Abstract
The bacterial blight (BB) disease of rice is a major disease that reduces yield heavily in susceptible varieties. Ranidhan is a late maturing popular rice variety but shows high susceptibility to the disease. Two BB resistance genes were transferred into the variety through a marker-assisted backcross breeding approach. Tightly linked molecular markers were deployed to track the BB resistance genes in the plants carrying the target genes in each backcross generation. Foreground screening detected 17, 16 and 15 progenies to carry the 3 BB resistance genes in BC1F1, BC2F1 and BC3F1 generations, respectively. The selected BC3F1 plant was selfed and three different combinations of BB resistance genes were tracked in homozygous state in seven BC3F2 plants. The pyramided lines carrying three resistance genes in homozygous conditions were evaluated for BB disease resistance by inoculating with eight virulent Xoo strains. Five pyramided lines carrying two resistance gene combinations (Xa21+xa13 and Xa21+xa5) exhibited enhanced resistance against the BB pathogens. The disease resistance was in the order of Xa21+xa5 < Xa21+xa13 < xa13+xa5 gene combinations in conferring the resistance. The developed pyramided lines were similar to the recipient parent for the majority of the important agro-morphologic and grain quality traits.
1. Introduction
Rice, the queen of cereals, serves as food and fodder for millions of people and domesticated animals around the world. The importance of the crop in India is not only as a staple food but also taken as a ceremonial crop [1]. Its dietary composition includes carbohydrates, proteins, oils, vitamins, antioxidants, fiber, minerals and a variety of plant chemicals [2]. Rice cultivation is possible in high elevations as well asbelow sea level due to its wide adaptability. It is cultivated nearly 45% as a rainfed crop out of a total of 160 mha of rice land in the world [3]. Contributing to the livelihood of about 400 crores of people constituting nearly 55% of the world population, rice generates nearly $206 billion annually, which is about 17% of the total value of the crop [4]. The crop is highly affected by adverse climatic effects in recent years, which is reflected in the production from the rainfed rice cultivation in the world in general and in India in particular [5]. At the juncture of an alarming population increase and resources such as land, labor, inputs and chemicals, future production should be obtained in an eco-friendly manner [6,7]. Therefore, popular varieties deficient in resistance to major diseases and pests should be developed for sustainable production. In this investigation, the improvement of BB resistance and grain yield were targeted in rice.
Many biotic and abiotic stresses are the main constraints for higher rice production in rainfed ecology. BB is the major constraint amongst the biotic stresses in reducing productivity in low land ecology. The ecology covers about 15 mha in India, of which >80% are located in the eastern states of the country [8]. BB disease is a serious yield-limiting factor in the late maturing group of rice which is further aggravated by the application of a high dose of nitrogenous fertilizer for higher yield that is favorable for this disease development. The yield losses due to this disease normally vary from 20% to 40% [9], but reduce the crop yield up to 50% [10] or even up to 80% in Asia [11].
Globally, a total of 45 BB resistance genes have been identified from diverse sources [12]. Many elite donor lines with different resistance genes are being used in the BB resistance improvement programs [13]. The resistance genes, namely Xa4, xa5, Xa7, xa13 and Xa21, have been extensively utilized in BB resistance improvement programs [14,15,16]. Suitable combinations of BB resistance genes in a variety of backgrounds are reported to be effective against the pathogen isolates. However, large acreage and long-term growth of few varieties with single resistance genes may break down the pathogen resistance. The resistance breakdown can be delayed by pyramiding suitable BB resistance genes into rice cultivars. It is difficult to transfer multiple resistance genes simultaneously through classical improvement programs. Recently, many closely linked molecular markers are reported for use in the BB resistance genes detection carrying two and three resistance genes together in a plant. The mutation in pathogen for virulence to two or more resistance genes in a suitable combination is much lower than for a single resistance gene in a cultivar. Multiple resistance gene pyramiding in a single genetic background provides a durable and broad-spectrum resistance against pathogens. Many closely linked molecular markers are available for BB resistance genes and are successfully deployed through the marker-assisted breeding approach [17,18,19,20,21,22,23,24,25,26,27,28,29]. The development of pyramided lines carrying two BB resistance genes, xa13+Xa21 and xa5+Xa21 through marker-assisted selection in the popular rice variety, Ranidhan is reported here. The results indicated an increased resistance to the BB disease against the eight Xoo strains populations.
2. Materials and Methods
2.1. Plant Materials Used in the Breeding Program
The elite parent, CR Dhan 800 carrying 3 resistance genes Xa21, xa13 and xa5 in the Swarna variety background for BB resistance was used as the male donor parent in the hybridization program. The recurrent parent, Ranidhan, is a late maturing variety but is highly susceptible to BB disease. The parental lines were obtained from the gene bank of the National Rice Research Institute (NRRI), Cuttack, India. Ranidhan, the recipient parent, was crossed with the donor variety, CR Dhan 800, during the wet season of 2017 as per the marker-assisted breeding scheme (Figure 1). A true F1 plant developed during the previous wet season was backcrossed with the recipient parent during the dry season of 2018 for the production of BC1F1 seeds. Three BB resistance genes were tracked in the BC1F1 plants using the closely linked markers (Table 1). BC1F1 plant with more similarity with recipient variety was hybridized to produce BC2F1 seeds during the wet season of 2018. BC2F1 generation plants were developed during the dry season of 2019. BC2F1 plant with more similarity with a recurrent parent was hybridized to get BC3F1 seeds. Similarly, BC3F1 plants were developed during the wet season of 2019. BC3F2 generation progenies were produced from the selected foreground positive plants during the dry season of 2020. Foreground selection was performed in BC3F2 progenies to select the lines carrying the suitable target gene combinations. Evaluation of the pyramided and parental lines for yield, agro-morphologic and grain quality traits were conducted during the wet seasons of 2020 and 2021.
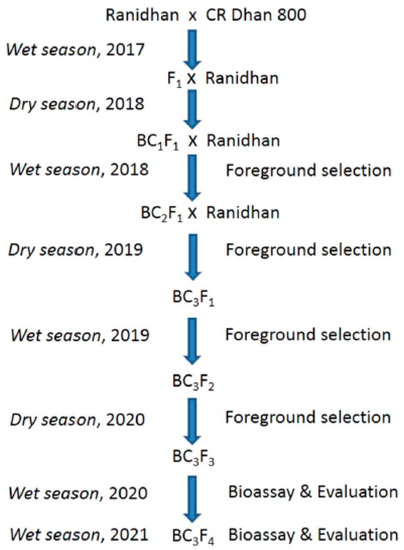
Figure 1.
Marker-assisted breeding scheme for transfer of BB resistance genes into the popular variety, Ranidhan.

Table 1.
Markers used for the three bacterial blight resistance genes for foreground selection in the marker-assisted backcross breeding program.
2.2. DNA Isolation, PCR and Marker Analysis
The protocol of Dellaporta et al. was used for DNA extraction [32]. PCR was performed following the standard procedure used in an earlier publication [33]. The markers used for the 3 BB resistance genes in the marker-assisted backcross breeding program are presented in Table 1. For the three target genes, four gene-specific and tightly linked markers were used in the foreground selection (Table 1). The PCR products were separated by using agarose gel electrophoresis. The gel documentation system (SynGene, Cambridge, UK) was used to capture the image of the bands obtained in the electrophoresis. The data analysis and dendrogram construction were done following the earlier publications [34,35,36].
2.3. Bioassay of Bacterial Blight
All the BC3F3 and BC3F4 pyramided lines of forty-five-days-old seedlings along with the parents were inoculated with eight isolates of Xoo during the wet seasons of 2020 and 2021. The Xoo isolates were maintained at ICAR-NRRI, and Cuttack was used for the bioassay study. The bacterial suspension is prepared by suspending the bacterial mass in sterile water to a concentration of approximately 109 cells/mL [37]. Five leaves from different five plants of each entry at the maximum tillering stage were taken for inoculation. The disease symptom and the lesion lengths (LL) were measured after 15 days. The mean lesion length was used for scoring and used as resistant (R) for LL ≤ 3.0 cm, moderately resistant (MR) for 3.0 cm < LL ≤ 6.0 cm and moderately susceptible (MS) for 6.0 cm < LL ≤ 9.0 cm or susceptible (S, LL > 9.0 cm) [38].
2.4. Evaluation of the Genotypes for Various Agro-Morphological Characters
Twenty-five-day-old genotypes carrying the BB resistance genes were transplanted along with the parents during the wet seasons, 2020 and 2021 at ICAR-NRRI, Cuttack. The materials were planted at a spacing of 20 × 15 cm2 in three replications in a randomized block design providing a plot size of 5.25 m2 to each entry with 35 plants per row. The data for ten plants/each entry/replication was recorded for agro-morphologic characters namely, plant height (cm), panicle weight (g), panicles per plant, number of grains per panicle, panicle length (cm), grain breadth (mm), grain length (mm) and 1000-grain weight while the data on days to 50% flowering and plot yield were estimated on the whole plot basis. SAS statistical software was used for the data analysis. Principal component analysis (PCA) for distributing the pyramided and parental lines was drawn by the multivariate analysis for the 10 morphologic and quality traits. A scatter plot was generated by plotting two major components: principal component 1 (PC1) and principal component 2 (PC2). The percentage of variance and eigen value were generated by the interaction of a variance–covariance matrix. All the plots and results of PCA were generated as per a standard procedure following the previous publications [39,40,41].
3. Results
3.1. Development of Pyramided Lines Carrying BB Resistance Genes
3.1.1. Validation of the Target Resistance Genesin the Parental Lines
The presence of the target genes in the parental lines was confirmed before the beginning of the hybridization and selection activities. Three BB resistance genes were confirmed in the parent, CR Dhan 800, whereas these genes were absent in the recipient parent, Ranidhan (Figure 1). Tightly linked molecular markers were used for the validation of the bacterial blight resistance genes in the parental lines (Table 1).
3.1.2. Screening of the BB Resistance Genes in BC1F1Progenies
The popular variety, Ranidhan, was hybridized with the donor parent, CR Dhan 800 and 57 F1 seeds were produced. These seeds were planted in the next season and the true hybridity was confirmed by genotyping the hybrid plants using the BB gene markers. A true hybrid plant (F1) was backcrossed with the recipient parent, Ranidhan, and 145 BC1F1 crossed seeds were produced. The crossed seeds were raised for BC1F1 generation, and foreground selection was done in all the plants using the linked markers for the bacterial blight resistance gene (Figure 2).
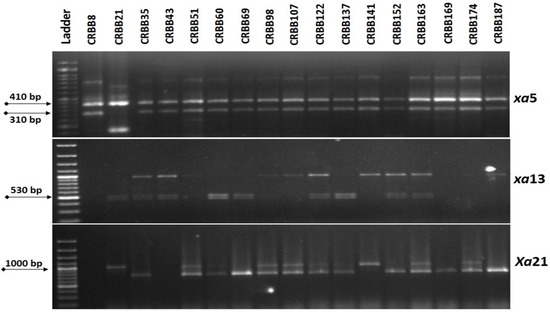
Figure 2.
Representative electropherogram generated from the tracking of Xa21, xa13 and xa5 BB resistance genes in BC1F1 progenies of resistance gene pyramiding of Ranidhan.
Out of a total of 145 BC1F1 plants, the screening results revealed the presence of the xa5 gene in 71 derivatives showing an expected band size of 440 bp. Tracking for the presence of the xa13 gene was confirmed in 35 derivatives with an expected band size of 530 bp. Finally, screening of the 35 positive plants resulted in 17 derivatives with the presence of the Xa21 gene detected with the expected band size of 530 bp (Figure 2). Out of these 17 plants, the progeny showing more similarity with Ranidhan was selected and backcrossed and 156 seeds were generated.
3.1.3. Screening of the BB Resistance Genes in BC2F1 Progenies
All the generated BC2F1 seeds were raised in the next season for selection. Foreground selection was performed in all the BC2F1 progenies using the closely linked markers with the target BB resistance genes. Out of 156 BC2F1 progenies, the screening revealed the presence of the Xa21 gene in 71 plants. Screening of those 71 plants for the xa5 gene revealed its presence in 33 plants. Final screening for the xa5 gene revealed the presence of Xa21+xa5+xa13 genes in 16 plants (Figure 3). The plant showing more similarity with Ranidhan was backcrossed and 148 seeds were produced.
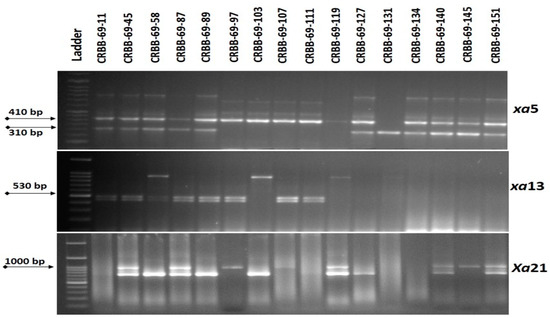
Figure 3.
Representative electropherogram generated from the tracking of Xa21, xa13 and xa5 BB resistance genes in BC2F1 progenies of resistance gene pyramiding of Ranidhan.
3.1.4. Marker-Assisted Selection in the BC3F1 and BC3F2 Progenies
A total of 148 BC3F1 seeds were raised in the next season for selection. Target resistance gene selection was performed in all the BC3F1 plants using the closely linked markers for the BB resistance genes through foreground screening. The genotyping selection for the target xa5 gene showed its presence in 67 progenies. Those 67 progenies were genotyped and screened for the xa13 gene. This analysis identified 32 plants positive for the xa13 gene and further genotyped for Xa21. Finally, 15 plants were positive for the Xa21 gene (Figure 4). The plant most similar to the Ranidhan plant was self-pollinated.
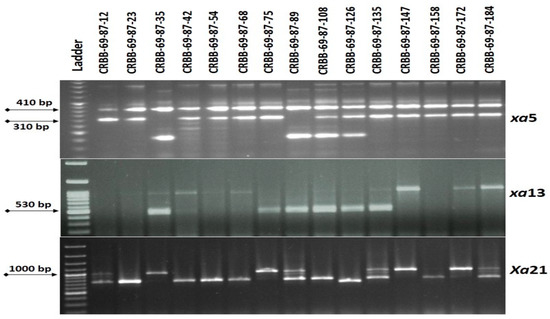
Figure 4.
Representative electropherogram generated from the tracking of Xa21, xa13 and xa5 BB resistance genes in B3F1 progenies of resistance gene pyramiding of Ranidhan.
The self pollinated seeds of the previous generation were raised during the next season and 225 BC3F2 generation progenies were tracked via foreground selection using the BB resistance gene markers. The genotyping screening of all the 225 BC3F2 progenies showed the presence of Xa21 gene in 53 plants. These 53 positive plants were checked for xa5 gene and found its presence in 13 plants. Out of the thirteen plants, finally three progenies carrying xa5+Xa21 resistance genes, two plants containing Xa21+xa13 genes and two plants containing xa5+xa13 genes were identified to be homozygous (Figure 5).
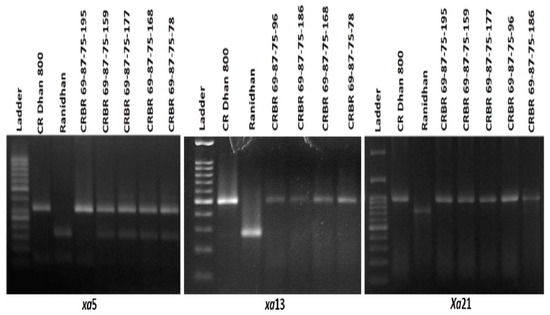
Figure 5.
Representative electropherogram generated from the tracking of Xa21, xa13 and xa5 BB resistance genes in BC3F2 progenies of resistance gene pyramiding of Ranidhan.
3.2. Bioassay of the Pyramided Lines against Bacterial Blight Pathogens
Twelve test entries including seven BC3F4 generation pyramided lines (three plants carrying xa5+Xa21 genes, two plants with Xa21+xa13 genes and two plants containing xa5+xa13 genes) along with one plant each carrying xa5/xa13/Xa21 genes and donor parent CR Dhan 800 and recipient parent Ranidhan were screened after 15 days of artificial inoculation using eight Xoo isolates. The bioassay results confirmed the resistance and susceptible reaction of the donor and the recurrent parents, respectively, based on the phenotyping results of BB disease reaction during wet seasons 2020 and 2021. The donor parent, CR Dhan 800, showed low average lesion lengths (2.2–2.6 cm) while the recurrent parent, Ranidhan, had longer lesion lengths (9.5–11.1 cm) (Table 2). The results based on the phenotypic data showed that the developed pyramided lines were better than the recurrent parent for disease resistance against the BB isolates. Gene combinations of xa5+Xa21 showed a range of 3.9–4.3 cm and xa13+Xa21 showed a range of 4.3–5.3 while xa5+xa13 gene combinations showed a range of 6.1–6.5 cm. The pyramided plants carrying xa13+Xa21 and xa5+Xa21 gene combinations showed higher levels of BB disease resistance with shorter lesion lengths against all the BB isolates. However, all the plants tested carrying different resistance gene combinations did not show any susceptible reaction to any of the eight BB strains tested. From this investigation, it may be inferred that that the of severity of the disease may be in the order of Xa21+xa5 < Xa21+xa13 < xa13+xa5 resistance genes combinations in conferring BB resistance.

Table 2.
BB disease reaction of parental, BC3F3, and BC3F4 pyramided lines against different Xoo strains.
3.3. Evaluation of the Pyramided Lines for Yield, Agro-Morphologic and Grain Quality Traits
Twelve genotypes including the sevenpyramided lines carrying two BB resistance genes and three lines containing one resistance gene at BC3F4 generations along with the parents were evaluated during wet seasons, 2020 and 2021 at ICAR-NRRI, Cuttack. Ten traits for yield, agro-morphologic and grain quality traits were recorded. The popular variety, Ranidhan, produced a mean grain yield of 5.82 t/ha. All test entries carrying two BB resistance target genes showed grain yield higher than the recipient parent, Ranidhan (Table 3). The majority of the two resistance genes containing pyramided lines were similar to the recipient parent, Ranidhan, for the main agro-morphologic and quality traits (Figure 6).

Table 3.
Grain yield, agro-morphologic and grain quality traits of parental and selected BC3F3 and BC3F4 pyramided lines.
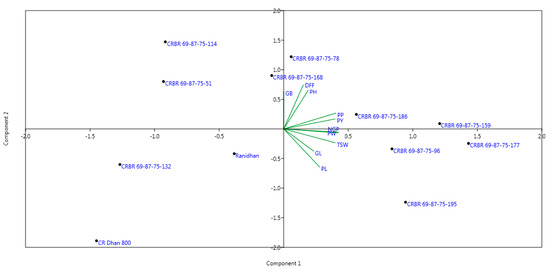
Figure 6.
Biplot diagram generated using the 10 traits for yield, agro-morpholic and grain quality characters of the pyramided and parental lines.
The pyramided line CRBR 69-87-75-177 showed the highest grain yield 67.1 q/ha followed by CRBR 69-87-75-159 (63.8 q/ha) and CRBR 69-87-75-96 (62.7 q/ha) (Table 3). A total of six pyramided lines showed more yield than the recipient parent. The best-pyramided line showed a higher yield of 15.29% than the recipient variety, Ranidhan. The best-pyramided line showed a 5.52 cm grain length, while the recipient parent had 5.62 cm. The popular variety and the best-pyramided line had the same grain breadth of 2.32 cm. Seven pyramided lines were observed to be at par with many of the agro-morphologic traits.
4. Discussion
The integration of molecular markers for the selection of desired plants in a backcross breeding program not only increases the precision in transferring the target genes into the recipient variety but also reduces the duration of the selection cycle compared to the classical breeding approach. The selection of progenies in three back cross generations followed by two successive selfing generations was sufficient for the transfer of the two gene combinations into the susceptible variety. Ranidhan is a popular variety in late maturing rice varieties but is highly susceptible to the BB disease. Under lowland rainfed ecology, controlling the BB disease is a relatively hard and difficult task. Developing BB resistant lines in the background of the popular high-yielding variety, Ranidhan, took less time and was easier by using fewer generations through the integration of molecular markers in the breeding program. The pyramided resistance genes in the developed lines showed a higher level of tolerance against the virulent BB isolates. The development and release of BB pyramided lines by employing marker-assisted backcross breeding has been successful, as reported in earlier publications. It illustrated less time, more precision and more eco-friendliness [1,15,24,25,26,27,29,41,42,43,44].
Two resistance genes carrying pyramided lines conferred higher BB resistance compared to the introgressed line carrying a single resistance gene. Results indicated that the two gene combinations (Xa21+xa5) pyramided lines showed relatively smaller lesion lengths followed by Xa21+xa13 gene combinations pyramided lines compared to the recipient variety, Ranidhan. The pyramided lines carrying the dominant resistance gene, Xa21 along with xa5 or xa13 showed higher resistance than single gene carrying lines. Similar findings were also reported by Sanchez et al. [19]; Singh et al. [17] and Pradhan et al. [45] for conferring higher resistance using the Xa21 gene in rice. The synergistic action of the resistance genes or quantitative complementation between the BB resistance genes might result in higher resistance in rice [19,45].
Twelve genotypes including seven BC3F4 pyramided lines (three plants carrying xa5+Xa21 genes, two plants containing xa13+Xa21 genes and two plants containing xa5+xa13 genes) along with one plant each of carrying xa5/xa13/Xa21 genes and donor parent, CR Dhan 800 and recipient parent, Ranidhan, were screened using eight Xoo isolates. Validation confirmed the resistance and susceptible reaction of the donor and the recurrent parents, respectively, through phenotyping the results of BB disease reaction. The BB donor parent showed lower average lesion lengths (2.2–2.6 cm) while the popular variety, Ranidhan, had higher lesion lengths (9.5–11.1 cm) (Table 2). The bioassay result confirmed that the pyramided lines were better in resisting BB disease compared to the recurrent parent. Gene combinations of xa5+Xa21 showed a range of 3.9–5.3 cm and xa13+Xa21 showed a range of 4.3–5.3, while xa5+xa13 gene combinations showed a range of 6.1–6.5 cm. Results of the bioassay study indicated that the pyramided lines carrying target gene combinations did not show a susceptible reaction to any of the eight isolates employed. The gene pyramids in Xa21+xa5 and Xa21+xa13 resistance gene combinations showed better BB disease resistance showing smaller mean lesion lengths against all BB pathotypes. From this investigation, it is concluded that gene combinations conferred resistance in the order of Xa21+xa5 < Xa21+xa13 < xa13+xa5.
The combined evaluation results of the two seasons revealed a higher grain yield of the pyramided lines than the recipient popular variety, Ranidhan (Table 3). The majority of the pyramided lines carrying resistance genes were similar to the recipient parent, Ranidhan, for important agro-morphologic and grain quality traits (Figure 6). The pyramided line CRBR 69-87-75-177 showed the highest grain yield 67.1qt/ha followed by CRBR 69-87-75-159 (63.8 q/ha) and CRBR 69-87-75-96 (62.7 q/ha) (Table 3). A total of six pyramided lines carrying resistance genes were higher yielders than the recipient parent. The best-pyramided line showed a yield advantage of 15.3% over the recurrent parent. The best-pyramided line showed a 5.52 cm grain length, while the recipient parent had 5.62 cm. The best-pyramided line and the recipient parent had the same grain breadth of 2.32 cm. Seven pyramided lines were observed to be at par with many of the agro-morphologic traits.
Integration of molecular markers in classical breeding program for the selection of the BB resistance in the pyramids conferred a higher level of resistance to the BB disease. Single resistance gene incorporation is broken down by the rich diversity of the genetically distinct virulent Xoo strains available in the different agro-climatic zones of the country. Development of the pyramided lines carrying two BB resistance genes in suitable combination compared to the single resistance gene carrying will provide alternative variety to the Ranidhan growers. These pyramided lines containing xa5+Xa21 and Xa21+xa13 can also be useful for the other BB endemic regions of the country. The investigation clearly indicates that the deployment of a suitable resistance gene combination through gene pyramiding will enhance the level of resistance against this disease.
5. Conclusions
Marker-assisted breeding is a precise breeding approach for pyramiding BB resistance genes in rice through backcross breeding. It was successful in developing superior BB resistant lines, which conferred a higher level of resistance in the background of a popular variety, Ranidhan. The developed pyramided lines can either be useful as promising donors for BB resistance or tested under multi-location testing for release as a variety in the state/country. BB resistant pyramided lines developed through marker assisted backcrossing will provide a solution in the BB endemic areas of the region/country. These pyramided lines are expected to give a high impact not only on yield stability but also the sustainability of the rainfed ecology, particularly ineastern India where the majority of rice areas are in rainfed lowlands.
Author Contributions
S.K.P. conceived the study; K.C.P., S.R.B., S.M., D.K.N., E.P., A.S., S.S. and B.K.J. performed the genotyping work.; S.K.P., J.M., A.K.M. and A.P. performed the phenotyping work.; L.B., D.P. analyzed the data. S.K.P. wrote the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
No externally aided fund was availed for this research work. However, Institute’s internal funding (Project 1.6) was used for this investigation.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data generated or analyzed in this study are included in this article.
Acknowledgments
The authors acknowledge the support of Director, ICAR-National Rice Research Institute, Cuttack for providing all the necessary facilities including the funding for conducting the experiment.
Conflicts of Interest
The authors declare that there is no competing interest and the article is submitted without any commercial or economic interest that could be generated as a potential conflict of interest.
Ethics Approval and Consent to Participate
The authors declare that this study complies with the current laws of our country in which the experiment was performed.
References
- Pandit, E.; Pawar, S.; Barik, S.R.; Mohanty, S.P.; Meher, J.; Pradhan, S.K. Marker-assisted backcross breeding for improvement of submergence tolerance and grain yield in the popular rice variety ‘Maudamani’. Agronomy 2021, 11, 1263. [Google Scholar] [CrossRef]
- Pradhan, S.K.; Pandit, E.; Pawar, S.; Baksh, S.Y.; Mukherjee, A.K.; Mohanty, S.P. Development of flash-flood tolerant and durable bacterial blight resistant versions of mega rice variety ‘Swarna’ through marker-assisted backcross Breeding. Sci. Rep. 2019, 9, 12810. [Google Scholar] [CrossRef]
- Mohapatra, S.; Bastia, A.K.; Meher, J.; Sanghamitra, P.; Pradhan, S.K. Development of submergence tolerant, bacterial blight resistant and high yielding near isogenic lines of popular variety ‘Swarna’ through marker-assisted breeding approach. Front. Plant Sci. 2021. [Google Scholar] [CrossRef] [PubMed]
- Food and Agriculture Organization. Food and Agriculture Organization of the United Nations. Rice Mark. Monit. 2017, 20, 1–38. [Google Scholar]
- Pradhan, S.K.; Barik, S.R.; Sahoo, J.; Pandit, E.; Nayak, D.K.; Pani, D.R.; Anandana, A. Comparison of Sub1 markers and their combinations for submergence tolerance and analysis of adaptation strategies of rice in rainfed lowland ecology. Comptes Rendus Biol. 2015, 338, 650–659. [Google Scholar] [CrossRef] [PubMed]
- Khush, G.S. What it will take to feed 5.0 billion rice consumers in 2030. Plant Mol. Biol. 2005, 59, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Mishra, A.; Barik, S.R.; Pandit, E.; Yadav, S.S.; Das, S.R.; Pradhana, S.K. Genetics, Mechanisms and Deployment of Brown Planthopper Resistance Genes in Rice. Crit. Rev. Plant Sci. 2022, 41, 91–127. [Google Scholar] [CrossRef]
- Ismail, A.M.; Singh, U.S.; Singh, S.; Dar, M.H.; Mackill, D.J. The contribution of submergence-tolerant (Sub1) rice varieties to food security in flood-prone rainfed lowland areas in Asia. Field Crops Res. 2013, 152, 83–93. [Google Scholar] [CrossRef]
- Sonti, R.V. Bacterial leaf blight of rice: New insights from molecular genetics. Curr. Sci. 1998, 74, 206–212. [Google Scholar]
- Khush, G.S.; Mackill, D.J.; Sidhu, G.S. Breeding rice for resistance to bacterial leaf blight. In Bacterial Blight of Rice; IRRI: Manila, Philippines, 1989; pp. 207–217. [Google Scholar]
- Singh, G.P.; Srivastava, M.K.; Singh, R.M.; Singh, R.V. Variation in quantitative and Xanthomonas oryzae. Plant Dis. Rep. 1977, 57, 537–541. [Google Scholar]
- Pradhan, S.K.; Barik, S.R.; Nayak, D.K.; Pradhan, A.; Pandit, E.; Nayak, P.; Das, S.R.; Pathak, H. Genetics, molecular mechanisms and deployment of bacterial blight resistance genes in rice. Crit. Rev. Plant Sci. 2020, 39, 360–385. [Google Scholar] [CrossRef]
- Bhasin, H.; Bhatia, D.; Raghuvanshi, S.; Lore, S.J.; Gurpreet, K.; Sahi, K.G.; Kaur, B.; Vikal, Y.; Singh, K. New PCR-based sequence-tagged site marker for bacterial blight resistance gene Xa38 of rice. Mol. Breed. 2012, 30, 607–611. [Google Scholar] [CrossRef]
- Nayak, D.K.; Pandit, E.; Mohanty, S.; Barik, D.P.; Pradhan, S.K. Marker assisted selection in back cross progenies for transfer of bacterial leaf blight resistance genes into a popular lowland rice cultivar. Agronomy 2015, 52, 163–168. [Google Scholar]
- Pradhan, K.C.; Pandit, E.; Mohanty, S.P.; Moharana, A.; Sanghamitra, P.; Meher, J.; Jena, B.K.; Dash, P.K.; Behera, L.; Mohapatra, P.M.; et al. Development of Broad Spectrum and Durable Bacterial Blight Resistant Variety through Pyramiding of Four Resistance Genes in Rice. Agronomy 2022, 12, 1903. [Google Scholar] [CrossRef]
- Singh, S.; Sidhu, J.S.; Huang, N.; Vikal, Y.; Li, Z.; Brar, D.S.; Dhaliwal, H.S.; Khush, G.S. Pyramiding three bacterial blight resistance genes (xa-5, xa-13 and Xa-21) using marker-assisted selection into indica rice cultivar PR-106. Theor. Appl. Genet. 2001, 102, 1011–1015. [Google Scholar] [CrossRef]
- Huang, N.; Angeles, E.R.; Domingo, J.; Magpantay, G.; Singh, S.; Zhang, G.; Kumaravadevil, N.; Bennett, J.; Khush, G.S. Pyramiding of bacterial blight resistance genes in rice: Marker assisted selection using RFLP and PCR. Theor. Appl. Genet. 1997, 95, 313–320. [Google Scholar] [CrossRef]
- Sanchez, A.C.; Brar, D.S.; Huang, N.; Khush, G.S. Sequence tagged site markers-assisted selection for three bacterial blight resistance genes in rice. Crop Sci. 2000, 40, 792–797. [Google Scholar] [CrossRef]
- Shanti, M.L.; George, M.L.C.; Vera Cruz, C.M.; Bernardo, M.A.; Nelson, R.J.; Leung, H.; Reddy, J.N.; Sridhar, R. Identification of resistance genes effective against bacterial leaf blight pathogen in eastern India. Plant Dis. 2001, 85, 506–512. [Google Scholar] [CrossRef]
- Joseph, M.; Gopalakrishnan, S.; Sharma, R.K. Combining bacterial blight resistance and basmati quality characteristics by phenotypic and molecular marker assisted selection in rice. Mol. Breed. 2004, 13, 377–387. [Google Scholar] [CrossRef]
- Pha, P.N.; Lang, N.T. Marker assisted selection in rice breeding for bacterial leaf blight. Omon Rice 2004, 12, 19–26. [Google Scholar]
- Bharat, K.S.; Paulraj, R.S.D.; Brindha, P.V.; Kavitha, S.; Gnanamanickam, S.S. Improvement of bacterial blight resistance in rice cultivars Jyothiand IR50 via marker-assisted backcross breeding. J. Crop Improve. 2008, 21, 101–116. [Google Scholar]
- Perez, L.M.; Redona, E.D.; Mendioro, M.S.; Vera Cruz, C.M.; Leung, H. Introgression of Xa4, Xa7 and Xa21 for resistance to bacterial blight in thermo-sensitivegenetic male sterile rice (Oryza sativa L.) for the development of two-line hybrids. Euphytica 2008, 164, 627–636. [Google Scholar] [CrossRef]
- Sundaram, R.M.; Vishnupriya, M.R.; Biradar, S.K.; Laha, G.S.; Reddy, G.A.; Rani, N.S.; Sarma, N.P.; Sonti, R.V. Marker assisted introgression of bacterial blight resistance in Samba Mahsuri, an elite indica rice variety. Euphytica 2008, 160, 411–422. [Google Scholar] [CrossRef]
- Rajpurohit, D.; Kumar, R.; Kumar, M.; Paul, P.; Awasthi, A.A.; Basha, P.O.; Puri, A.; Jhang, T.; Singh, K.; Dhaliwal, H.S. Pyramiding of two bacterial blight resistance and a semi dwarfing gene in Type 3 Basmati using marker-assisted selection. Euphytica 2011, 178, 111–126. [Google Scholar] [CrossRef]
- Dokku, P.; Das, K.M.; Rao, G.J.N. Pyramiding of four resistance genes of bacterial blight in Tapaswini, an elite rice cultivar, through marker-assisted selection. Euphytica 2013, 192, 87–96. [Google Scholar] [CrossRef]
- Suh, J.P.; Jeung, J.U.; Noh, T.H.; Cho, Y.C.; Park, S.H.; Park, H.S.; Shin, M.S.; Kim, C.K.; Jena, K.K. Development of breeding lines with three pyramided resistance genes that confer broad-spectrum bacterial blight resistance and their molecular analysis in rice. Rice 2013, 6, 5. [Google Scholar] [CrossRef]
- Pradhan, S.K.; Nayak, D.K.; Mohanty, S.; Behera, L.; Barik, S.R.; Pandit, E.; Lenka, S. Pyramiding of three bacterial blight resistance genes for broad-spectrum resistance in deepwater rice variety, Jalmagna. Rice 2015, 8, 19. [Google Scholar] [CrossRef]
- Tyagi, J.P.; Singh, T.; Singh, S.; Nitika, G.; Pradhan, S.K.; Singh, V.P. Identification of rice genotypes with high resistance to bacterial leaf blight caused by Xanthomonas oryzae pv oryzae. Indian J.Agric.Sci. 2010, 80, 63–68. [Google Scholar]
- Mohapatra, S.; Bastia, A.K.; Panda, A.K.; Pradhan, S.K. Marker-assisted selection for transfer of submergence tolerance, bacterial blight resistance and yield enhancement in the rice backcross derivatives. Aust.J.Crop Sci. 2020, 14, 1288–1294. [Google Scholar] [CrossRef]
- Chu, Z.; Fu, B.; Yang, H.; Xu, C.; Li, Z.; Sanchez, A.; Park, Y.J.; Bennetzen, J.L.; Zhang, Q.; Wang, S. Targeting xa13, a recessive gene for bacterial blight resistance in rice. Theor. Appl. Genet. 2006, 112, 455–461. [Google Scholar] [CrossRef]
- Dellaporta, S.L.; Wood, J.; Hicks, J.B. A plant DNA mini preparation: Version II. Plant Mol. Biol. Rep. 1983, 1, 19–21. [Google Scholar] [CrossRef]
- Pradhan, S.K.; Pandit, E.; Pawar, S.; Naveenkumar, R.; Barik, S.R.; Mohanty, S.P.; Nayak, D.K.; Ghritlahre, S.K.; Rao, D.S.; Reddy, J.N.; et al. Linkage disequilibrium mapping for grain Fe and Zn enhancing QTLs useful for nutrient dense rice breeding. BMC Plant Biol. 2020, 20, 57. [Google Scholar] [CrossRef] [PubMed]
- Perrier, X.; Jacquemoud-Collet, J.P. DARwin Software. 2006. Available online: http://darwin.cirad.fr/darwin (accessed on 20 July 2022).
- Singh, S.; Pradhan, S.K.; Singh, A.K.; Singh, O.N. Marker validation in recombinant inbred lines and random varieties of rice for drought tolerance. Aust. J. Crop Sci. 2012, 6, 606–612. [Google Scholar]
- Pandit, E.; Sahoo, A.; Panda, R.K.; Mohanty, D.P.; Pani, D.R.; Anandan, A.; Pradhan, S.K. Survey of rice cultivars and landraces of upland ecology for phosphorous uptake 1 (pup1) QTL using linked and gene specific molecular markers. Oryza 2016, 53, 1–9. [Google Scholar]
- Kauffman, H.E.; Reddy, A.P.K.; Hsien, S.P.Y.; Merca, S.D. An improved technique for evaluating resistance of rice varieties to Xanthomonas oryzae. Plant Dis. Rep. 1973, 57, 537–541. [Google Scholar]
- Amante-Bordeos, A.; Sitch, L.; Nelson, R.; Dalmacio, R.; Oliva, N.; Aswidinnoor, H.; Leung, H. Transfer of bacterial blight and blast resistance from the tetraploid wild rice Oryzaminuta to cultivated rice, Oryzasativa. Theor. Appl. Genet. 1992, 84, 345–354. [Google Scholar] [CrossRef]
- Pandit, E.; Tasleem, S.; Nayak, D.K.; Barik, S.R.; Mohanty, D.P.; Das, S.; Pradhan, S.K. Genome-wide association mapping reveals multiple QTLs governing tolerance response for seedling stage chilling stress in indica rice. Front. Plant Sci. 2017, 8, 552. [Google Scholar] [CrossRef]
- Pandit, E.; Panda, R.K.; Sahoo, A.; Pani, D.R.; Pradhan, S.K. Genetic relationship and structure analyses of root growth angle for improvement of drought avoidance in early and mid-early maturing rice genotypes. Rice Sci. 2020, 27, 124–132. [Google Scholar] [CrossRef]
- Pradhan, S.K.; Pandit, E.; Pawar, S.; Bharati, B.; Chatopadhyay, K.; Singh, S.; Dash, P.; Reddy, J.N. Association mapping reveals multiple QTLs for grain protein content in rice useful for biofortification. Mol. Genet. Genom. 2019, 294, 963–983. [Google Scholar] [CrossRef]
- Pradhan, S.K.; Pandit, E.; Barik, S.R.; Mohanty, S.P.; Nayak, D.K.; Sah, R.P.; Behera, L.; Sanghamitra, P.; Bose, L.K.; Das, S.R. Climate-Smart Rice Breeding: Progress and Challenges for the Rain-fed ecologies in India. In Advances in Rice Breeding: Stress Tolerance, Climate Resilience, Quality and High Yield; Pradhan, S.K., Das, S.R., Patra, B.C., Eds.; ICAR: Odisha, India, 2021; pp. 144–162. Available online: https://icar-nrri.in/wp-content/uploads/2022/05/Book-Advances-in-Rice-Breeding-SKP.pdf (accessed on 12 August 2022).
- Narayanan, N.N.; Baisakh, N.; Oliva, N.P.; Vera-Cruz, C.M.; Gnanamanickam, S.S.; Datta, K.; Datta, S.K. Molecular breeding: Marker-assisted selection combined with biolistic transformation for blast and bacterial blight resistance in indica rice (cv. CO39). Mol. Breed. 2004, 14, 61–71. [Google Scholar] [CrossRef]
- Pradhan, S.K.; Nayak, D.K.; Pandit, E.; Barik, S.R.; Mohanty, S.P.; Anandan, A.; Reddy, J.N. Characterization of morpho-quality traits and validation of bacterial blight resistance in pyramided rice genotypes under various hotspots of India. Aust. J. Crop Sci. 2015, 9, 127–134. [Google Scholar]
- Pradhan, S.K.; Nayak, D.K.; Pandit, E.; Behera, L.; Anandan, A.; Mukherjee, A.K.; Lenka, S.; Barik, D.P. Incorporation of bacterial blight resistance genes into lowland rice cultivar through marker-assisted backcross breeding. Phytopathology 2016, 106, 710–718. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).