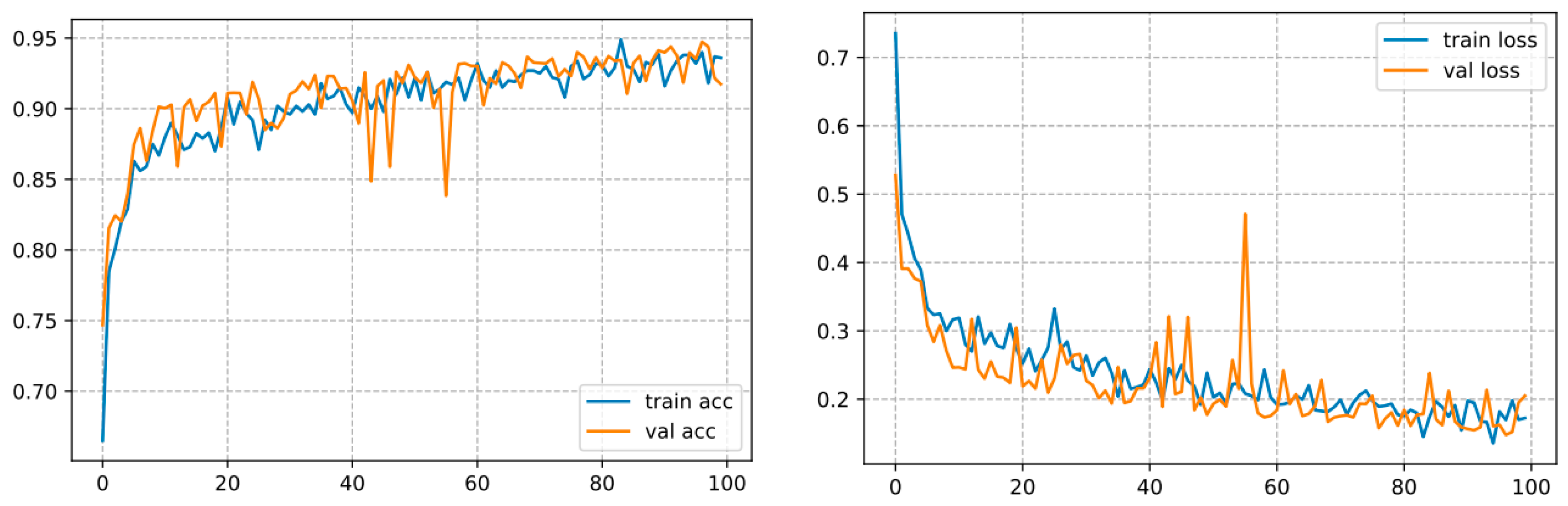
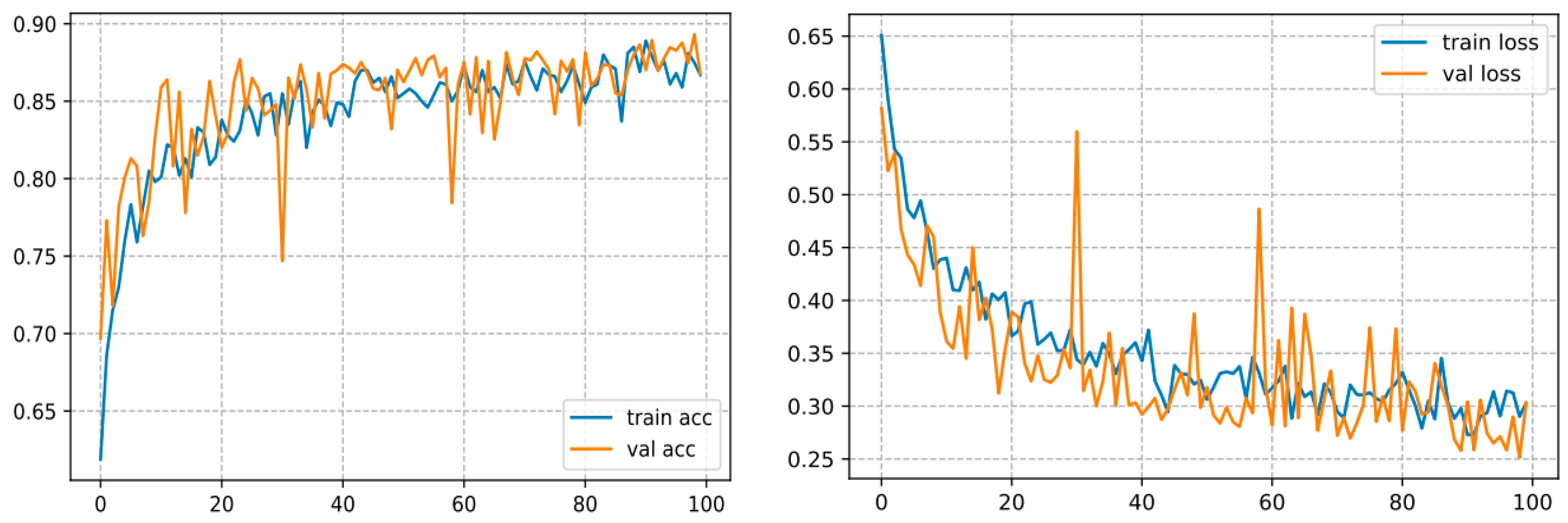
This one converges over 100 epochs and reaches an accuracy of 93.57%, as shown in Figure 11.
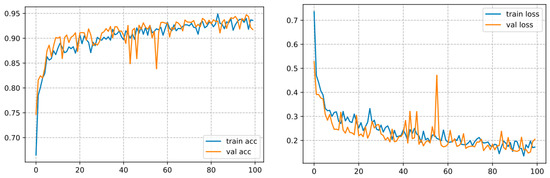
Figure 11.
Curve represents the loss and accuracy value for the ResNet50 model.
2.4.4. InceptionV3
Inception vN was firstly presented by Szegedy et al. [59] in the GoogLeNet architecture with N pointing to the version number. Szegedy et al. proposed the InceptionV3 network, which provides upgrades to the module of Inception to similarly increase the classification accuracy of ImageNet. This Inception module consists of basic asymmetric and symmetric components, including convolutions, maximum pooling, medium pooling, dropping, fully connected layers, and concatenations. Batch normalization is widely used in InceptionV3 and applied to the activation inputs. Loss is primarily calculated via Softmax. We employed the InceptionV3 architecture with pre-trained weights on the ImageNet dataset.
Following are the parameters used:
- DL Optimizer: Adam
- Shear range: 0.2
- Activation function: Sigmoid
- Loss function: Binary-cross-entropy
- Number of epochs: 100
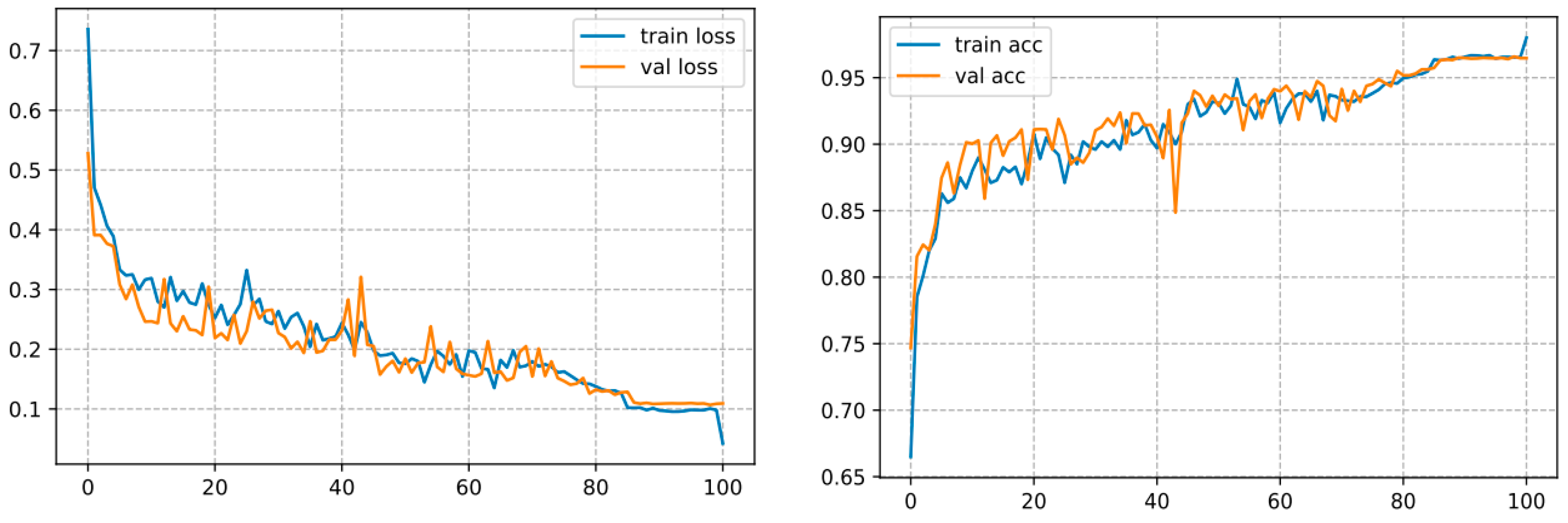
This architecture converges over 100 epochs and reaches an accuracy of 98.01%, as shown in Figure 12.
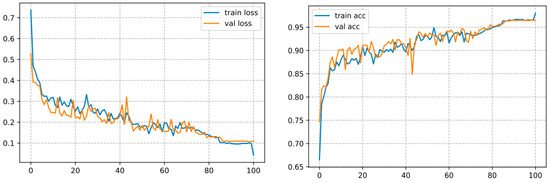
Figure 12.
Curve represents the loss and accuracy value for the InceptionV3 model.
2.4.5. VGG16
VGGNet is a CNN architecture introduced by Simonyan et al. [60] for the ILSVRC-2014 competition. The model achieved an accuracy of 98.09% with an error rate of 7.5%, placing it in the top five in ImageNet. It is a dataset including over than 14 million images pertaining to 1000 classes. The VGG16 model was trained over a period of weeks using NVIDIA Titan Black GPUs. This is an improvement over AlexNet, which uses numerous 3 × 3 kernel filters instead of large kernel filters. Typically, this network is described by its simplicity, as described by Simonyan et al. [60] along with just 3 × 3 convolutional layers piled on top of each other in ascending profundity. The max-pooling reduces the volume size. Furthermore, two fully connected layers, each containing 4096 nodes and a Softmax function. The finetuning of VGG16 was performed by removing the original Softmax classifier and replacing it with our own. We employed the VGG16 network with pre-trained weights on the ImageNet dataset.
Following are the parameters used:
- DL Optimizer: RMS prop.
- Shear range: 0.2.
- Activation function: Sigmoid.
- Loss function: Categorical-cross-entropy.
- Number of epochs: 100.
This architecture converges over 100 epochs and achieves an accuracy of 87.50%, as shown in Figure 13.
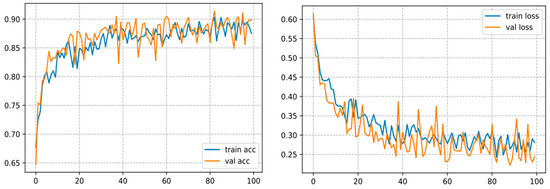
Figure 13.
Curve represents the loss and accuracy value for the InceptionV3 model.
2.4.6. VGG19
VGG19 is an alternative variant of VGGNet consisting of 19 weight layers and includes sixteen convolutional layers, three fully connected layers, five MaxPool layers, and one SoftMax layer. It was developed for large-scale visual recognition. The primary upside of this network is that its coding script is open source, and we could feasibly implement transfer learning and run the architecture for further networks. Moreover, the network can learn small collective kernels instead of a single large kernel, as including multiple small kernels enables the network to learn intricate features. We employed the VGG19 network with pre-trained weights on the ImageNet dataset.
Following are the parameters used:
- Optimizer: RMS prop.
- Shear range: 0.2
- Activation function: Sigmoid.
- Loss function: Binary-cross-entropy.
- Number of epochs: 100.
It converges over 100 epochs and reaches an accuracy of 86.70%, as shown in Figure 14.
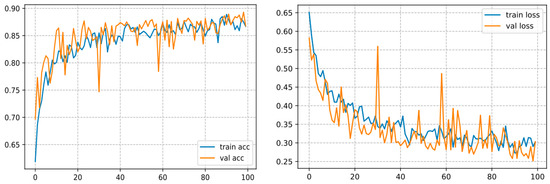
Figure 14.
Curve represents the loss and accuracy value for the VGG19 model.
3. Experimental Results
This part firstly reports on the comparative analysis of six classical ML algorithms and five architectures based on the CNN model to select the best algorithm, and then the findings regarding the performance improvement of the best fitting model (InceptionV3) using a variety of activation functions and DL optimization algorithms.
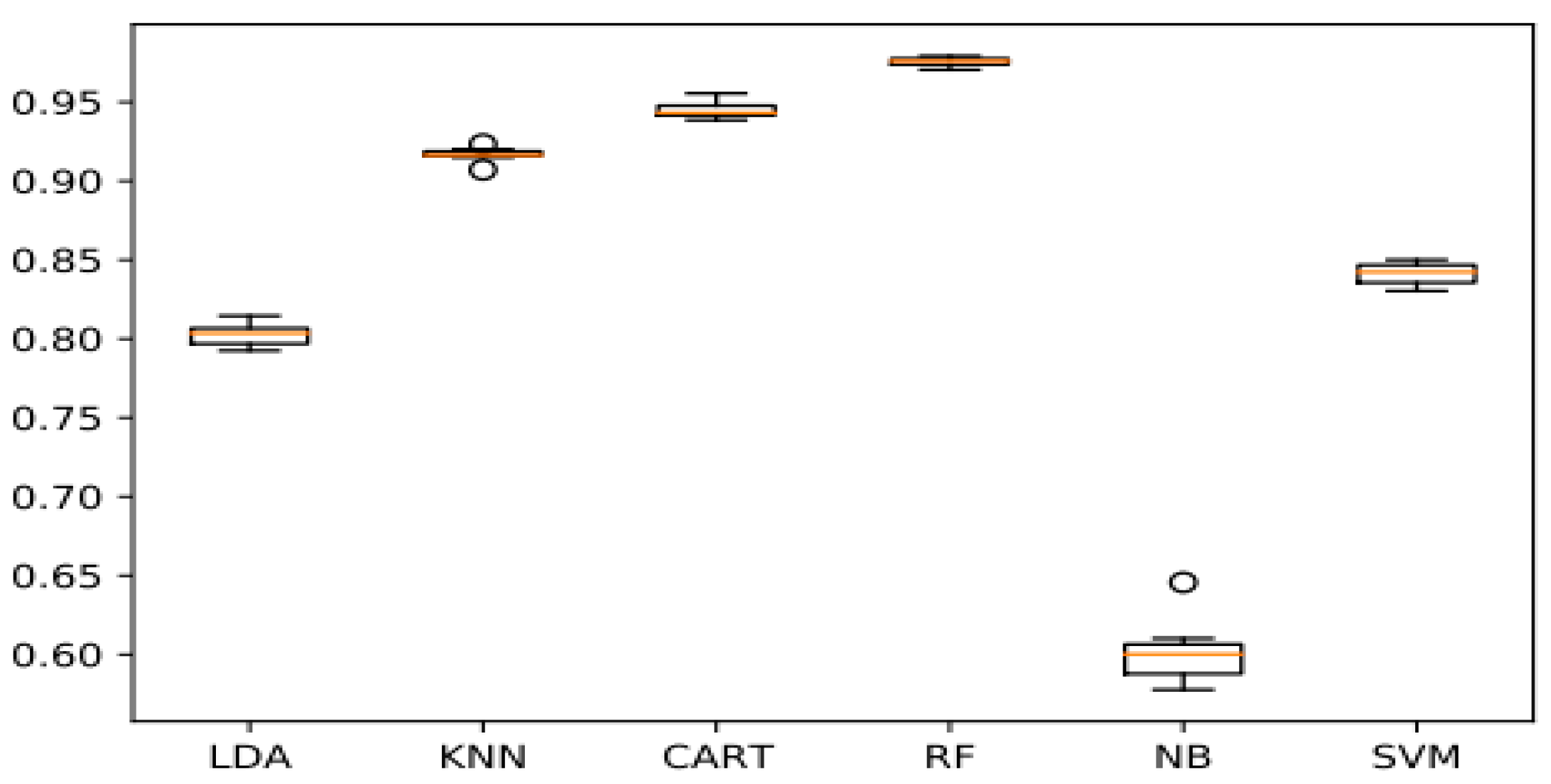
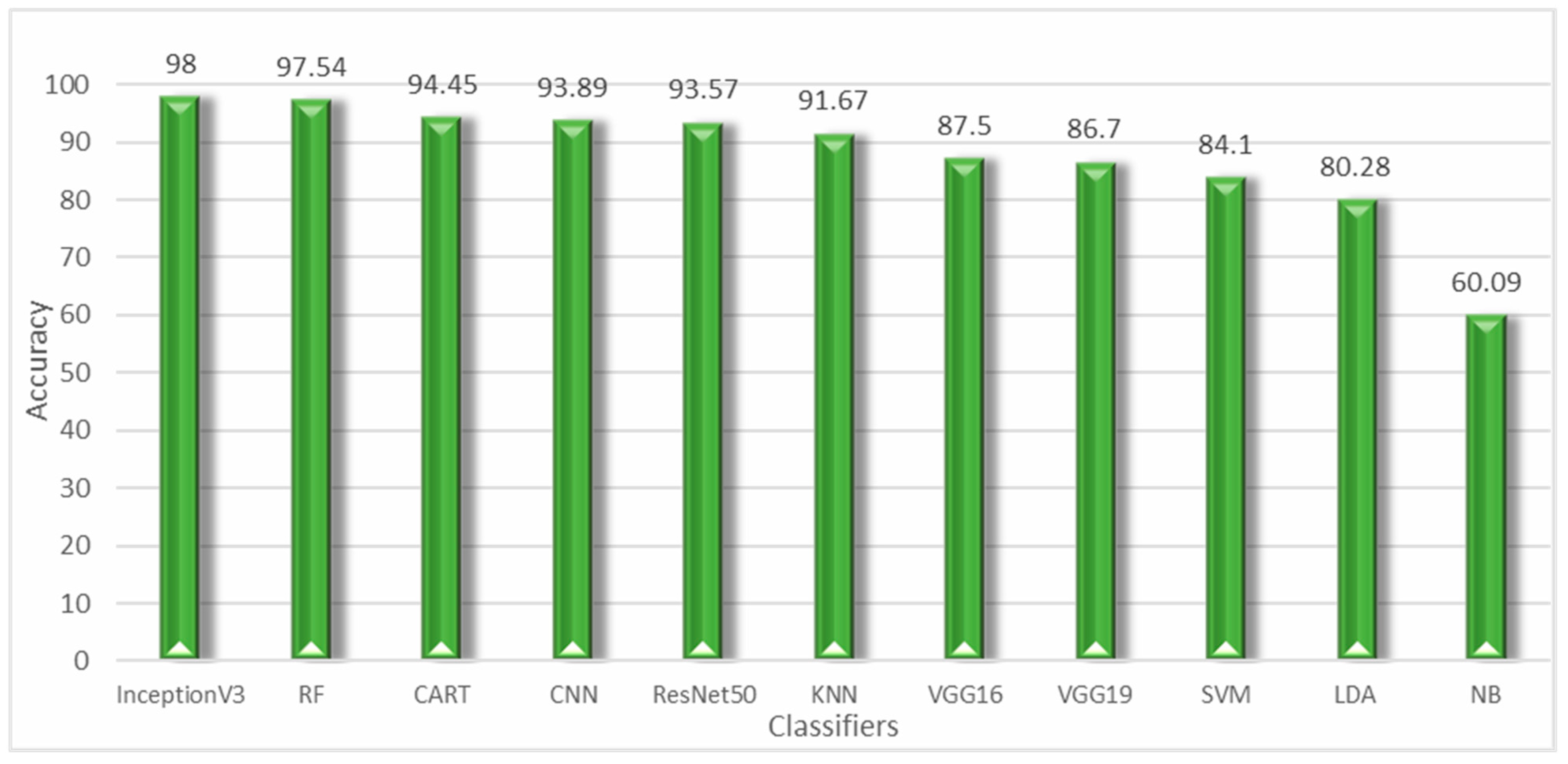
A detailed description of the parameters used has been given in the prior section. The ML and DL models were implemented in Python, through the scikit-learn library for the conventional algorithms and Keras as well as TensorFlow for the DL model. The program was run on the Google Colaboratory notebook, which provides freely available GPU, TPU, and CPU resources. The classical algorithms were trained on the GPU, while the DL model was trained on the TPU. The dataset was split into training and testing sets in an 80–20 ratio (We utilized 80% of the data for training and 20% for testing). For performance metrics, we employed f1-score, accuracy, recall, precision, the ROC curve, and the confusion matrix. Thus, the results are shown below in Figure 15. From the experimental findings, we found that the NB classifier achieved an accuracy of 60,09%, much lower than other classical ML algorithms. Unlike the RF model, with an accuracy of 97.54%, it was able to far outperform all other ML algorithms. Whereas CART and KNN produced results that were roughly comparable, ranging between 94.45% (for the CART) and 91.67% (for the KNN). On the other hand, for models based on deep transfer learning. The InceptionV3 model is the most efficient, with a precision of 98.01%, so it was able to outperform CNN from scratch and other pretrained models. This is due to its ability to handle both missing and unbalanced data without forgetting that its execution time is quite fast compared to other DL models.
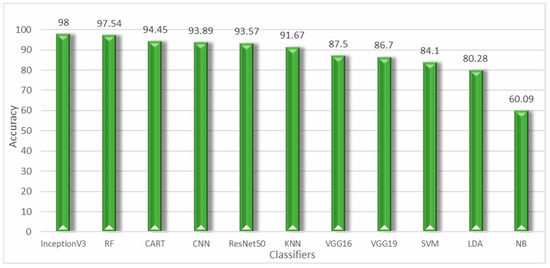
Figure 15.
Chart of machine learning and deep transfer learning models.
3.1. Performance Measures
The evaluation of the trained models’ output was elaborated using several performance measures, namely f1-score, accuracy, recall, and precision, as reported in Table 4, and AUC (Area Under ROC), which is determined by the true negatives (TN), true positives (TP), false negatives (FN), and false positives (FP) obtained from the confusion matrix.
- Precision: It depicts in binary classifications all the positive classes predicted correctly by the model; how many of them are positive. It is computed by dividing the number of correctly classified positive samples by the number of predicted positive examples. The formula is expressed as follows:
- b.
- Recall/or sensitivity [61]: It sets the number of correctly predicted samples among all the positive classes. The equation is stated as below:
- c.
- F1-score [62]: The F1 score yields a global estimate of a test subject’s recall and accuracy. It refers to the harmonic average of recall and precision. Formally, the F1 score is determined by the following:
- d.
- Accuracy: This is a metric for assessing classification models. It is a fraction of all correct predictions. Formally, it is expressed as below:
These various performance metrics are applied to predict the outcome of the different models, and a list of them is presented in Table 6. From this exhibit Table, we can see that the metrics of the InceptionV3 network outperformed all the measured machine learning algorithms. For instance, the recall of the tested machine learning algorithms was 92.26% (KNN), 84.37% (SVM), and 97.47% (RF), while it was 97.53% (InceptionV3), 91.64% (ResNet50), 91.39% (CNN), and 90.17 (VGG16) for the evaluated deep learning networks.

Table 6.
The testing set performance of 12 algorithms is addressed in this benchmarking study.
To be noticed, we are highly concerned about the sensitivity/recall, which calculates the percentage of true positives (diseased leaves). For example, if a leaf infected (true positive) is predicted to be healthy (predicted negative), the consequences will be extremely onerous. All evaluated metric values were superior to 60.09% of the tested machine learning algorithms and superior to 86.70% of the tested deep learning models. The classification outcomes of the RF classifier were the best among the six machine learning algorithms examined, followed by the CART classifier and lastly the KNN classifier. Concurrently, the ranking order of the classification outcomes of the tested deep learning models, from top to bottom, is InceptionV3, CNN from scratch, ResNet50, VGG16, and finally VGG19.
- e.
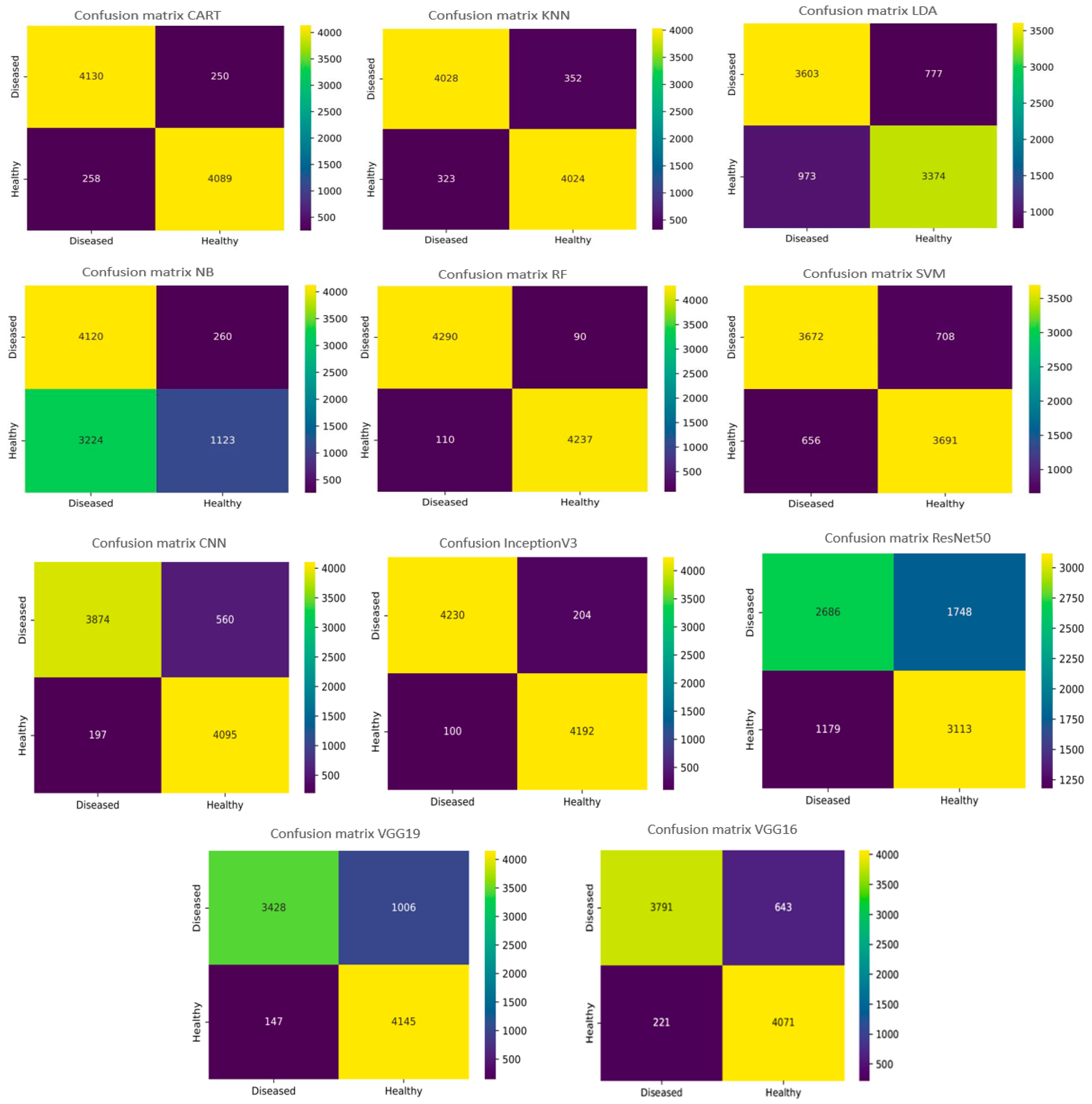
- The confusion or error matrix [63]: It describes the classifier’s performance on the test data. It helps to identify and pinpoint each cluster that might have been misclassified by the classifier and to further improve the proposed classification model in the future. Each row represents the predicted class examples, and each column of the matrix represents the actual class examples. Thus, we computed the confusion matrix of each model to compare the different algorithms, as it allows us to measure the degree of classification model accuracy for each category. As we are dealing with a binary classification of leaf disease, we are interested in false negatives (FN). This is also called “type 2 error”, which represents the rate of misclassified crop leaves that appear healthy but are affected by diseases, thereby presenting a severe threat to the crop, especially if it concerns a viral disease that will spread swiftly over the field. To gain clearer perspective of classification findings, we employed confusion matrix plots and ROC curves (receiver operator characteristic) to depict the distinct crop binary classification results with distinct ML and DL algorithms.
Figure 16 illustrates the confusion matrix graphs of the six machine learning models, as well as the five DL networks. In a confusion matrix chart, the abscissa represents the predicted label, and the ordinate represents the actual label. The confusion matrix diagonal contains the correctly classified instance data, and the values over/under the diagonal are the misclassified instances. As depicted in Figure 16, with the eleven machine and deep learning models, there were many instances (<300) where diseased leaves were misclassified as healthy (False Negative) on a test set of 8727. For the RF algorithm, the number of diseased crop leaves misclassified as healthy is 90. On the other hand, the number of healthy crop leaves misclassified as diseased is 110. Thus, the number of misclassified leaves is 200 (FN + FP) with an accuracy reaching 97.54%, which demonstrates its reliability in plant disease classification compared to other ML algorithms such as the NB algorithm, where 260 diseased leaves were misclassified as healthy. The results revealed that the NB algorithm is the least recommended for leaf disease classification regarding accuracy and FN. Regarding the deep learning networks, the InceptionV3 network was found on top as the number of leaves that were diseased but predicted to be healthy (FN) was 204, and otherwise for the FP was 100 so the total number of misclassified leaves (FP + FN) was 304 out of a dataset of 8727.
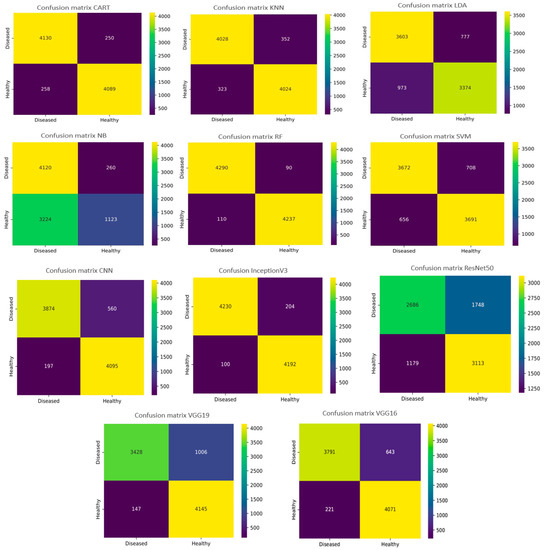
Figure 16.
Confusion matrixes for the ML and DL algorithms.
So, we are keen to minimize the incidence of misclassifying diseased leaves, since it is a genuine threat when the classifier classes them as healthy, especially in the presence of a contagious disease that spreads swiftly through the field. Respectively for the DL models, we found that the InceptionV3 model is the best one for classifying diseases since not only does it produce a very high accuracy of 98.01% but it can also properly classify the leaves wherein the number of misclassified diseased leaves is 204. In contrast, the ResNet50 model yielded a great accuracy of 93.57% except that the number of misclassified diseased leaves (1748 incorrect predictions) is extremely high compared to the InceptionV3 architecture. This implies that studies based only on prediction are insufficiently reliable to assess the efficiency of a classifier. Then, what might be the root cause of this phenomenon? Looking at the experimental dataset, we observed that the characteristics of crop leaf diseases, including leaf mold and bacterial spot and downy mildew diseases, were quite close to each other, which could explain this phenomenon.
- f.
- The ROC curve (Receiver Operating Characteristics Curve) [64] is a powerful tool for evaluating the performance of an ML/DL model. The ROC is applied to binary classification tasks where the output is composed of two distinct classes, and it represents a probability curve plotting the TPR against FPR at different threshold values and basically separates “signal” from “noise”. Otherwise, it expresses sensitivity as a function of 1- specificity for all possible threshold values of the marker studied. So, it demonstrates a trade-off phenomenon between specificity and sensitivity. Indeed, Sensitivity is the ability of the test to detect diseased leaves correctly, and specificity is the capacity of the test to detect healthy leaves correctly. This underscores the affectability of the classifier model. Where False Positive Rate (FPR) indicates the ratio of the negative class that was incorrectly classified by the classifier. Formally, it is expressed as below:where True Positive Rate (TPR) indicates the proportion of the positive class that was properly classified by the classifier. It is formally stated as follows:
The ideal classifier will have a ROC curve where the contour reaches a rate of 100% certified positives with zero improvements in the center x point (true positives). The area under the curve (AUC) measures the ability of a classifier to distinguish between clusters and serves as a summary of the ROC curve across all cutoffs, which is invariant to disease prevalence and diagnosis threshold choice. Generally, there are 4 cases: When the AUC is equal to 1, then the classifier is perfectly and correctly capable of discriminating all negative and positive class items. If the AUC corresponds to 0, then the classifier would predict all positives as negative, and all negatives as positive.
- -
- When AUC = 0.5, the classifier is unable to differentiate between positive and negative class points. This means that it predicts a random or a steady class for all data points.
- -
- When 0.5 < AUC < 1, it is likely that the classifier can discriminate positive class values from negative ones. This stems from the ability of the classifier to detect a greater number of true negatives and true positives than false negatives and false positives.
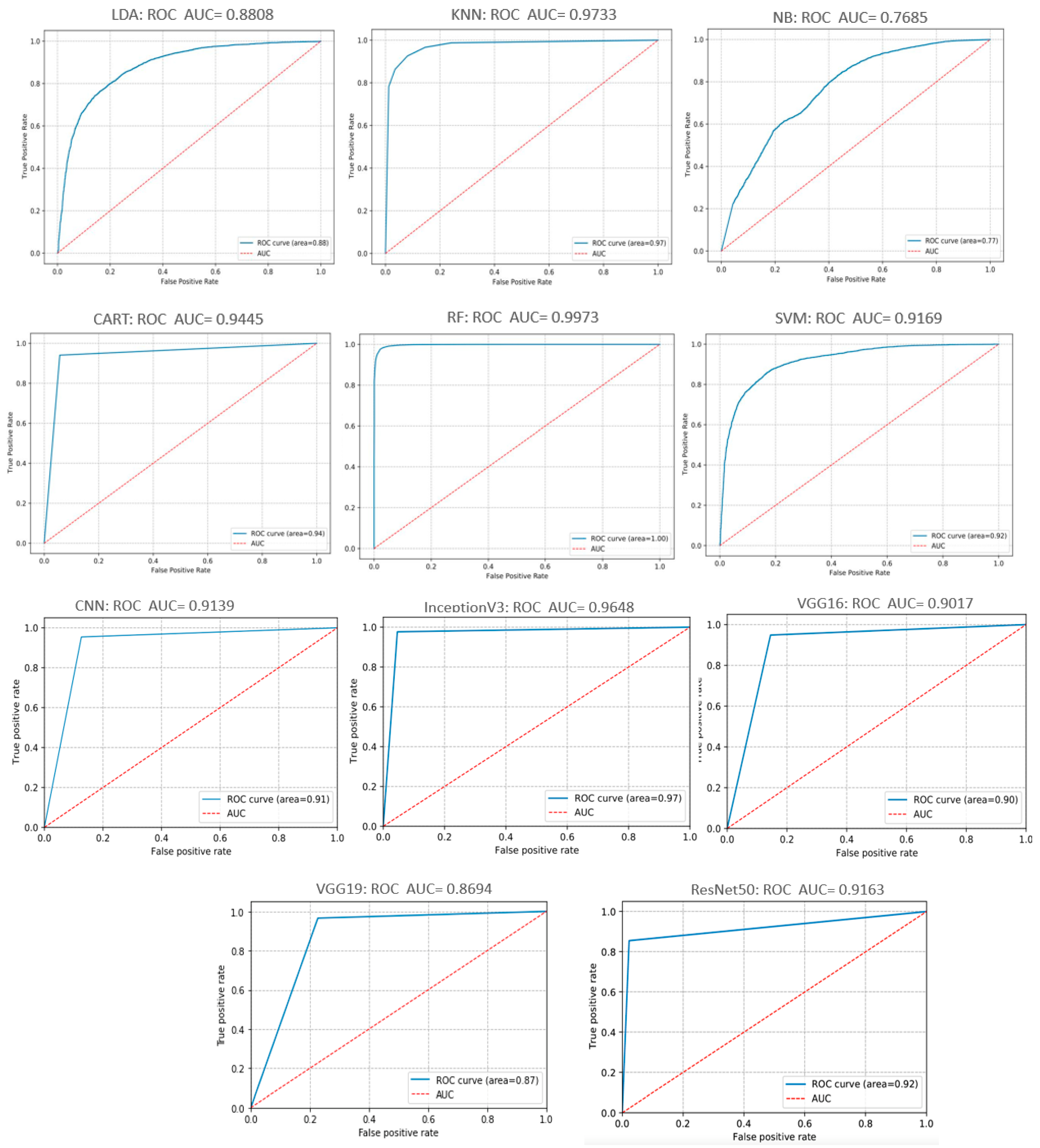
In general, the wider the AUC, the better the model’s ability in discriminating between negative and positive classes. Figure 17 illustrates the ROC curves of the ML/DL algorithms tested. As illustrated in the Figure, the areas under the curve (AUC) of the different algorithms were greater than 76.85%; some even reached 99.73%. In Figure 17, The RF algorithm scored the best performance, with the AUC of the crop leaves hitting. While the ReNet50 network and the NB algorithm performed poorly compared to the other ML/ DL models tested.
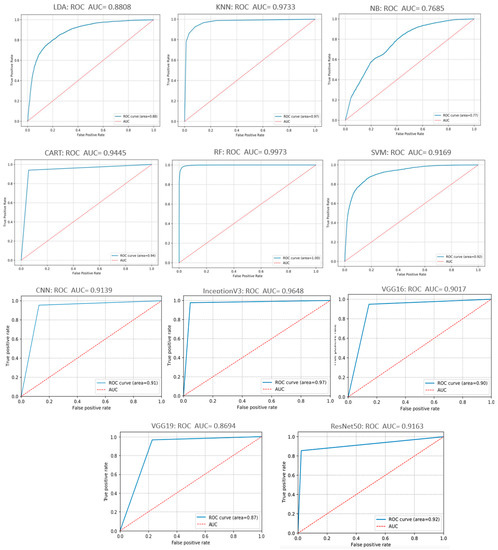
Figure 17.
ROC and AUC curves of tested DL and ML models.
3.2. Improvement in Classification Outcomes by DL Optimizers
In this work, a performance enhancement of the CNN architectures has been attempted by training the best model (obtained from the prior phase of algorithm comparison) over several deep learning optimization functions. Table 7 resumes the outcomes found by using different optimization algorithms. Some significant observations can be stated as following:

Table 7.
Performance of the applied deep learning optimizers to train the InceptionV3 model.
Noticeable variations were seen in training/validation accuracy, loss, recall, precision, and F1 score when training the InceptionV3 model using various deep learning optimizers. SGD and Adam were the most performing optimizers for the chosen architecture.
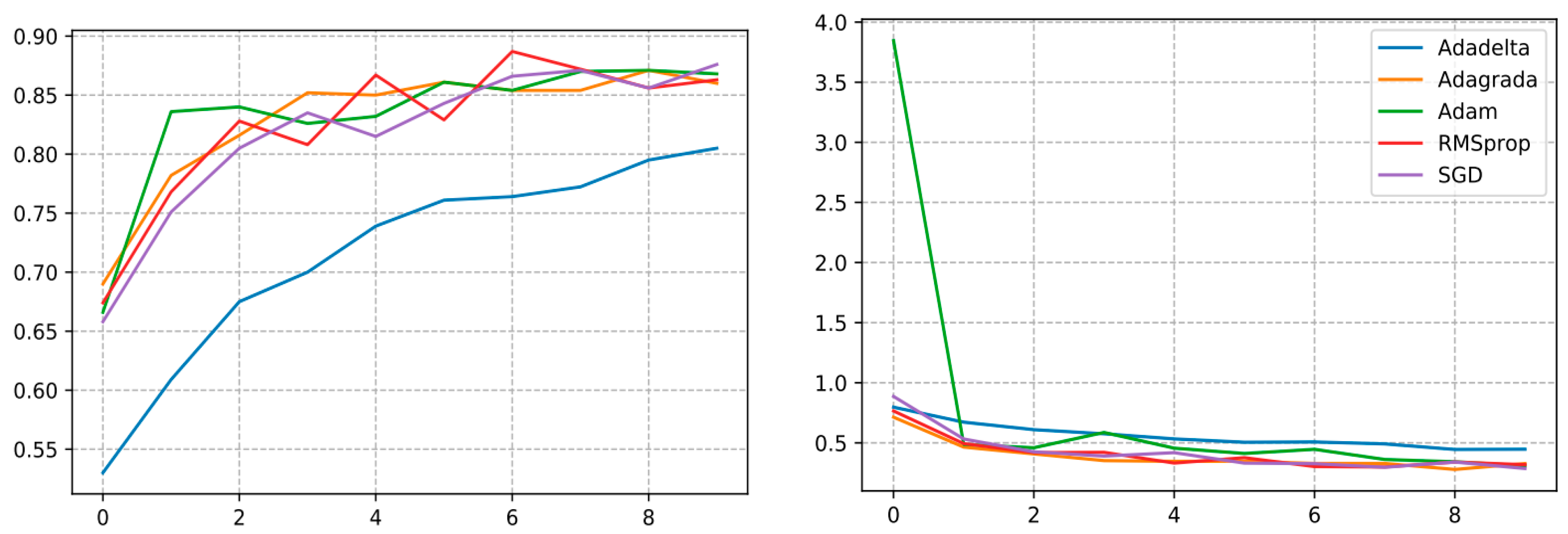
The InceptionV3 architecture trained with the SGD optimizer reached the highest validation accuracy and F1 score of 87.60% and 0.8941, respectively, which clearly shows the effectiveness of the proposed optimizer. Therefore, it could be used for various other agricultural operations. Yet, a decrement in performance was also noticed once the optimization functions were switched from SGD to Adadelta and Adagrada, as illustrated in Figure 18 where the optimizer showed its lowest validation accuracy.
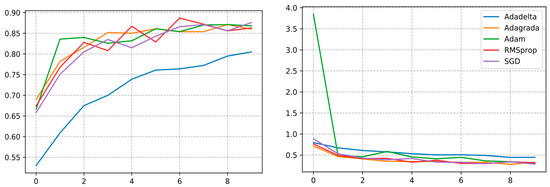
Figure 18.
Curve represents the loss and accuracy value of different DL optimizers applied for training the InceptionV3 model.
3.3. Effectiveness of Deep Learning with Different Activation Functions
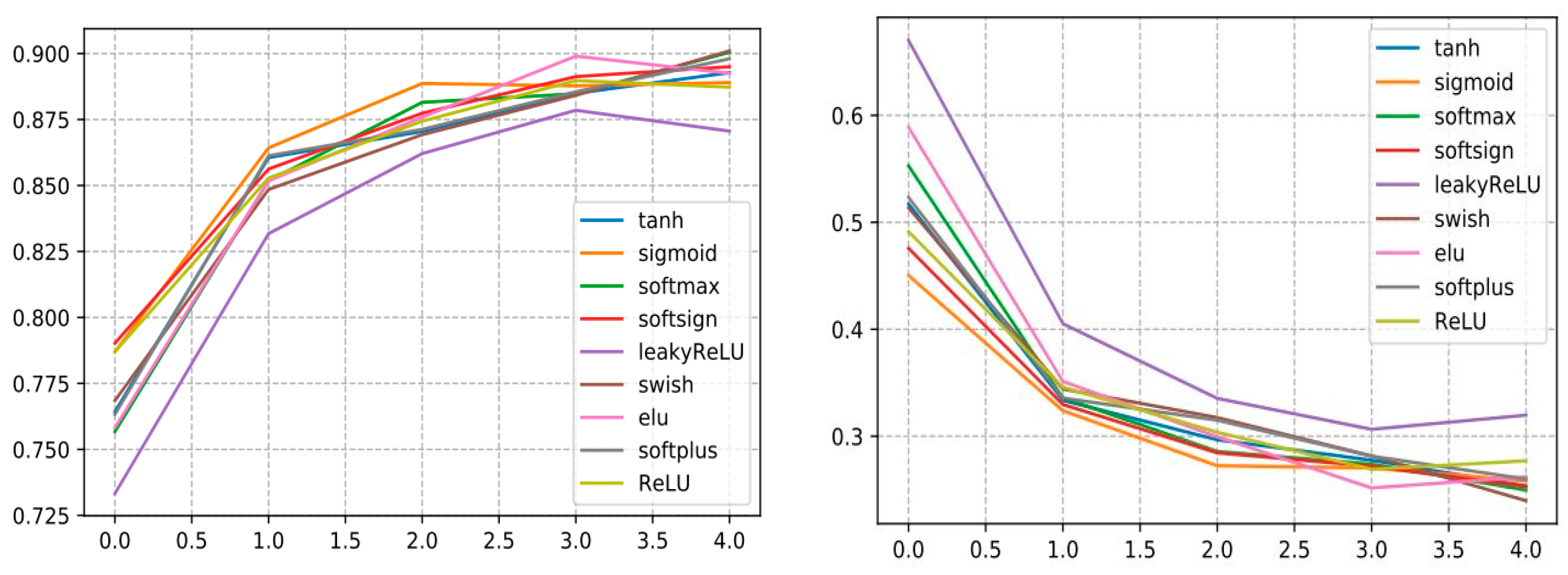
To build an optimized and efficient DL model based on the CNN architecture, we performed several experiments by varying the hyperparameters and activation functions to assess the DL performance using various activation functions, as shown in Table 8. From the findings, we remark that InceptionV3 with swish surpasses all other AFs and reveals a very good performance and reaches an accuracy of 0.9010 with a precision that amounts to 0.9135, a recall that comes to 0.9088. and the F1-score stands at 0.9077.

Table 8.
Performance of the different activation functions applied to train the InceptionV3 architecture.
Additionally, it is noticed that the InceptionV3 with LeakyReLU model shows its lowest validation accuracy amounting to 87.06, so according to Figure 19, it is remarkable in the loss curve that the InceptionV3 with LeakyReLu model marks a high loss value compared to the other AF curves.
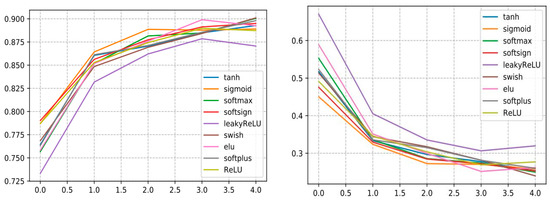
Figure 19.
Curve represents the loss and accuracy value of different activation function applied for training the InceptionV3 model.
3.4. Computational Time Spent for Building Each Model
We have seen the performance of these eleven algorithms in crop disease classification. Indeed, the computational complexity of ResNet50 as well as the training of the VGG19 architecture, is more significant than that of machine learning. The training over 100 epochs took more than 32 h in ResNet50 with TPU. This makes the network more complex and challenging to train on standard computers. On the other hand, validation and testing are much faster. Based on the comparison of processing time presented in Table 9 and the classification accuracy of the deep and machine learning techniques employed in our analysis, the InceptionV3 pre-trained model produced better classification accuracy and consumed lower time compared to other DL models such as ResNet 50 and VGG19 and CNN from scratch, which are very time consuming. On the other hand, RF is the best classifier compared to other ML counterparts, as it provides better performance. Even if its training time is slightly longer than KNN and CART, but it remains much less time-consuming than SVM. In summary, the leading architectures such as VGG16 and VGG19 produce better performance during training but not perfect results during crop disease classification tests. Furthermore, these architectures reduce the overhead of feature extraction, which is unlike machine learning algorithms. We discovered through these comparative experiments that the quality of the retrieved features has a substantial influence on the final classification outcomes for machine learning models.

Table 9.
The comparison of execution time for deep and machine learning techniques.
4. Discussion
Typically, research in this area is conducted by following a specific architecture or classifier on a single crop species using only a few performance measures. It might be challenging to gather several architectures and then compare them to determine which one is most suitable for a given task and yields the highest accuracy. Thus, we were motivated to perform an extensive comparison using multiple ML and DL algorithms on a large dataset containing multiple crops.
What distinguishes our work from prior ones is that we have employed a wider array of performance evaluation metrics to properly appraise the model’s reliability, while developing a comparison between various deep learning optimizers and activation functions to improve the results obtained from the comparison of machine learning algorithms with DL, with the aim of finding the most appropriate combination of the model, which can also be applied to further advance research on other agricultural applications, such as weed classification, grass/crop discrimination, and plant recognition. Since we are working on a binary classification where there are two classes: diseased/healthy leaves. The stakes of confounding a diseased leaf with a healthy one, or even missing a diseased leaf, might be highly raised, especially for viral diseases that are spreading rapidly in the field. Accordingly, our main objective is to prevent this confusion and to this end, we grant great importance to recall measuring the model’s performance in disease classification. Moreover, we strive not only for high accuracy or recall. However, we aim at overcoming the models’ weaknesses (fitting of hyperparameters, proper choice of parameters such as the loss function) and determining whether they have misclassified the crop leaves. Indeed, there is no point in deploying a model that provides the highest accuracy but misclassifies diseased leaves as healthy.
Based on this comparison, we found that the InceptionV3 network performs better in terms of accuracy, precision, and recall. Nevertheless, we are seeking for decreasing FN rate (diseased leaves being misclassified as healthy) which is a bit high compared to the RF classifier. On the other hand, the RF classifier offers lower accuracy than the InceptionV3 model, but its FN rate is significantly lower than any other model, as shown in Figure 16 and Figure 17, the RF has a wider area under the curve which means that the classifier has more ability to distinguish classes. It is noteworthy that the CNN-based models have key advantages over other machine learning algorithms, which are reflected in their outstanding ability to autonomously perform feature engineering. The DL parses the data for associating features and will combine them to facilitate faster learning without being explicitly prompted to do so. Conversely, the hand-crafted feature extractor requires elaborate features and captures only low-level edge information. This is why neural structures are of such relevance and is exemplified by the InceptionV3 model’s successful use of the receptive field concept to acquire local visual features to depict the topology of the image.
In fact, since the theoretical learning process of the InceptionV3 is similar to MLP, it represents an extension model of the MLP. A constraint of MLP is that it has a tendency to allocate a large value (almost +1) to one neuron in the output layer while all remaining neurons possess a low value (almost −1). This poses challenges in dismissing errors in real-time applications [65]. Conversely, the Random Forest considers the estimated probability when making a classification decision. This likelihood information gives a reliable and accurate grading list of label predictions. Moreover, using these likelihood values can facilitate the design of an effective rejection mechanism.
The Random Forest method is highly promising for classification, as it is not only non-parametric [66], but also offers the ability to estimate the significance of individual variables in the classification and to process high-dimensional data simultaneously with high computational efficiency. Even in cases of multiple noisy features, Random Forest performs perfectly, so there is no need for feature selection. Most interestingly, it is known to have high ROC and classification accuracy compared to the existing classification algorithms [67]. Therefore, it can enhance the classification performance of the hybrid architecture after changing the output units in the InceptionV3 model. As such, this idea of combining this novel hybrid InceptionV3 architecture with the RF classifier will constitute a new avenue to explore so as to benefit from both higher accuracy and recall with a much lower FN rate. Moreover, the tweaking of hyperparameters, as well as the choice of parameters and activation function, are tedious tasks that drastically impact the result obtained from the DL, its computational efficiency and the model’s learning accuracy. The employment of non-linear FAs can aid the model in discovering complex data, processing them, and learning by producing correct predictions. This urged us to elaborate comparison between these different AFs on our dataset, and the latter ones yielded dissimilar results, we remarked that the combination of the InceptionV3 with the Swish function outperformed the other models, providing better accuracy of up to 90.10% and a recall up to 0.9188. Furthermore, it was observed that when the InceptionV3 architecture was trained using different deep learning optimizers, the Inception network trained by the SGD optimizer achieved the highest Recall of 0.9753, suggesting that this combination of CNN architecture and optimization algorithms. The details are displayed in Figure 18 and Figure 19 and Table 7 and Table 8.
Limitations of Machine Learning and Deep Learning
In the following, we explore several limitations of the machine learning and deep learning models employed in our present work, in order to tackle the challenges encountered during our experimental work and propose new strategies that could be undertaken. One of them is that the present datasets do not include images collected and annotated from real-life scenarios. Thus, training is performed by using images captured in a controlled environment. Another constraint is that the existing proposed techniques cannot recognize multiple diseases in an image or several occurrences of the same disease in an image. Additionally, we faced many problems while building the ML models. For example, during the training of the KNN classifier, the number of neighbors must be defined by the user, and the model is highly sensitive to noise. Moreover, the main problem with the SVM algorithm is the choice of the right kernel function. Because for each dataset, a different kernel function produces different results.
Meanwhile, DL models demand more learning time and computational resources, but they may attain higher prediction accuracy and higher generalization ability, indicating that deep learning has superior learning ability. Compared with conventional machine learning, it may automatically and efficiently retrieve features from an image. DL models can clearly differentiate the image with similar features that are troublesome for the conventional machine learning algorithms to recognize. As the computing cost of deep learning models increases sharply with the dataset size, the trade-off between the accuracy of the results and the computing cost remains challenging. Furthermore, traditional machine learning algorithms require subjective feature extraction to transform binary vectors into one-dimensional vectors. In contrast, deep learning models can objectively extract features and directly process two-dimensional image data. In a nutshell, deep learning model is better suited for modeling image data. Its robust characteristic extraction capability and learning ability are not acquired by conventional machine learning algorithms. Moreover, the problem of class imbalance in classification typically causes the learning algorithm to be dominated by the majority classes, and the minority class features are sometimes ignored, resulting in misclassification bias. As a result, careful consideration must be given to the learning algorithm to improve minority class accuracy, as well as resort to hybrid techniques based on misclassification analysis.
Another challenge encountered during the application of the DL optimizers for example is that the SGD is consistently slow to converge as it requires a forward and backward propagation for each record. Then, the path to attain the global minima gets very noisy. Moreover, it is mandatory to tune the learning rate manually, by running several experiments since the suggested value is often not appropriate for every task. This way, the Adagrad optimizer surmounts the drawbacks of SGD as there is no necessity to manually tweak the learning rate; however, there is a tremendous downside, due to the monotonic decline of the learning rates, at some point in the time step, the model will cease learning because the learning rate is approaching 0. A major drawback of using the sigmoid is the vanishing gradient problem. For a very high or low value of x sample, the sigmoid derivative is very small. This may lead to a failure of the network in learning more information, as well as it is computationally demanding since it contains an exponential term.
In light of these stated limitations, we briefly recap all the restrictions and benefits of deep learning applied during model training in Table 10.

Table 10.
Strengths and weaknesses of deep learning optimization.
Other guidelines might also be considered to address these shortcomings as we mentioned in our prior work [68], such as the use of a new dataset, which contains many labeled images of leaves captured in a real environment; or as an alternative to create synthesized and generated images by hybrid data augmentation techniques based on the Generative Adversarial Network (GAN) architecture [69] to enhance crop leaf disease detection in real-world images. Finally, the comparison results reveal that there is no single tailored technique to meet all research challenges. Therefore, the appropriate method is selected based upon the applications and the dataset size.
5. Conclusions
In the present research paper, an exhaustive comparative study has been carried out between several cutting-edge deep learning models and machine learning ones. Moreover, the performance of the best-resulting network has been improved by using various activation functions and deep learning optimization algorithms. The findings are very promising and strongly indicate the dominance of the DL models over classical ML algorithms. Thus, the recall, the obtained accuracy and above all the simplicity of the approach confirm that the DL method is the best way forward for image classification problems with relatively large datasets. Nevertheless, DL algorithms also have some constraints, namely that a very powerful GPU/TPU is mandatory for training, as CNN models are time-consuming to train and might take hours to weeks depending on the dataset size.
Therefore, to lessen the learning time, we employed pre-trained models. Moreover, merging the machine learning algorithms and deep networks requires much fewer CPU resources and will use about half of the memory bandwidth while generating better models. So, going forward, a web application can be implemented with a complete system composed of server-side elements containing a hybrid model based on the best-found functions (SGD optimizer+ swish activation function + RF+ InceptionV3) trained with features such as a display of the crop’s recognized diseases that can be applied in the field for validation and testing. Moreover, the application may provide a forum for agronomists and farmers to discuss treatments and precautions for diseases they have experienced. Moreover, we will strive to decrease the learning time, computational complexity, and deep model size to run them on embedded or mobile platforms.
Author Contributions
Conceptualization, H.O. and M.S.; methodology, H.O.; software, H.O.; validation, M.S., M.K. and E.S.; formal analysis, M.S.; investigation, M.S.; resources, M.K.; data curation, M.K.; writing—original draft preparation, H.O.; writing—review and editing, M.K.; visualization, E.S.; supervision, E.S.; project administration, M.S. and M.K.; funding acquisition, M.S. All authors have read and agreed to the published version of the manuscript.
Funding
This research k was done within the framework “Agrometeorological Stations Platform” project funded by the Moroccan Ministry of Higher Education and Scientific Research-National Centre for Scientific and Technical Research (NCSTR) (PPR2 project).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data presented in this study are openly available in [PlantVillage Dataset] at [https://www.plantvillage.org/], reference number [20].
Conflicts of Interest
The authors declare no conflict of interest.
Abbreviations
The following abbreviations are used throughout this manuscript.
| AI | Artificial Intelligence |
| AF | Activation function |
| ANN | Artificial Neural Networks |
| AUC | Area Under Curve |
| Adam | Adaptive moment estimation method |
| CA | Classification Accuracy |
| CA | Channel-wise Attention |
| CART | Classification And Regression Trees |
| CNN | Convolutional Neural Networks |
| DL | Deep Learning |
| ELM | Extreme Learning Machine |
| ELU | Exponential Linear Unit |
| FLS | Few-Short learning |
| FC | Fully Connected |
| IMPS | Importance Sampling |
| KNN | K-Nearest Neighbor |
| LDA | Latent Dirichlet Allocation |
| ML | Machine Learning |
| MLP | Multilayer Perceptron |
| NB | Naive Bayes |
| NN | Neural Networks |
| ResNet | Residual Network |
| ReLU | Rectified Linear Unit |
| RNN | Recurrent Neural Network |
| RF | Random Forest |
| ROS | Random Over Sampling |
| ROC | Receiver Operating Characteristics |
| RUS | Random Under Sampling |
| SGD | Stochastic Gradient Descent |
| SMOTE | Synthetic Minority Over-sampling Technique |
| SNN | Spiking Neural Networks |
| SVM | Support Vector Machine |
| TPMD | Tomato Powdery Mildew Disease |
| VGG | Visual Geometry Group |
References
- Savary, S.; Willocquet, L.; Pethybridge, S.J.; Esker, P.; McRoberts, N.; Nelson, A. The global burden of pathogens and pests on major food crops. Nat. Ecol. Evol. 2019, 3, 430–439. [Google Scholar] [CrossRef] [PubMed]
- Dhingra, G.; Kumar, V.; Joshi, H.D. Study of digital image processing techniques for leaf disease detection and classification. Multimedia Tools Appl. 2018, 77, 19951–20000. [Google Scholar] [CrossRef]
- Singh, V.; Sharma, N.; Singh, S. A review of imaging techniques for plant disease detection. Artif. Intell. Agric. 2020, 4, 229–242. [Google Scholar] [CrossRef]
- Mojjada, R.K.; Kumar, K.K.; Yadav, A.; Prasad, B.S.V. WITHDRAWN: Detection of plant leaf disease using digital image processing. Mater. Today Proc. 2020. [Google Scholar] [CrossRef]
- Vishnoi, V.K.; Kumar, K.; Kumar, B. Plant disease detection using computational intelligence and image processing. J. Plant Dis. Prot. 2021, 128, 19–53. [Google Scholar] [CrossRef]
- Applalanaidu, M.V.; Kumaravelan, G. A Review of Machine Learning Approaches in Plant Leaf Disease Detection and Classification. In Proceedings of the 2021 Third International Conference on Intelligent Communication Technologies and Virtual Mobile Networks (ICICV), Tirunelveli, India, 4–6 February 2021. [Google Scholar]
- Prathusha, P.; Murthy, K.; Srinivas, K. Plant Disease Detection Using Machine Learning Algorithms. In International Conference on Computational and Bio Engineering; Springer International Publishing: Berlin/Heidelberg, Germany, 2019. [Google Scholar]
- Pradhan, S.S.; Patil, R. Comparison of Deep Learning Approaches for Plant Disease Detection. In Proceedings of International Conference on Wireless Communication; Springer: Singapore, 2020. [Google Scholar] [CrossRef]
- Sachdeva, G.; Singh, P.; Kaur, P. Plant leaf disease classification using deep Convolutional neural network with Bayesian learning. Mater. Today Proc. 2021, 45, 5584–5590. [Google Scholar] [CrossRef]
- Devaraj, A.; Rathan, K.; Jaahnavi, S.; Indira, K. Identification of plant disease using image processing technique. In Proceedings of the 2019 International Conference on Communication and Signal Processing (ICCSP), Chennai, India, 4–6 April 2019. [Google Scholar]
- Iniyan, S.; Jebakumar, R.; Mangalraj, P.; Mohit, M.; Nanda, A. Plant Disease Identification and Detection Using Support Vector Machines and Artificial Neural Networks. In Artificial Intelligence and Evolutionary Computations in Engineering Systems; Springer: Berlin/Heidelberg, Germany, 2020; pp. 15–27. [Google Scholar] [CrossRef]
- Orchi, H.; Sadik, M.; Khaldoun, M. A General Survey on Plants Disease Detection Using Image Processing, Deep Transfer Learning and Machine Learning Techniques. In International Symposium on Ubiquitous Networking; Springer: Cham, Switzerland, 2021; pp. 210–224. [Google Scholar] [CrossRef]
- Argüeso, D.; Picon, A.; Irusta, U.; Medela, A.; San-Emeterio, M.G.; Bereciartua, A.; Alvarez-Gila, A. Few-Shot Learning approach for plant disease classification using images taken in the field. Comput. Electron. Agric. 2020, 175, 105542. [Google Scholar] [CrossRef]
- Pantazi, X.; Moshou, D.; Tamouridou, A. Automated leaf disease detection in different crop species through image features analysis and One Class Classifiers. Comput. Electron. Agric. 2019, 156, 96–104. [Google Scholar] [CrossRef]
- Arora, J.; Agrawal, U.; Sharma, P. Classification of Maize leaf diseases from healthy leaves using Deep Forest. J. Artif. Intell. Syst. 2020, 2, 14–26. [Google Scholar] [CrossRef]
- Tang, Z.; Yang, J.; Li, Z.; Qi, F. Grape disease image classification based on lightweight convolution neural networks and channelwise attention. Comput. Electron. Agric. 2020, 178, 105735. [Google Scholar] [CrossRef]
- Bhatia, A.; Chug, A.; Singh, A.P. Application of extreme learning machine in plant disease prediction for highly imbalanced dataset. J. Stat. Manag. Syst. 2020, 23, 1059–1068. [Google Scholar] [CrossRef]
- Shamsudin, H.; Yusof, U.K.; Jayalakshmi, A.; Khalid, M.N.A. Combining oversampling and undersampling techniques for imbalanced classification: A comparative study using credit card fraudulent transaction dataset. In Proceedings of the 2020 IEEE 16th International Conference on Control & Automation (ICCA), Singapore, 9–11 October 2020. [Google Scholar] [CrossRef]
- Chawla, N.V.; Bowyer, K.W.; Hall, L.O.; Kegelmeyer, W.P. SMOTE: Synthetic Minority Over-sampling Technique. J. Artif. Intell. Res. 2002, 16, 321–357. [Google Scholar] [CrossRef]
- Faust, J.; Hanelt, P.H.P.; Bhat, S.A. PlantVillage Dataset: A Dataset of 5539 Training and Validation Images for 26 Crop Species. 2016. Available online: https://www.plantvillage.org/ (accessed on 20 October 2022).
- Carneiro, T.; Da Nobrega, R.V.M.; Nepomuceno, T.; Bian, G.-B.; De Albuquerque, V.H.C.; Filho, P.P.R. Performance Analysis of Google Colaboratory as a Tool for Accelerating Deep Learning Applications. IEEE Access 2018, 6, 61677–61685. [Google Scholar] [CrossRef]
- Lukic, M.; Tuba, E.; Tuba, M. Leaf recognition algorithm using support vector machine with Hu moments and local binary patterns. In Proceedings of the 2017 IEEE 15th International Symposium on Applied Machine Intelligence and Informatics (SAMI), Herl’any, Slovakia, 26–28 January 2017; pp. 000485–000490. [Google Scholar] [CrossRef]
- Hu, M.-K. Visual pattern recognition by moment invariants. IEEE Trans. Inf. Theory 1962, 8, 179–187. [Google Scholar] [CrossRef]
- Basavaiah, J.; Anthony, A.A. Tomato Leaf Disease Classification using Multiple Feature Extraction Techniques. Wirel. Pers. Commun. 2020, 115, 633–651. [Google Scholar] [CrossRef]
- Karthickmanoj, R.; Sasilatha, T.; Padmapriya, J. Automated machine learning based plant stress detection system. Mater. Today Proc. 2021, 47, 1887–1891. [Google Scholar] [CrossRef]
- Haralick, R.M.; Shanmugam, K.; Dinstein, I.H. Textural Features for Image Classification. IEEE Trans. Syst. Man Cybern. 1973, SMC-3, 610–621. [Google Scholar] [CrossRef]
- Bankar, S.; Dube, A.; Kadam, P.; Deokule, S. Plant disease detection techniques using canny edge detection & color histogram in image processing. Int. J. Comput. Sci. Inf. Technol 2014, 5, 1165–1168. [Google Scholar]
- Koranne, S. Hierarchical data format 5: HDF5. In Handbook of Open Source Tools; Springer: Berlin/Heidelberg, Germany, 2011; pp. 191–200. [Google Scholar]
- Cortes, C.; Vapnik, V. Support-vector networks. Mach. Learn. 1995, 20, 273–297. [Google Scholar] [CrossRef]
- Hsu, W.-C.; Lin, L.-F.; Chou, C.-W.; Hsiao, Y.-T.; Liu, Y.-H. EEG Classification of Imaginary Lower Limb Stepping Movements Based on Fuzzy Support Vector Machine with Kernel-Induced Membership Function. Int. J. Fuzzy Syst. 2017, 19, 566–579. [Google Scholar] [CrossRef]
- Cunningham, P.; Delany, S.J. k-Nearest Neighbour Classifiers—A Tutorial. ACM Comput. Surv. 2021, 54, 1–25. [Google Scholar] [CrossRef]
- Liu, Y.; Wang, Y.; Zhang, J. New machine learning algorithm: Random forest. In International Conference on Information Computing and Applications; Springer: Berlin/Heidelberg, Germany, 2012; pp. 246–252. [Google Scholar]
- Breiman, L. Random forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef]
- Rish, I. An empirical study of the naive Bayes classifier. In IJCAI 2001 Workshop on Empirical Methods in Artificial Intelligence; IBM Research: New York, NY, USA, 2001; pp. 41–46. [Google Scholar]
- Blei, D.M.; Ng, A.Y.; Jordan, M.I. Latent dirichlet allocation. J. Mach. Learn. Res. 2003, 3, 993–1022. [Google Scholar]
- Trendowicz, A.; Jeffery, R. Classification and regression trees. In Software Project Effort Estimation; Springer: Berlin/Heidelberg, Germany, 2014; pp. 295–304. [Google Scholar]
- LeCun, Y.; Bengio, Y.; Hinton, G. Deep Learning. Nature 2015, 521, 436–444. [Google Scholar] [CrossRef]
- Schmidhuber, J. Deep Learning in Neural Networks: An Overview. Neural Netw. 2015, 61, 85–117. [Google Scholar] [CrossRef]
- Mercioni, M.A.; Holban, S. The most used activation functions: Classic versus current. In Proceedings of the 2020 International Conference on Development and Application Systems (DAS), Suceava, Romania, 21–23 May 2020. [Google Scholar]
- Nwankpa, C.; Ijomah, W.; Gachagan, A.; Marshall, S. Activation functions: Comparison of trends in practice and research for deep learning. arXiv 2018, arXiv:1811.03378, 124–133. [Google Scholar] [CrossRef]
- Szandała, T. Review and Comparison of Commonly Used Activation Functions for Deep Neural Networks. In Bio-Inspired Neurocomputing; Springer: Berlin/Heidelberg, Germany, 2021; pp. 203–224. [Google Scholar]
- Ramachandran, P.; Zoph, B.; Le, Q.V. Searching for activation functions. arXiv 2017, arXiv:1710.05941. [Google Scholar]
- Maas, A.L.; Hannun, A.Y.; Ng, A.Y. Rectifier nonlinearities improve neural network acoustic models. In Proceedings of the ICML, Atlanta, GA, USA, 16–21 June 2013. [Google Scholar]
- Clevert, D.-A.; Unterthiner, T.; Hochreiter, S. Unterthiner, and S. Hochreiter, Fast and accurate deep network learning by exponential linear units (elus). arXiv Preprint 2015, arXiv:1511.07289. [Google Scholar]
- Trottier, L.; Giguere, P.; Chaib-Draa, B. Parametric exponential linear unit for deep convolutional neural networks. In Proceedings of the 2017 16th IEEE International Conference on Machine Learning and Applications (ICMLA), Cancun, Mexico, 18–21 December 2017. [Google Scholar]
- Shin, H.-C.; Roth, H.R.; Gao, M.; Lu, L.; Xu, Z.; Nogues, I.; Yao, J.; Mollura, D.; Summers, R.M. Deep Convolutional Neural Networks for Computer-Aided Detection: CNN Architectures, Dataset Characteristics and Transfer Learning. IEEE Trans. Med. Imaging 2016, 35, 1285–1298. [Google Scholar] [CrossRef]
- Ruder, S. An overview of gradient descent optimization algorithms. arXiv 2016, arXiv:1609.04747. [Google Scholar]
- Duchi, J.; Hazan, E.; Singer, Y. Adaptive subgradient methods for online learning and stochastic optimization. J. Mach. Learn. Res. 2011, 12, 2121–2159. [Google Scholar]
- Hinton, G.; Srivastava, N.; Swersky, K. Neural Networks for Machine Learning. 5 October 2020. Available online: http://www.cs.toronto.edu/~hinton/coursera/lecture6/lec6.pdf (accessed on 20 October 2022).
- Zeiler, M.D. Adadelta: An adaptive learning rate method. arXiv 2012, arXiv:1212.5701. [Google Scholar]
- Kingma, D.P.; Ba, J. Adam: A method for stochastic optimization. arXiv 2014, arXiv:1412.6980. [Google Scholar]
- Bergstra, J.; Bengio, Y. Random search for hyper-parameter optimization. J. Mach. Learn. Res. 2012, 13, 281–305. [Google Scholar]
- Ioffe, S.; Szegedy, C. Batch normalization: Accelerating deep network training by reducing internal covariate shift. In International Conference on Machine Learning; PMLR: London, UK, 2015. [Google Scholar]
- Brahimi, M.; Arsenovic, M.; Laraba, S.; Sladojevic, S.; Boukhalfa, K.; Moussaoui, A. Deep Learning for Plant Diseases: Detection and Saliency Map Visualisation. In Human and Machine Learning, Human–Computer Interaction Series; Springer: Cham, Switzerland, 2018; pp. 93–117. [Google Scholar]
- Krizhevsky, A.; Sutskever, I.; Hinton, G.E. Imagenet classification with deep convolutional neural networks. Adv. Neural Inf. Process. Syst. 2012, 25. [Google Scholar] [CrossRef]
- Alzubaidi, L.; Zhang, J.; Humaidi, A.J.; Al-Dujaili, A.; Duan, Y.; Al-Shamma, O.; Santamaría, J.; Fadhel, M.A.; Al-Amidie, M.; Farhan, L. Review of deep learning: Concepts, CNN architectures, challenges, applications, future directions. J. Big Data 2021, 8, 1–74. [Google Scholar] [CrossRef]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep residual learning for image recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 26 June–1 July 2016. [Google Scholar]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Identity Mappings in Deep Residual Networks. In European Conference on Computer Vision; Springer: Cham, Switzerland, 2016; pp. 630–645. [Google Scholar]
- Szegedy, C.; Vanhoucke, V.; Ioffe, S.; Shlens, J.; Wojna, Z. Rethinking the inception architecture for computer vision. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 26 June–1 July 2016. [Google Scholar]
- Simonyan, K.; Zisserman, A. Very deep convolutional networks for large-scale image recognition. arXiv 2014, arXiv:1409.1556. [Google Scholar]
- Powers, D.M. Evaluation: From precision, recall and F-measure to ROC, informedness, markedness and correlation. arXiv 2020, arXiv:2010.16061. [Google Scholar]
- Sasaki, Y. The truth of the F-measure. Teach Tutor Mater 2007, 1, 5. [Google Scholar]
- Fawcett, T. An introduction to ROC analysis. Pattern Recognit. Lett. 2006, 27, 861–874. [Google Scholar] [CrossRef]
- Brown, C.D.; Davis, H.T. Receiver operating characteristics curves and related decision measures: A tutorial. Chemom. Intell. Lab. Syst. 2006, 80, 24–38. [Google Scholar] [CrossRef]
- Niu, X.-X.; Suen, C.Y. A novel hybrid CNN–SVM classifier for recognizing handwritten digits. Pattern Recognit. 2012, 45, 1318–1325. [Google Scholar] [CrossRef]
- Gislason, P.O.; Benediktsson, J.A.; Sveinsson, J.R. Random Forests for land cover classification. Pattern Recognit. Lett. 2006, 27, 294–300. [Google Scholar] [CrossRef]
- Guo, L.; Ma, Y.; Cukic, B.; Singh, H. Robust prediction of fault-proneness by random forests. In Proceedings of the 15th International Symposium on Software Reliability Engineering, Washington, DC, USA, 2–5 November 2004. [Google Scholar]
- Orchi, H.; Sadik, M.; Khaldoun, M. On Using Artificial Intelligence and the Internet of Things for Crop Disease Detection: A Contemporary Survey. Agriculture 2021, 12, 9. [Google Scholar] [CrossRef]
- Goodfellow, I.; Pouget-Abadie, J.; Mirza, M.; Xu, B.; Warde-Farley, D.; Ozair, S.; Courville, A.; Bengio, Y. Generative adversarial networks. Commun. ACM 2020, 63, 139–144. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).