Changes in Allele Frequencies and Genetic Diversity in Red Clover after Selection for Cold Tolerance Using SSR Markers
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Material
2.2. DNA Isolation and SSR Amplification
2.3. Data Analysis
2.3.1. Waples Neutrality Test
2.3.2. H-W Test, Observed and Expected Heterozygosity
3. Results
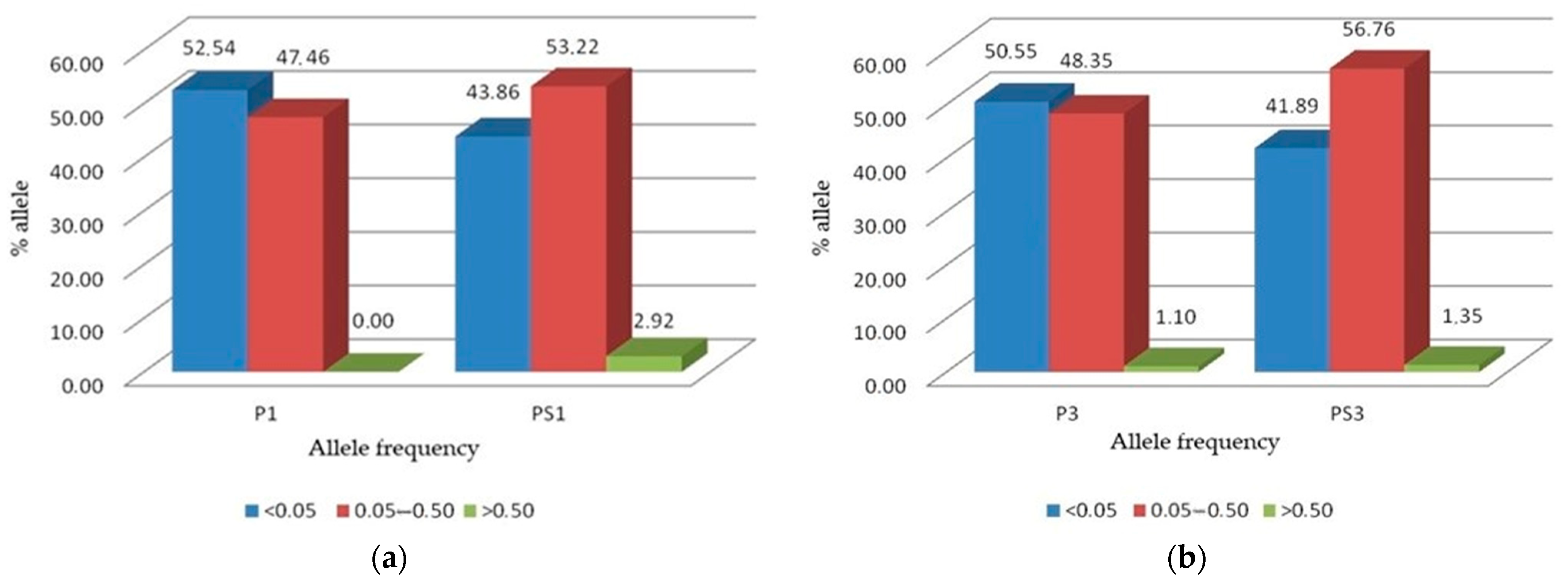
3.1. Allele Frequencies and Waples Neutrality Test
3.2. Genetic Diversity Measures and Hardy–Weinberg Test
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- IPCC. Summary for Policymakers. In Climate Change 2023: Synthesis Report; Core Writing Team, Lee, H., Romero, J., Eds.; Contribution of Working Groups I, II and III to the Sixth Assessment Report of the Intergovernmental Panel on Climate Change; IPCC: Geneva, Switzerland, 2023; pp. 1–34. [Google Scholar]
- Porter, J.R.; Challinor, A.J.; Henriksen, C.B.; Howden, S.M.; Martre, P.; Smith, P. Invited review: Intergovernmental Panel on Climate Change, agriculture, and food—A case of shifting cultivation and history. Glob. Chang. Biol. 2019, 25, 2518–2529. [Google Scholar] [CrossRef] [PubMed]
- Parween, A.; Singh, V.; Bajpai, M. Abiotic Stress and Red Clover: A Less Explored Area of Research. Adv. Res. 2020, 21, 1–5. [Google Scholar] [CrossRef]
- Xu, Y.; Qin, F.; Chu, C.; Varshney, R.K. Abiotic stress tolerance: Genetics, genomics, and breeding. Crop J. 2023, 11, 969–974. [Google Scholar] [CrossRef]
- Sustek-Sánchez, F.; Rognli, O.A.; Rostoks, N.; Sõmera, M.; Jaškūnė, K.; Kovi, M.R.; Statkevičiūtė, G.; Sarmiento, C. Improving abiotic stress tolerance of forage grasses—Prospects of using genome editing. Front. Plant Sci. 2023, 14, 1127532. [Google Scholar] [CrossRef] [PubMed]
- Roberts, D.P.; Mattoo, A.K. Sustainable Agriculture—Enhancing Environmental Benefits, Food Nutritional Quality and Building Crop Resilience to Abiotic and Biotic Stresses. Agriculture 2018, 8, 8. [Google Scholar] [CrossRef]
- Hossain, A.; Skalicky, M.; Brestic, M.; Maitra, S.; Ashraful Alam, M.; Syed, M.A.; Hossain, J.; Sarkar, S.; Saha, S.; Bhadra, P.; et al. Consequences and Mitigation Strategies of Abiotic Stresses in Wheat (Triticum aestivum L.) under the Changing Climate. Agronomy 2021, 11, 241. [Google Scholar] [CrossRef]
- Li, W.; Zheng, C.; Zhou, J.; Zhang, Z.; Zhou, G.; Xie, X. Characterization of a naturally occurring early-flowering rice mutant resulting from a novel variation in the Ghd7 locus. Czech J. Genet. Plant Breed. 2021, 57, 166–169. [Google Scholar] [CrossRef]
- Adhikari, L.; Baral, R.; Paudel, D.; Min, D.; Makaju, S.O.; Poudel, H.P.; Acharya, J.P.; Missaoui, A.M. Cold stress in plants: Strategies to improve cold tolerance in forage species. Plant Stress 2022, 4, 100081. [Google Scholar] [CrossRef]
- Sanghera, G.S.; Wani, S.H.; Hussain, W.; Singh, N.B. Engineering cold stress tolerance in crop plants. Curr. Genom. 2011, 12, 30–43. [Google Scholar] [CrossRef]
- Casler, M.D.; van Santen, E. Breeding Objectives in Forages. In Fodder Crops and Amenity Grasses. Handbook of Plant Breeding; Boller, B., Posselt, U.K., Veronesi, F., Eds.; Springer: New York, NY, USA, 2010; Volume 5, pp. 115–136. [Google Scholar]
- Taylor, N.L. A Century of Clover Breeding Developments in the United States. Crop Sci. 2008, 48, 1–13. [Google Scholar] [CrossRef]
- Riday, H. Progress Made in Improving Red Clover (Trifolium pratense L.) through Breeding. Int. J. Plant Breed. 2010, 4, 22–29. [Google Scholar]
- Bertrand, A.; Bipfubusa, M.; Castonguay, Y.; Rocher, S.; Szopinska-Morawska, A.; Papadopoulos, Y.; Renaut, J. A Proteome Analysis of Freezing Tolerance in Red Clover (Trifolium pratense L.). BMC Plant Biol. 2016, 16, 65. [Google Scholar] [CrossRef] [PubMed]
- Beljo, J. Oplemenjivanje Bilja; Sveučilište u Mostaru: Mostar, Bosnia, 2006; p. 169. [Google Scholar]
- Verwimp, C.; Ruttink, T.; Muylle, H.; Glabeke, S.V.; Cnops, G.; Quataert, P.; Honnay, O.; Roldán-Ruiz, I. Temporal Changes in Genetic Diversity and Forage Yield of Perennial Ryegrass in Monoculture and in Combination with Red Clover in Swards. PLoS ONE 2018, 13, e0206571. [Google Scholar] [CrossRef] [PubMed]
- Ergon, Å.; Skøt, L.; Sæther, V.E.; Rognli, O.A. Allele Frequency Changes Provide Evidence for Selection and Identification of Candidate Loci for Survival in Red Clover (Trifolium pratense L.). Front. Plant. Sci. 2019, 10, 718. [Google Scholar] [CrossRef]
- Taylor, N.L.; Quesenberry, K.H. Breeding Methodology. In Red Clover Science; Kluwer Academic Publishers: Dordrecht, The Netherlands, 1996; Volume 28, pp. 141–160. [Google Scholar]
- Sjodin, J.; Ellerstrom, A. Autopolyploid forage crops. In Svalöf 1886–1986. Research and Results in Plant Breeding; Olsson, G., Ed.; LTs Förla: Stockholm, Sweden, 1986; pp. 102–113. [Google Scholar]
- Petrauskas, G.; Norkevičienė, E.; Baistruk-Hlodan, L. Genetic Differentiation of Red Clover (Trifolium pratense L.) Cultivars and Their Wild Relatives. Agriculture 2023, 13, 1008. [Google Scholar] [CrossRef]
- Collins, R.P.; Helgadóttir, Á.; Frankow-Lindberg, B.E.; Skøt, L.; Jones, C.; Skøt, K.P. Temporal Changes in Population Genetic Diversity and Structure in Red and White Clover Grown in Three Contrasting Environments in Northern Europe. Ann. Bot. 2012, 110, 1341–1350. [Google Scholar] [CrossRef]
- Göransson, M.; Kristjánsdóttir, T.A.; Dalmannsdottir, S.; Helgadottir, A. Genetic Shift in White Clover (Trifolium repens) after Natural Selection in a Marginal Area. Icel. Agric. Sci. 2012, 25, 41–50. [Google Scholar]
- Grljusic, S.; Bolaric, S.; Popovic, S.; Cupic, T.; Tucak, M.; Kozumplik, V. Assessment of Morphological and RAPD Variation among and within Red Clover Cultivars after Natural Selection. Die Bodencultur. 2005, 56, 183–188. [Google Scholar]
- Amiteye, S. Basic concepts and methodologies of DNA marker systems in plant molecular breeding. Heliyon 2021, 7, e08093. [Google Scholar] [CrossRef]
- Choudhury, A.; Deb, S.; Kharbyngar, B.; Rajpal, V.R.; Rao, S.R. Dissecting the plant genome: Through new generation molecular markers. Genet. Resour. Crop. Evol. 2022, 69, 2661–2698. [Google Scholar] [CrossRef]
- Vieira, M.L.C.; Santini, L.; Diniz, A.L.; Munhoz, C.d.F. Microsatellite Markers: What They Mean and Why They Are so Useful. Genet. Mol. Biol. 2016, 39, 312–328. [Google Scholar] [CrossRef]
- Gupta, M.; Sharma, V.; Singh, S.K.; Chahota, R.K.; Sharma, T.R. Analysis of Genetic Diversity and Structure in a Genebank Collection of Red Clover (Trifolium pratense L.) Using SSR Markers. Plant Genet. Resour. 2017, 15, 376–379. [Google Scholar] [CrossRef]
- Dugar, Y.N.; Popov, V.N. Genetic Structure and Diversity of Ukrainian Red Clover Cultivars Revealed by Microsatellite Markers. Open J. Genet. 2013, 3, 235–242. [Google Scholar] [CrossRef]
- Ahsyee, S.R.; Vasiljević, S.; Ćalić, I.; Zorić, M.; Karagić, Đ.; Šurlan-Momirović, G. Genetic Diversity in Red Clover (Trifolium pratense L.) Using SSR Markers. Genetika 2014, 46, 949–961. [Google Scholar] [CrossRef]
- Berzina, I.; Zhuk, A.; Veinberga, I.; Rashal, I.; Rungis, D. Genetic Fingerprinting of Latvian Red Clover (Trifolium pratense L.) Varieties Using Simple Sequence Repeat (SSR) Markers: Comparisons over Time and Space. Latv. J. Agron. 2008, 11, 28–32. [Google Scholar]
- Dias, P.M.B.; Julier, B.; Sampoux, J.-P.; Barre, P.; Dall’Agnol, M. Genetic Diversity in Red Clover (Trifolium pratense L.) Revealed by Morphological and Microsatellite (SSR) Markers. Euphytica 2008, 160, 189–205. [Google Scholar] [CrossRef]
- Radinović, I.; Vasiljević, S.; Branković, G.; Salem-Ahsyee, R.; Momirović, U.; Perović, D.; Surlan-Momirović, G. Molecular characterization of red clover genotypes utilizing microsatellite markers. Chilean J. Agric. Res. 2017, 77, 41–47. [Google Scholar] [CrossRef]
- Kolliker, R.; Enkerli, J.; Widmer, F. Characterization of novel microsatellite loci for red clover (Trifolium pratense L.) from enriched genomic libraries. Mol. Ecol. Notes 2006, 6, 50–53. [Google Scholar] [CrossRef]
- Sato, S.; Isobe, S.; Asamizu, E.; Ohmido, N.; Kataoka, R.; Nakamura, Y.; Kaneko, T.; Sakurai, N.; Okumura, K.; Klimenko, I.; et al. Comprehensive Structural Analysis of the Genome of Red Clover (Trifolium pratense L.). DNA Res. 2005, 12, 301–364. [Google Scholar] [CrossRef]
- Istvanek, J.; Dluhosova, J.; Dluhos, P.; Patkova, L.; Nedelnik, J.; Repkova, J. Gene Classification and Mining of Molecular Markers Useful in Red Clover (Trifolium pratense) Breeding. Front. Plant Sci. 2017, 8, 367. [Google Scholar] [CrossRef]
- Jones, C.; De Vega, J.; Lloyd, D.; Hegarty, M.; Ayling, S.; Powell, W.; Skøt, L. Population structure and genetic diversity in red clover (Trifolium pratense L.) germplasm. Sci. Rep. 2020, 10, 8364. [Google Scholar] [CrossRef] [PubMed]
- Nay, M.M.; Grieder, C.; Frey, L.A.; Amdahl, H.; Radovic, J.; Jaluvka, L.; Palme, A.; Skøt, L.; Ruttink, T.; Kölliker, R. Multilocation trials and population-based genotyping reveal high diversity and adaptation to breeding environments in a large collection of red clover. Front. Plant Sci. 2023, 14, 1128823. [Google Scholar] [CrossRef] [PubMed]
- Zanotto, S.; Ruttink, T.; Pegard, M.; Skøt, L.; Grieder, C.; Kölliker, R.; Ergon, Å. A genome-wide association study of freezing tolerance in red clover (Trifolium pratense L.) germplasm of European origin. Front. Plant Sci. 2023, 14, 1189662. [Google Scholar] [CrossRef] [PubMed]
- Singh, N.; Choudhury, D.R.; Singh, A.K.; Kumar, S.; Srinivasan, K.; Tyagi, R.K.; Singh, N.K.; Singh, R. Comparison of SSR and SNP Markers in Estimation of Genetic Diversity and Population Structure of Indian Rice Varieties. PLoS ONE 2013, 8, e84136. [Google Scholar] [CrossRef] [PubMed]
- Waples, R.S. Temporal Variation in Allele Frequencies: Testing the Right Hypothesis. Evolution 1989, 43, 1236–1251. [Google Scholar] [CrossRef]
- Šarčević, H. Genetske Promjene u M3 Sintetičkoj Populaciji Kukuruza (Zea mays L.) Izloženoj Rekurentnoj Selekciji. Ph.D. Thesis, University of Zagreb, Zagreb, Croatia, 2003. [Google Scholar]
- Weir, B.S. Diversity. In Genetic Data Analysis II: Methods for Discrete Population Genetic Data; Sinauer Associates, Inc.: Sunderland, MA, USA, 1996; pp. 141–160. [Google Scholar]
- Hartl, D.L.; Clark, A.G. Principles of Population Genetics, 3rd ed.; Sinauer Associates: Sunderland, MA, USA, 1997; p. 118. [Google Scholar]
- Nei, M. Analysis of Gene Diversity in Subdivided Populations. Proc. Natl. Acad. Sci. USA 1973, 70, 3321–3323. [Google Scholar] [CrossRef]
- Shannon, C.; Weaver, W. The Mathematical Theory of Communication; University of Illinois Press: Urbana, IL, USA, 1949; p. 87. [Google Scholar]
- Yeh, F.C.; Boyle, T.J.B. Population Genetic Analysis of Co-Dominant and Dominant Markers and Quantitative Traits. Belg. J. Bot. 1997, 129, 157. [Google Scholar]
- Fernández-Otero, C.I.; Ramos-Cabrer, A.M.; López-Díaz, J.E.; Pereira-Lorenzo, S. Evaluating the Diversity of Ecotypes of Red Clover (Trifolium pratense L.) from Northwestern Spain by Phenotypic Traits and Microsatellites. Agronomy 2021, 11, 2275. [Google Scholar] [CrossRef]
- Osterman, J.; Hammenhag, C.; Ortiz, R.; Geleta, M. Insights Into the Genetic Diversity of Nordic Red Clover (Trifolium pratense) Revealed by SeqSNP-Based Genic Markers. Front. Plant Sci. 2021, 12, 748750. [Google Scholar] [CrossRef]

| Parent Populations | Low Temperature Conditions | Reselections |
|---|---|---|
| P1 (cultivar ‘Reichersberger’) | One cycle | PS1 |
| P3 (cultivar ‘Croatia’) | One cycle | PS3 |
| SSR Locus | LG | Forward Primer Sequence | Reverse Primer Sequence | SSR Motif | Size (bp) |
|---|---|---|---|---|---|
| RCS1411 | LG4 | F: GGGTGTTTCATCGGAACAAT | R:CAACGAAACTAAACCCTAACCAA | AG | 187 |
| RCS1501 | LG2 | F: AGAAGGAAGCGTCGGTACAA | R:CAAGACGTCCACGAGCAATA | AAG | 180 |
| RCS1285 | LG2 | F: GATCCCCAACATCACCAATC | R:CAGACAAGGGTTGAGTTCAGTG | AG | 164 |
| RCS1952 | LG3 | F: CATGGGCTGTGTTGATTGAG | R:CTGCAGCAACAGCAACAACT | AAC | 210 |
| RCS3095 | LG2 | F: GTGTTCCATTAGAGGCGGAA | R:AGCGGCTCGTTTTAATGCTA | AAAT | 208 |
| RCS2408 | LG6 | F: GCGGAATCCCAAGATGAATA | R:CATCAGCAACACCAATGACC | AAG | 274 |
| RCS1667 | LG3 | F: AAGAGGCGAAGAAGCACAAC | R:TCTTTCTCCACGCTGTTCCT | GGA | 229 |
| RCS2010 | LG3 | F: GCTTCCACAGTTTTTGCTCC | R:GAACCTGCACAACCAAAGGT | ATC | 137 |
| RCS1866 | LG3 | F: GCAGCTTCCAGTAAAATCGC | R:GAGAGGAATCGGAGTGGTGA | AAG | 294 |
| RCS1897 | LG7 | F: CATGTCAGCATATCCATTTTCC | R:ATGAGCACCTTCACCAATCC | AAAG | 280 |
| RCS0907 | LG1 | F: ATTTGAGCACAAGGCCTCAC | R:TGGGGAAGTGAAGGATGTTC | AAC | 206 |
| RCS3681 | LG5 | F: AAAGCACGTGAAGAAAATGGA | R CCCTTCATCAATGGCTTTCT | ATC | 141 |
| RCS0428 | LG6 | F: GAATGCCAAGACACCTGTGA | R:TCTCATCAAGGGAGGTGGTC | ATC | 175 |
| RCS2185 | LG7 | F: AAACAATCAAAAACCGACAACA | R:TGCTGTTCCATCACCAATTT | AAG | 151 |
| RCS0035 | LG1 | F: CATTGTAGGTTATGTTTATCAGG | R:CCCAAAGCCTACAAGGAAAG | AC | 162 |
| RCS3709 | LG4 | F: TTCATCTTTCTCAACTTCATAATCA | R: CTGGGCTTGAATGAATTGGT | AAC | 278 |
| Population | Number of Alleles | Mean Allele Frequency | S.E. |
|---|---|---|---|
| P1 | 177 | 0.090 | ±0.007 |
| PS1 | 171 | 0.094 | ±0.009 |
| P3 | 182 | 0.088 | ±0.008 |
| PS3 | 148 | 0.108 | ±0.010 |
| Cycle | Ne 1 | Number of Significant Loci | |
|---|---|---|---|
| Number of Loci | Number of Alleles | ||
| P1 → PS1 | 38 | 12 | 27 |
| P3 → PS3 | 38 | 9 | 13 |
| Diversity Measure | Population | Population | |||||
|---|---|---|---|---|---|---|---|
| P1 | PS1 | p (T ≤ t) | P3 | PS3 | p (T ≤ t) | ||
| No alleles /locus | Mean | 11.06 | 10.69 | 0.4054 | 11.38 | 9.25 | 0.0961 |
| Min | 6 | 5 | 4 | 3 | |||
| Max | 20 | 17 | 19 | 17 | |||
| Hl | Mean | 0.714 | 0.705 | 0.4465 | 0.706 | 0.724 | 0.3583 |
| Min | 0.170 | 0.277 | 0.458 | 0.370 | |||
| Max | 0.917 | 0.917 | 0.938 | 0.938 | |||
| Dl | Mean | 0.796 | 0.767 | 0.2243 | 0.786 | 0.762 | 0.5435 |
| Min | 0.655 | 0.479 | 0.509 | 0.458 | |||
| Max | 0.944 | 0.913 | 0.932 | 0.897 | |||
| I | Mean | 1.885 | 1.824 | 0.3632 | 1.884 | 1.725 | 0.1969 |
| Min | 1.226 | 0.948 | 1.069 | 1.726 | |||
| Max | 2.833 | 2.615 | 2.724 | 2.468 | |||
| Locus | P1 | PS1 | P3 | PS3 | ||||
|---|---|---|---|---|---|---|---|---|
| FIS | p | FIS | p | FIS | p | FIS | p | |
| RCS-1411 | 0.13 | 0.998 | 0.03 | 0.731 | −0.02 | 0.999 | 0.03 | 0.997 |
| RCS-1866 | −0.07 | 0.690 | −0.16 | 0.756 | −0.06 | 0.983 | −0.15 | 0.666 |
| RCS-1667 | 0.06 | 1.000 | 0.06 | 0.997 | 0.00 | 0.888 | −0.06 | 0.635 |
| RCS-1897 | −0.01 | 0.967 | 0.05 | 0.977 | 0.08 | 0.879 | −0.12 | 0.437 |
| RCS-0907 | 0.77 | 0.000 | 0.57 | 0.000 | 0.23 | 0.021 | 0.48 | 0.000 |
| RCS-1501 | 0.44 | 0.492 | 0.35 | 0.404 | 0.20 | 0.875 | 0.21 | 0.600 |
| RCS-3095 | −0.03 | 0.703 | 0.21 | 0.415 | 0.29 | 0.048 | 0.15 | 0.004 |
| RCS-2010 | 0.08 | 0.173 | −0.04 | 0.739 | 0.09 | 0.077 | −0.08 | 0.944 |
| RCS-0428 | 0.02 | 0.928 | −0.05 | 0.999 | 0.08 | 1.000 | −0.06 | 0.837 |
| RCS-1285 | −0.02 | 0.999 | −0.01 | 0.999 | 0.13 | 1.000 | 0.04 | 0.986 |
| RCS-3681 | 0.07 | 0.253 | 0.06 | 0.373 | 0.07 | 0.695 | −0.32 | 0.073 |
| RCS-2185 | 0.01 | 0.966 | −0.04 | 1.000 | 0.10 | 1.000 | 0.12 | 0.600 |
| RCS-1952 | 0.00 | 1.000 | 0.05 | 0.993 | 0.00 | 0.999 | 0.08 | 0.078 |
| RCS-2408 | −0.10 | 0.281 | 0.05 | 0.462 | 0.16 | 0.052 | 0.15 | 0.451 |
| RCS-0035 | 0.19 | 0.107 | 0.17 | 0.185 | 0.11 | 0.130 | −0.02 | 0.251 |
| RCS-3709 | −0.02 | 0.999 | −0.10 | 0.986 | 0.00 | 0.995 | 0.03 | 0.992 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Primorac, J.; Šarčević, H.; Knezović, Z.; Vokurka, A.; Mandić, A.; Bolarić, S. Changes in Allele Frequencies and Genetic Diversity in Red Clover after Selection for Cold Tolerance Using SSR Markers. Agriculture 2023, 13, 2019. https://doi.org/10.3390/agriculture13102019
Primorac J, Šarčević H, Knezović Z, Vokurka A, Mandić A, Bolarić S. Changes in Allele Frequencies and Genetic Diversity in Red Clover after Selection for Cold Tolerance Using SSR Markers. Agriculture. 2023; 13(10):2019. https://doi.org/10.3390/agriculture13102019
Chicago/Turabian StylePrimorac, Jurica, Hrvoje Šarčević, Zrinka Knezović, Aleš Vokurka, Ana Mandić, and Snježana Bolarić. 2023. "Changes in Allele Frequencies and Genetic Diversity in Red Clover after Selection for Cold Tolerance Using SSR Markers" Agriculture 13, no. 10: 2019. https://doi.org/10.3390/agriculture13102019
APA StylePrimorac, J., Šarčević, H., Knezović, Z., Vokurka, A., Mandić, A., & Bolarić, S. (2023). Changes in Allele Frequencies and Genetic Diversity in Red Clover after Selection for Cold Tolerance Using SSR Markers. Agriculture, 13(10), 2019. https://doi.org/10.3390/agriculture13102019






