Influence of Incremental Short Term Salt Stress at the Seedling Stage on Root Plasticity, Shoot Thermal Profile and Ion Homeostasis in Contrasting Wheat Genotypes
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Material and Growth
2.2. Plant Height and Biomass
2.3. Chlorophyll Content (CCM)
2.4. Relative Water Content (RWC) and Membrane Stability Index (MSI)
2.5. Shoot Thermography
2.6. Stomatal Density
2.7. Analysis of Root Traits
2.8. Localization of Reactive Oxygen Species
2.9. Superoxide Radicals (SOR) Content
2.10. Estimation of Na+/K+ Ratio
2.11. Biochemical Parameters
2.12. Assay of Lipid Peroxidation
2.13. Assay of Super Oxide Dismutase Activity
2.14. Assay of Ascorbate Peroxidase Activity
2.15. Assay of Catalase Activity
2.16. Glutathione Reductase (GR)
2.17. Salt-Tolerance Index (STI)
2.18. Statistical Analysis
3. Results
3.1. Biomass, Relative Water Content (RWC), Membrane Stability Index (MSI), Chlorophyll Content Index (CCM)
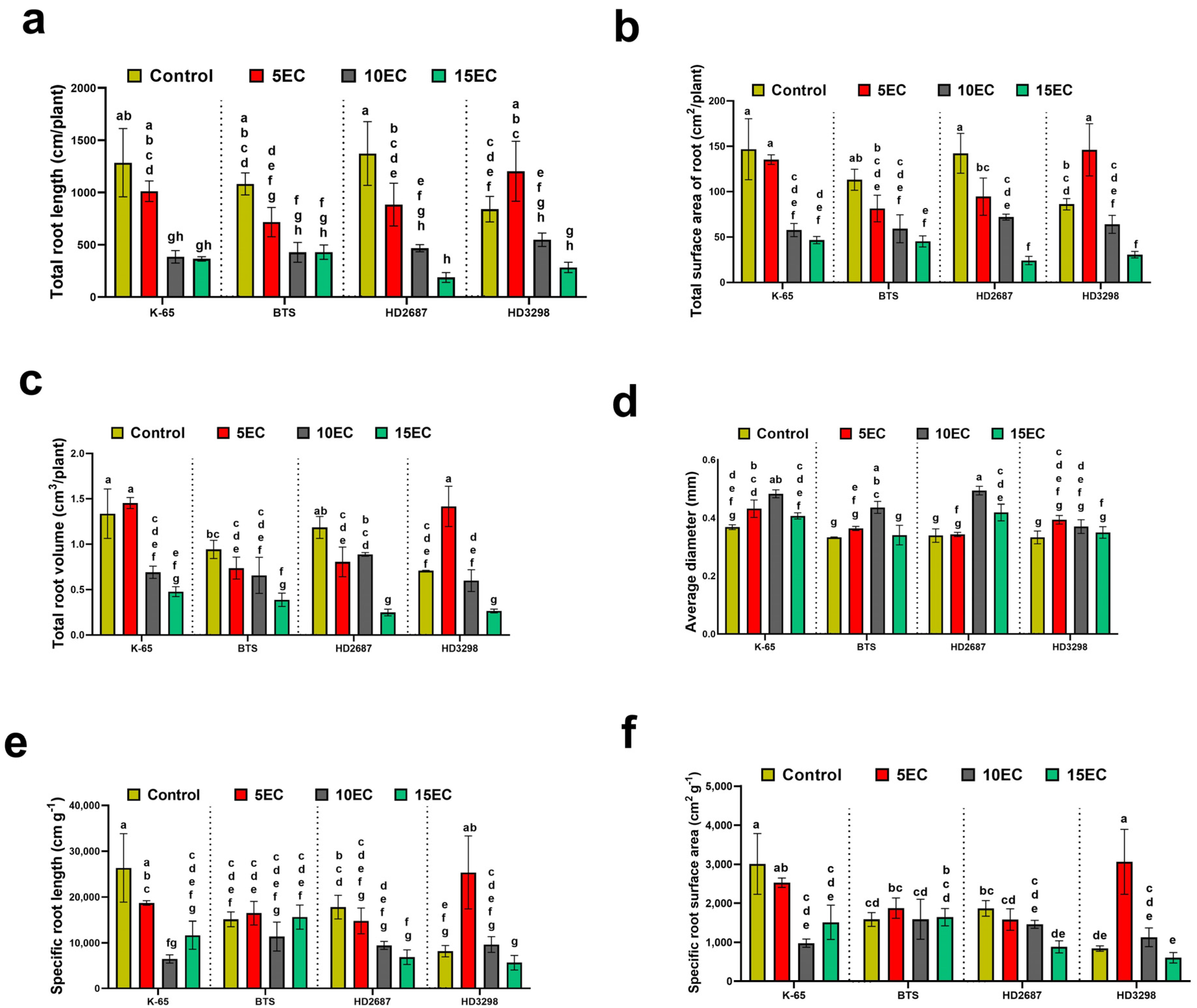
3.2. Root Traits
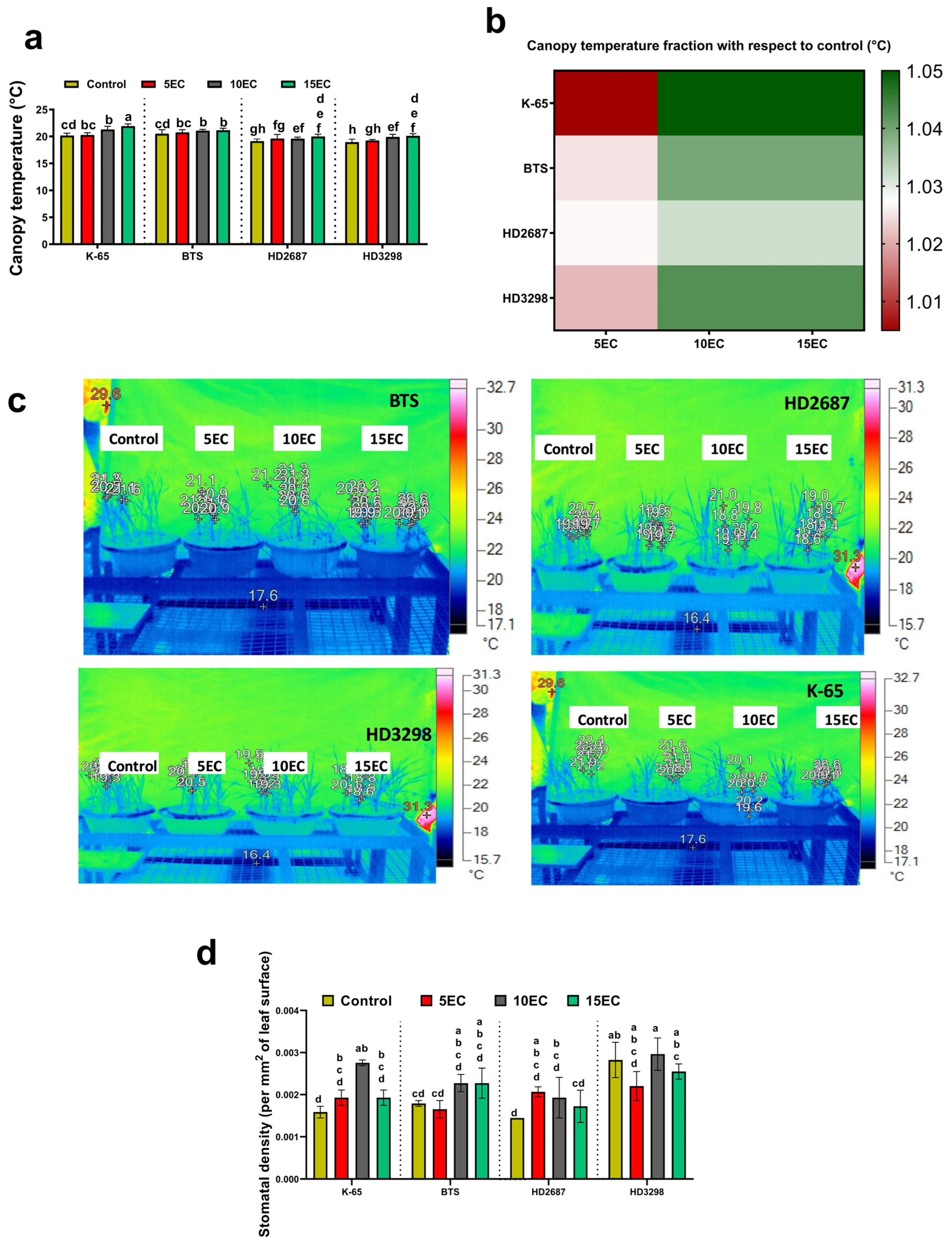
3.3. Shoot Thermal Image and Stomatal Density
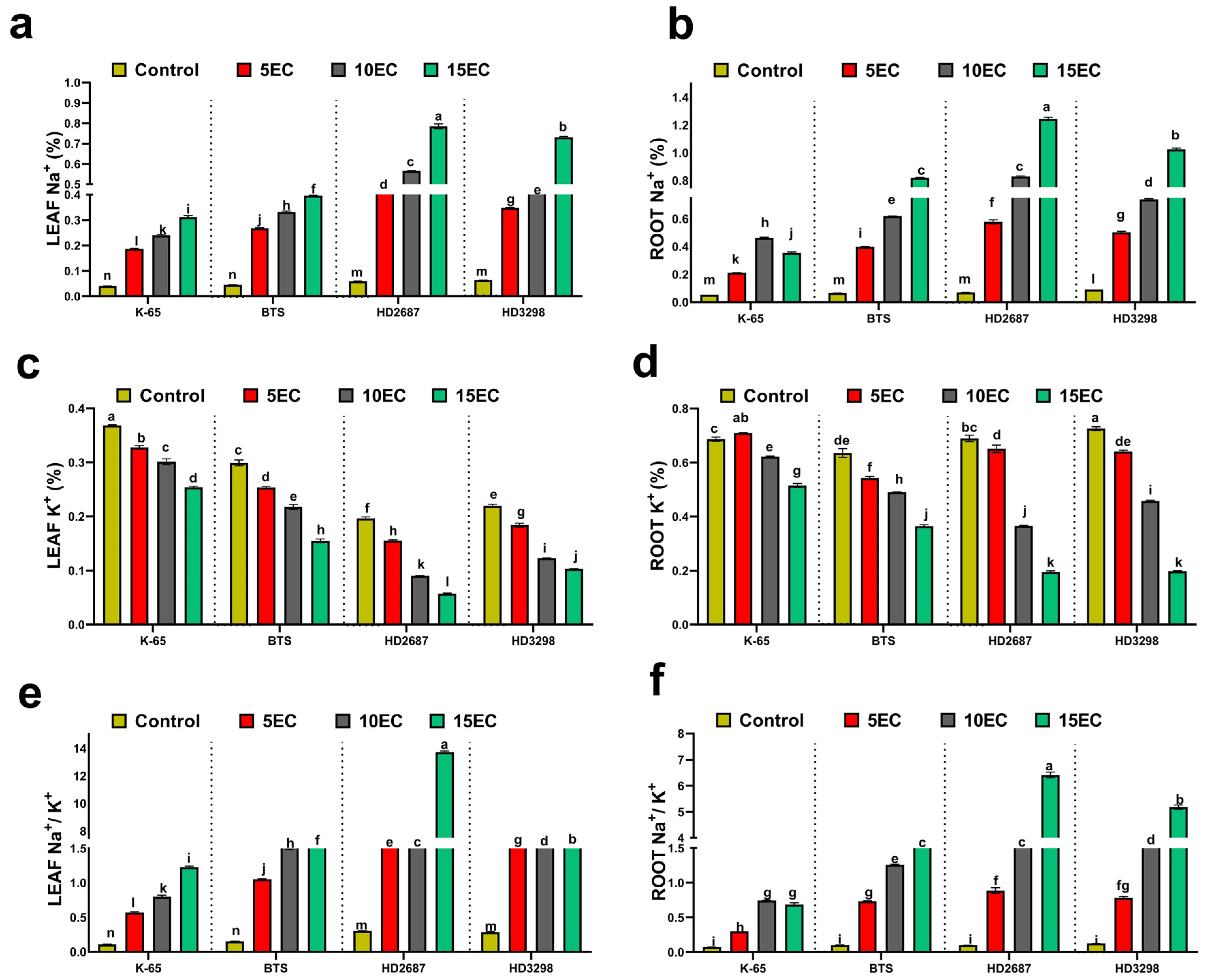
3.4. Tissue Na+ and K+ Content
3.5. Oxidative Stress Status of Tissues
3.6. Antioxidant Enzymes Activity
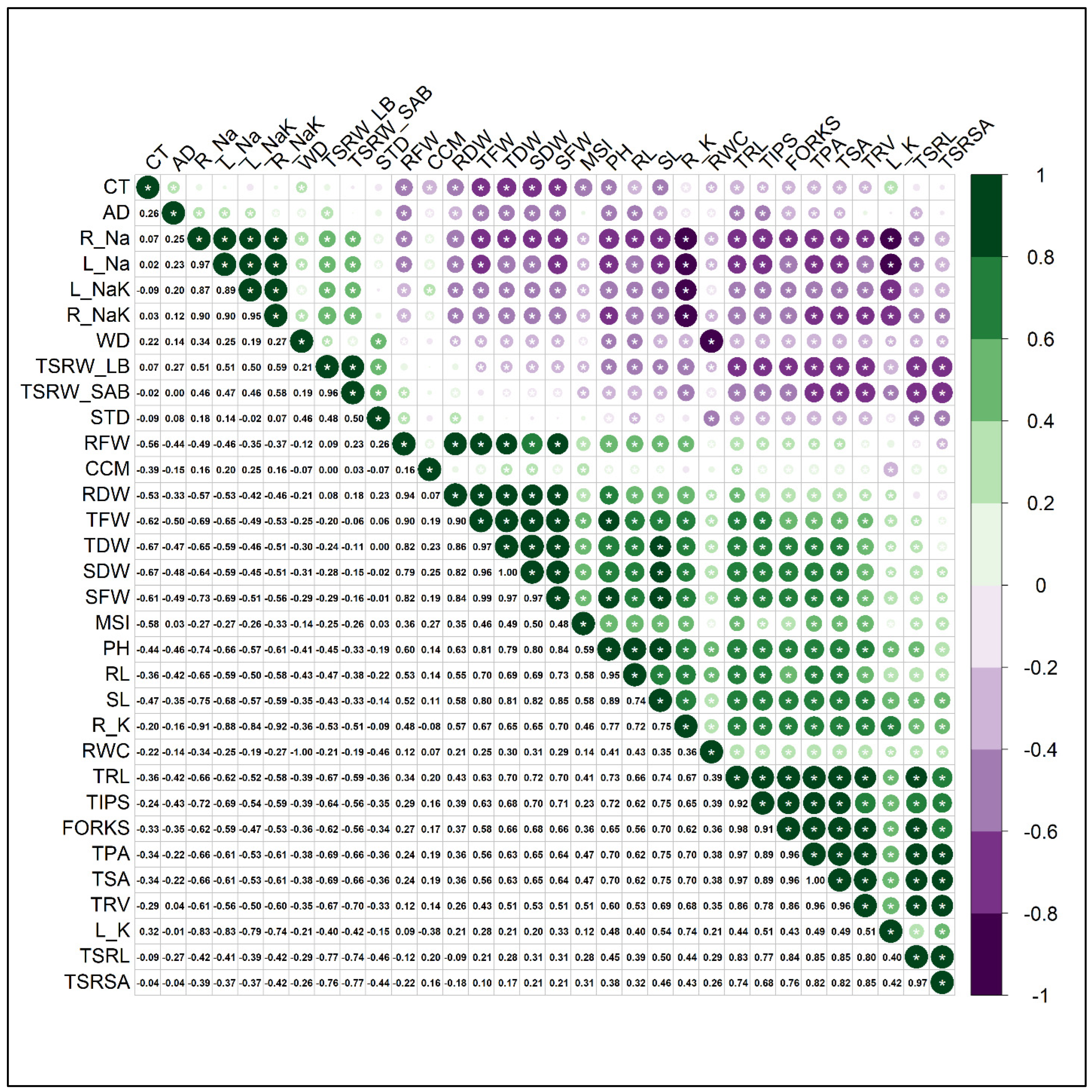
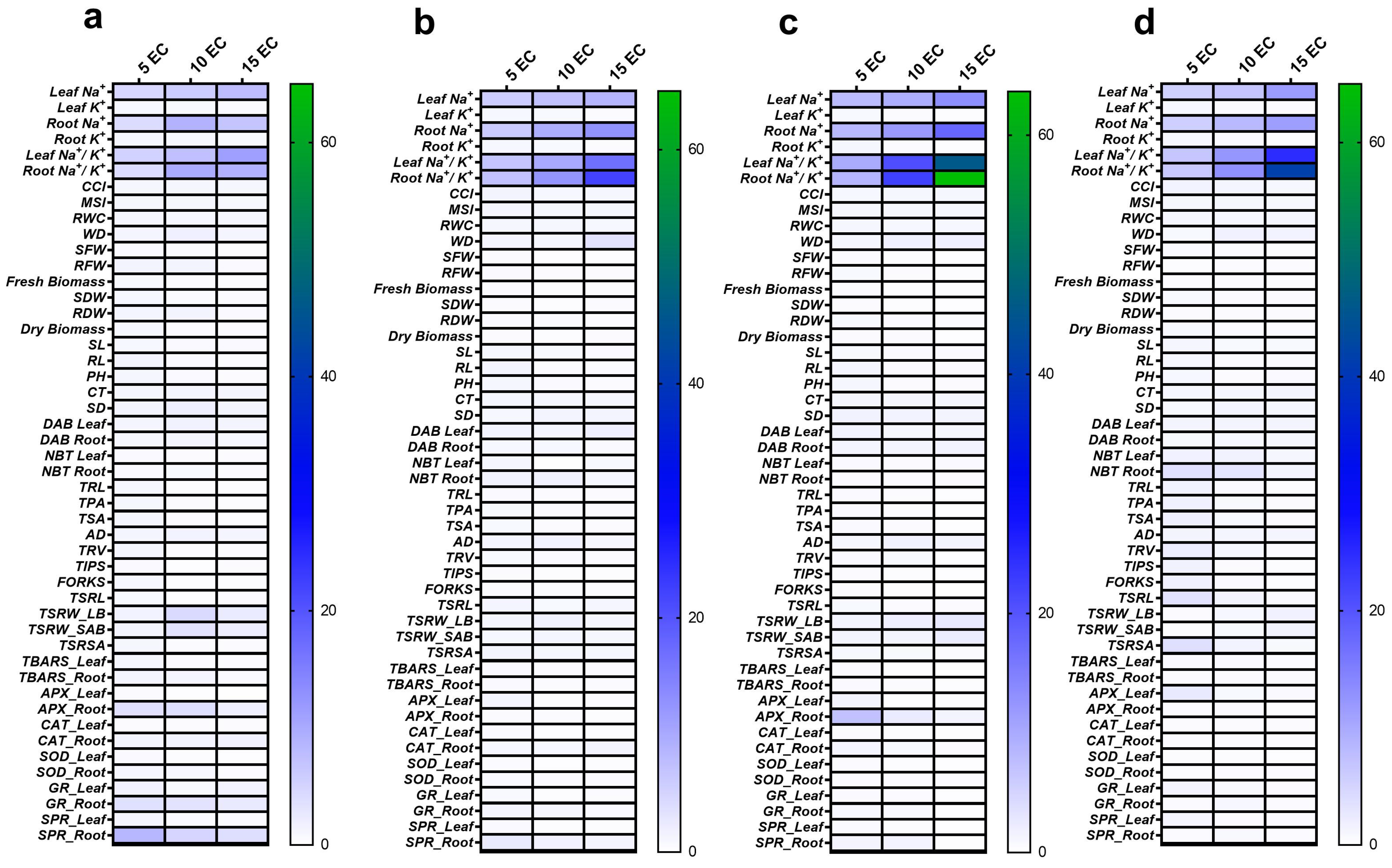
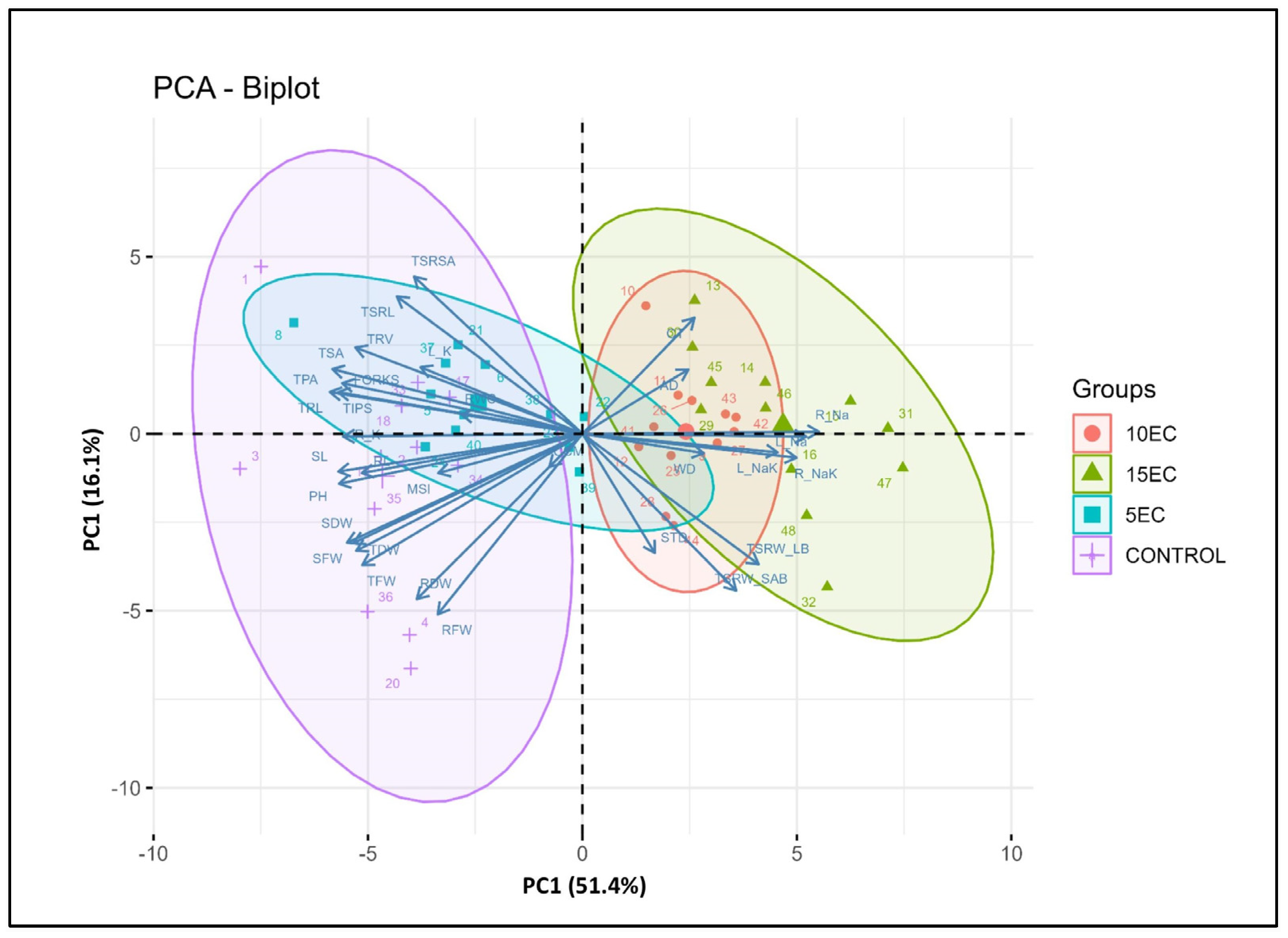
3.7. Correlation of Different Traits, STI and Principal Component Analysis (PCA)
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bhardwaj, S.C.; Prashar, M.; Kumar, S.; Jain, S.K.; Datta, D. Lr19 Resistance in Wheat Becomes Susceptible to Puccinia triticina in India. Plant Dis. 2005, 89, 1360. [Google Scholar] [CrossRef] [PubMed]
- Zörb, C.; Geilfus, C.M.; Dietz, K.J. Salinity and Crop Yield. Plant Biol. 2019, 21, 31–38. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Vinocur, B.; Altman, A. Plant Responses to Drought, Salinity and Extreme Temperatures: Towards Genetic Engineering for Stress Tolerance. Planta 2003, 218, 1–14. [Google Scholar] [CrossRef]
- Julkowska, M.M.; Testerink, C. Tuning Plant Signaling and Growth to Survive Salt. Trends Plant Sci. 2015, 20, 586–594. [Google Scholar] [CrossRef]
- Ashraf, M.; Harris, P.J.C. Photosynthesis under Stressful Environments: An Overview. Photosynthetica 2013, 51, 163–190. [Google Scholar] [CrossRef]
- Sathee, L.; Jha, S.K.; Rajput, O.S.; Singh, D.; Kumar, S.; Kumar, A. Expression Dynamics of Genes Encoding Nitrate and Ammonium Assimilation Enzymes in Rice Genotypes Exposed to Reproductive Stage Salinity Stress. Plant Physiol. Biochem. 2021, 165, 161–172. [Google Scholar] [CrossRef]
- Rengasamy, P. World Salinization with Emphasis on Australia. J. Exp. Bot. 2006, 57, 1017–1023. [Google Scholar] [CrossRef]
- Qadir, M.; Quillérou, E.; Nangia, V.; Murtaza, G.; Singh, M.; Thomas, R.J.; Drechsel, P.; Noble, A.D. Economics of Salt-Induced Land Degradation and Restoration. Nat. Resour. Forum 2014, 38, 282–295. [Google Scholar] [CrossRef]
- Singh, S.; Sengar, R.S.; Kulshreshtha, N.; Datta, D.; Tomar, R.S.; Rao, V.P.; Garg, D.; Ojha, A. Assessment of Multiple Tolerance Indices for Salinity Stress in Bread Wheat (Triticum aesticum L.). J. Agric. Sci. 2015, 7, 49–57. [Google Scholar]
- Goyal, E.; Amit, S.K.; Singh, R.S.; Mahato, A.K.; Chand, S.; Kanika, K. Transcriptome Profiling of the Salt-Stress Response in Triticum Aestivum Cv. Kharchia Local. Sci. Rep. 2016, 6, 27752. [Google Scholar] [CrossRef]
- Lekshmy, S.; Sairam, R.K.; Chinnusamy, V.; Jha, S.K. Differential Transcript Abundance of Salt Overly Sensitive (SOS) Pathway Genes Is a Determinant of Salinity Stress Tolerance of Wheat. Acta Physiol. Plant. 2015, 37, 1–10. [Google Scholar] [CrossRef]
- Sairam, R.K.; Rao, K.; Srivastava, G.C. Differential Response of Wheat Genotypes to Long Term Salinity Stress in Relation to Oxidative Stress, Antioxidant Activity and Osmolyte Concentration. Plant Sci. 2002, 163, 1037–1046. [Google Scholar] [CrossRef]
- Chaurasia, S.; Kumar, A.; Singh, A.K. Comprehensive Evaluation of Morpho-Physiological and Ionic Traits in Wheat (Triticum aestivum L.) Genotypes under Salinity Stress. Agriculture 2022, 12, 1765. [Google Scholar] [CrossRef]
- Munns, R.; James, R.A.; Läuchli, A. Approaches to Increasing the Salt Tolerance of Wheat and Other Cereals. J. Exp. Bot. 2006, 57, 1025–1043. [Google Scholar] [CrossRef]
- Munns, R.; Gilliham, M. Salinity Tolerance of Crops—What Is the Cost? New Phytol. 2015, 208, 668–673. [Google Scholar] [CrossRef]
- Lekshmy, S.; Vanita, J.; Sangeeta, K.; Pandey, R.; Rajendra, S. Effect of Elevated Carbon Dioxide on Kinetics of Nitrate Uptake in Wheat Roots. Indian J. Plant Physiol. 2009, 14, 16–22. [Google Scholar]
- Weatherley, P. Studies in Water Relations of Cotton Plants. I. The Field Measurement of Water Deficit in Leaves. New Phytol. 1950, 49, 81–87. [Google Scholar] [CrossRef]
- Sairam, R.K.; Deshmukh, P.S.; Shukla, D.S. Tolerance of Drought and Temperature Stress in Relation to Increased Antioxidant Enzyme Activity in Wheat. J. Agron. Crop Sci. 1997, 178, 171–178. [Google Scholar] [CrossRef]
- Naguib, W.B.; Divte, P.R.; Chandra, A.; Sathee, L.; Singh, B.; Mandal, P.K.; Anand, A. Raffinose Accumulation and Preferential Allocation of Carbon (14C) to Developing Leaves Impart Salinity Tolerance in Sugar Beet. Physiol. Plant. 2021, 173, 1421–1433. [Google Scholar] [CrossRef]
- Sathee, L.; Jain, V. Interaction of Elevated CO2 and Form of Nitrogen Nutrition Alters Leaf Abaxial and Adaxial Epidermal and Stomatal Anatomy of Wheat Seedlings. Protoplasma 2021, 2021, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Kusumi, K.; Hirotsuka, S.; Kumamaru, T.; Iba, K. Increased Leaf Photosynthesis Caused by Elevated Stomatal Conductance in a Rice Mutant Deficient in SLAC1, a Guard Cell Anion Channel Protein. J. Exp. Bot. 2012, 63, 5635–5644. [Google Scholar] [CrossRef]
- Jagadhesan, B.; Sathee, L.; Meena, H.S.; Jha, S.K.; Chinnusamy, V.; Kumar, A.; Kumar, S. Genome Wide Analysis of NLP Transcription Factors Reveals Their Role in Nitrogen Stress Tolerance of Rice. Sci. Rep. 2020, 10, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Kumar, D.; Yusuf, M.; Singh, P.; Sardar, M.; Sarin, N. Histochemical Detection of Superoxide and H2O2 Accumulation in Brassica Juncea Seedlings. Bio-Protocol 2014, 4, e1108. [Google Scholar] [CrossRef]
- Chaitanya, K.S.K.; Naithani, S.C. Role of Superoxide, Lipid Peroxidation and Superoxide Dismutase in Membrane Perturbation during Loss of Viability in Seeds of Shorea Robusta Gaertn.F. New Phytol. 1994, 126, 623–627. [Google Scholar] [CrossRef]
- Heath, R.L.; Packer, L. Photoperoxidation in Isolated Chloroplasts: I. Kinetics and Stoichiometry of Fatty Acid Peroxidation. Arch. Biochem. Biophys. 1968, 125, 189–198. [Google Scholar] [CrossRef] [PubMed]
- Dhindsa, R.S.; Plumb-Dhindsa, P.; Thorpe, T.A. Leaf Senescence: Correlated with Increased Levels of Membrane Permeability and Lipid Peroxidation, and Decreased Levels of Superoxide Dismutase and Catalase. J. Exp. Bot. 1981, 32, 93–101. [Google Scholar] [CrossRef]
- Nakano, Y.K.A. Hydrogen Peroxide Is Scavenged by Ascorbate Specific Peroxidase in Spinach Chloroplasts. Plant Cell Physiol. 1981, 22, 867–880. [Google Scholar]
- Aebi, H. Catalase. In Methods in Enzymatic Analysis; Bergmeyer, H.V., Ed.; Academic Press Inc.: New York, NY, USA, 1974; pp. 673–686. [Google Scholar]
- Smith, I.K.; Vierheller, T.L.; Thorne, C.A. Assay of Glutathione Reductase in Crude Tissue Homogenates Using 5,5′-Dithiobis(2-Nitrobenzoic Acid). Anal. Biochem. 1988, 175, 408–413. [Google Scholar] [CrossRef]
- Flowers, T.J.; Troke, P.F.; Yeo, A.R. The Mechanism of Salt Tolerance in Halophytes. Annu. Rev. Plant Physiol. 1977, 28, 89–121. [Google Scholar] [CrossRef]
- Munns, R.; Tester, M. Mechanisms of Salinity Tolerance. Annu. Rev. Plant Biol. 2008, 59, 651–681. [Google Scholar] [CrossRef]
- Katori, T.; Ikeda, A.; Iuchi, S.; Kobayashi, M.; Shinozaki, K.; Maehashi, K.; Sakata, Y.; Tanaka, S.; Taji, T. Dissecting the Genetic Control of Natural Variation in Salt Tolerance of Arabidopsis thaliana Accessions. J. Exp. Bot. 2010, 61, 1125–1138. [Google Scholar] [CrossRef]
- Sirault, X.R.R.; James, R.A.; Furbank, R.T.; Sirault, X.R.R.; James, R.A.; Furbank, R.T. A New Screening Method for Osmotic Component of Salinity Tolerance in Cereals Using Infrared Thermography. Funct. Plant Biol. 2009, 36, 970–977. [Google Scholar] [CrossRef] [PubMed]
- Epstein, E.; Norlyn, J.D.; Rush, D.W.; Kingsbury, R.W.; Kelley, D.B.; Cunningham, G.A.; Wrona, A.F. Saline Culture of Crops: A Genetic Approach. Science 1980, 210, 399–404. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Schmidhalter, U. Opportunity and Challenges of Phenotyping Plant Salt Tolerance. Trends Plant Sci. 2023, 28, 552–566. [Google Scholar] [CrossRef] [PubMed]
- Achard, P.; Cheng, H.; De Grauwe, L.; Decat, J.; Schoutteten, H.; Moritz, T.; van de Straeten, D.; Peng, J.; Harberd, N.P. Integration of Plant Responses to Environmentally Activated Phytohormonal Signals. Science 2006, 311, 91–94. [Google Scholar] [CrossRef]
- Geng, Y.; Wu, R.; Wee, C.W.; Xie, F.; Wei, X.; Chan, P.M.Y.; Tham, C.; Duan, L.; Dinneny, J.R. A Spatio-Temporal Understanding of Growth Regulation during the Salt Stress Response in Arabidopsis. Plant Cell 2013, 25, 2132–2154. [Google Scholar] [CrossRef] [PubMed]
- West, G.; Inze, D.; Beemster, G.T. Cell Cycle Modulation in the Response of the Primary Root of Arabidopsis to Salt Stress. Plant Physiol. 2004, 135, 1050–1058. [Google Scholar] [CrossRef]
- Essa, T.A. Effect of Salinity Stress on Growth and Nutrient Composition of Three Soybean (Glycine Max L. Merrill) Cultivars. J. Agron. Crop Sci. 2002, 188, 86–93. [Google Scholar] [CrossRef]
- Benito, B.; Haro, R.; Amtmann, A.; Cuin, T.A.; Dreyer, I. The Twins K+ and Na+ in Plants. J. Plant Physiol. 2014, 171, 723–731. [Google Scholar] [CrossRef]
- Julkowska, M.M.; Hoefsloot, H.C.; Mol, S.; Feron, R.; de Boer, G.J.; Haring, M.A.; Testerink, C. Capturing Arabidopsis Root Architecture Dynamics with ROOT-FIT Reveals Diversity in Responses to Salinity. Plant Physiol. 2014, 166, 1387–1402. [Google Scholar] [CrossRef]
- Zolla, G.; Heimer, Y.M.; Barak, S. Mild Salinity Stimulates a Stress-Induced Morphogenic Response in Arabidopsis Thaliana Roots. J. Exp. Bot. 2010, 61, 211–224. [Google Scholar] [CrossRef] [PubMed]
- McLoughlin, F.; Galvan-Ampudia, C.S.; Julkowska, M.M.; Caarls, L.; Van Der Does, D.; Laurière, C.; Munnik, T.; Haring, M.A.; Testerink, C. The Snf1-Related Protein Kinases SnRK2.4 and SnRK2.10 Are Involved in Maintenance of Root System Architecture during Salt Stress. Plant J. 2012, 72, 436–449. [Google Scholar] [CrossRef] [PubMed]
- Faiyue, B.; Al-Azzawi, M.J.; Flowers, T.J. The Role of Lateral Roots in Bypass Flow in Rice (Oryza sativa L.). Plant Cell Environ. 2010, 33, 702–716. [Google Scholar] [PubMed]
- Faiyue, B.; Vijayalakshmi, C.; Nawaz, S.; Nagato, Y.; Taketa, S.; Ichii, M.; Al-Azzawi, M.J.; Flowers, T.J. Studies on Sodium Bypass Flow in Lateral Rootless Mutants Lrt1and Lrt2, and Crown Rootless Mutant Crl1of Rice (Oryza sativa L.). Plant Cell Environ. 2010, 33, 687–701. [Google Scholar] [PubMed]
- Faiyue, B.; Al-Azzawi, M.J.; Flowers, T.J. A New Screening Technique for Salinity Resistance in Rice (Oryza Sativa L.) Seedlings Using Bypass Flow. Plant Cell Environ. 2012, 35, 1099–1108. [Google Scholar] [CrossRef] [PubMed]
- Julkowska, M.M.; McLoughlin, F.; Galvan-Ampudia, C.S.; Rankenberg, J.M.; Kawa, D.; Klimecka, M.; Haring, M.A.; Munnik, T.; Kooijman, E.E.; Testerink, C. Identification and Functional Characterization of the Arabidopsis Snf1-Related Protein Kinase SnRK2.4 Phosphatidic Acid-Binding Domain. Plant Cell Environ. 2014, 38, 614–624. [Google Scholar] [CrossRef]
- Wu, H.; Guo, J.; Wang, C.; Li, K.; Zhang, X.; Yang, Z.; Li, M.; Wang, B. An Effective Screening Method and a Reliable Screening Trait for Salt Tolerance of Brassica napus at the Germination Stage. Front. Plant Sci. 2019, 10, 530. [Google Scholar] [CrossRef]
- Huqe, M.A.S.; Haque, M.S.; Sagar, A.; Uddin, M.N.; Hossain, M.A.; Hossain, A.Z.; Rahman, M.M.; Wang, X.; Al-Ashkar, I.; Ueda, A.; et al. Characterization of Maize Hybrids (Zea mays L.) for Detecting Salt Tolerance Based on Morpho-Physiological Characteristics, Ion Accumulation and Genetic Variability at Early Vegetative Stage. Plants 2021, 10, 2549. [Google Scholar] [CrossRef]
- Sivakumar, J.; Prashanth, J.E.P.; Rajesh, N.; Reddy, S.M.; Pinjari, O.B. Principal component analysis approach for comprehensive screening of salt stress-tolerant tomato germplasm at the seedling stage. J. Biosci. 2020, 45, 141. [Google Scholar] [CrossRef]
- Sivakumar, J.; Sridhar Reddy, M.; Sergeant, K.; Hausman, J.F.; ShaValli Khan, P.S.; Osman Basha, P. Principal component analysis-assisted screening and selection of salt-tolerant tomato genotypes. Plant Physiol. Rep. 2023, 28, 272–288. [Google Scholar] [CrossRef]
- Kose, A.; Oguz, O.; Ozlem, B.; Ferda, K. Application of multivariate statistical analysis for breeding strategies of spring safflower (Carthamus tinctorius L.). Turkish J. F. Crop. 2018, 23, 12–19. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Boopal, J.; Sathee, L.; Ramasamy, R.; Pandey, R.; Chinnusamy, V. Influence of Incremental Short Term Salt Stress at the Seedling Stage on Root Plasticity, Shoot Thermal Profile and Ion Homeostasis in Contrasting Wheat Genotypes. Agriculture 2023, 13, 1946. https://doi.org/10.3390/agriculture13101946
Boopal J, Sathee L, Ramasamy R, Pandey R, Chinnusamy V. Influence of Incremental Short Term Salt Stress at the Seedling Stage on Root Plasticity, Shoot Thermal Profile and Ion Homeostasis in Contrasting Wheat Genotypes. Agriculture. 2023; 13(10):1946. https://doi.org/10.3390/agriculture13101946
Chicago/Turabian StyleBoopal, Jagadhesan, Lekshmy Sathee, Ramesh Ramasamy, Rakesh Pandey, and Viswanathan Chinnusamy. 2023. "Influence of Incremental Short Term Salt Stress at the Seedling Stage on Root Plasticity, Shoot Thermal Profile and Ion Homeostasis in Contrasting Wheat Genotypes" Agriculture 13, no. 10: 1946. https://doi.org/10.3390/agriculture13101946
APA StyleBoopal, J., Sathee, L., Ramasamy, R., Pandey, R., & Chinnusamy, V. (2023). Influence of Incremental Short Term Salt Stress at the Seedling Stage on Root Plasticity, Shoot Thermal Profile and Ion Homeostasis in Contrasting Wheat Genotypes. Agriculture, 13(10), 1946. https://doi.org/10.3390/agriculture13101946






