Modulation of Growth Duration, Grain Yield and Nitrogen Recovery Efficiency by EMS Mutagenesis under OsNRT2.3b Overexpression Background in Rice
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Materials and Growth Conditions
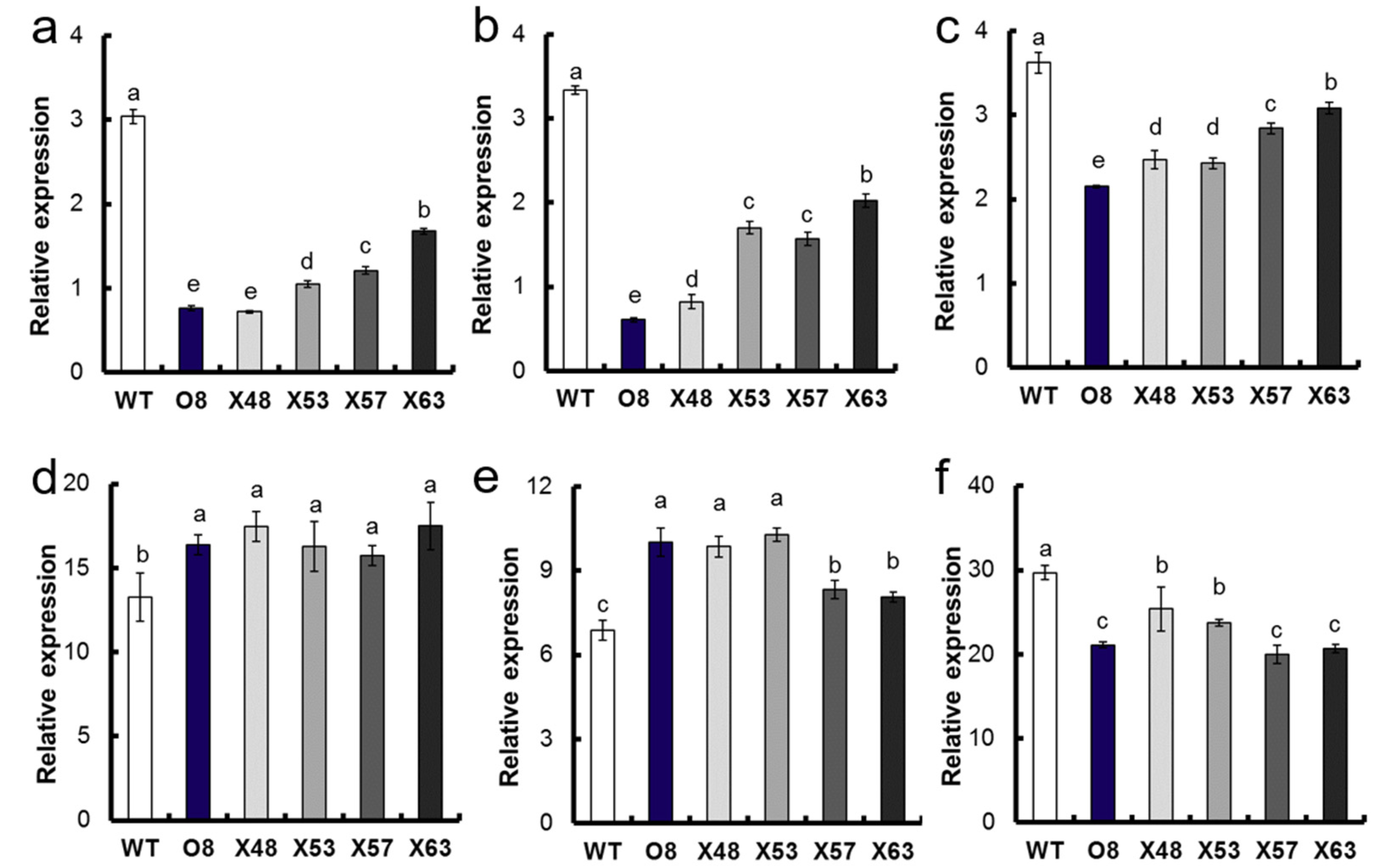
2.2. The qRT-PCR Assay
2.3. Sequence Analysis of OsNRT2.3b and Growth Duration Genes in O8 and EMS Mutants
2.4. Determination of Chlorophyll Content (SPAD Value)
2.5. NUE and N Translocation Efficiency
2.6. Statistical Analysis
3. Results
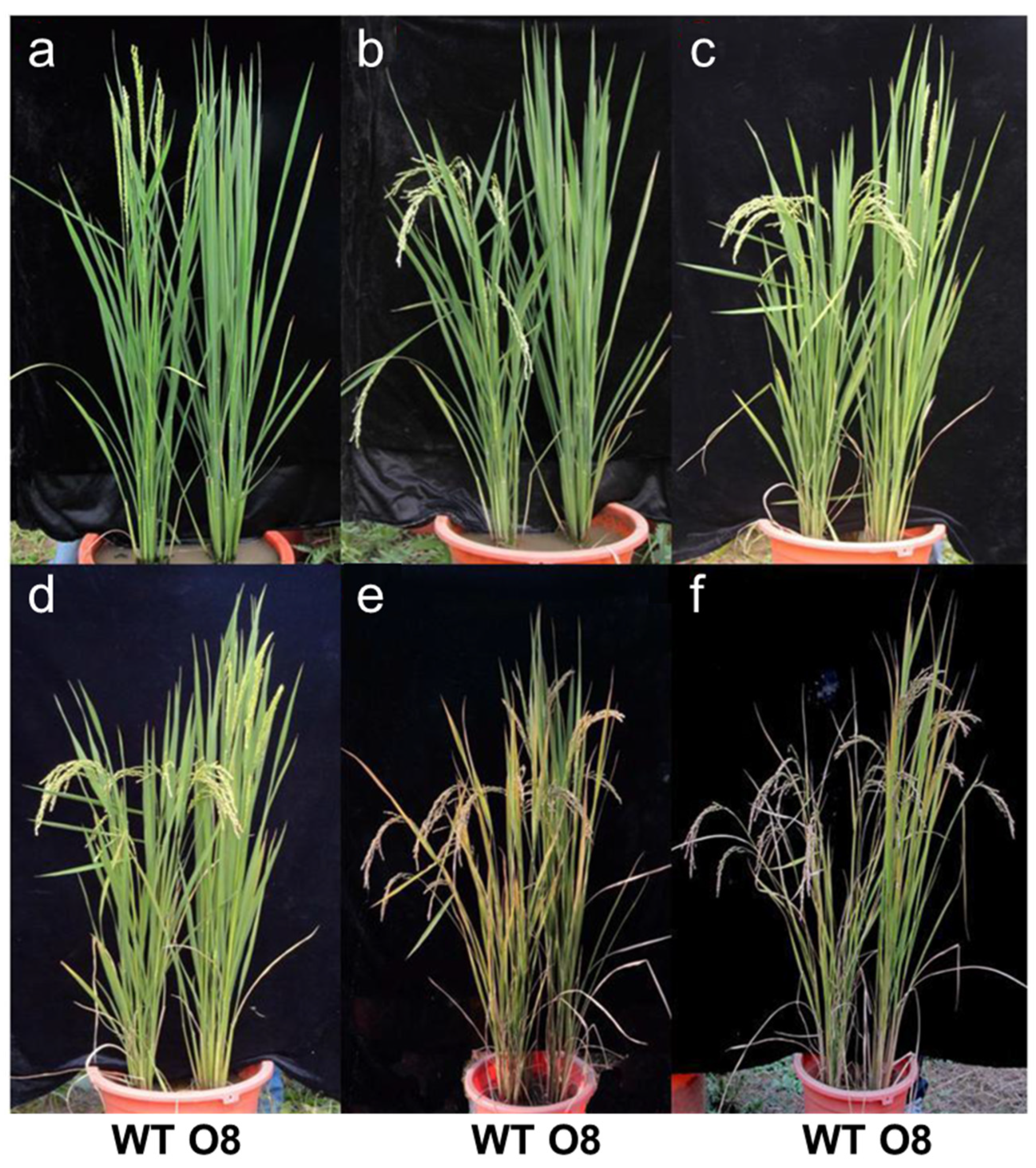
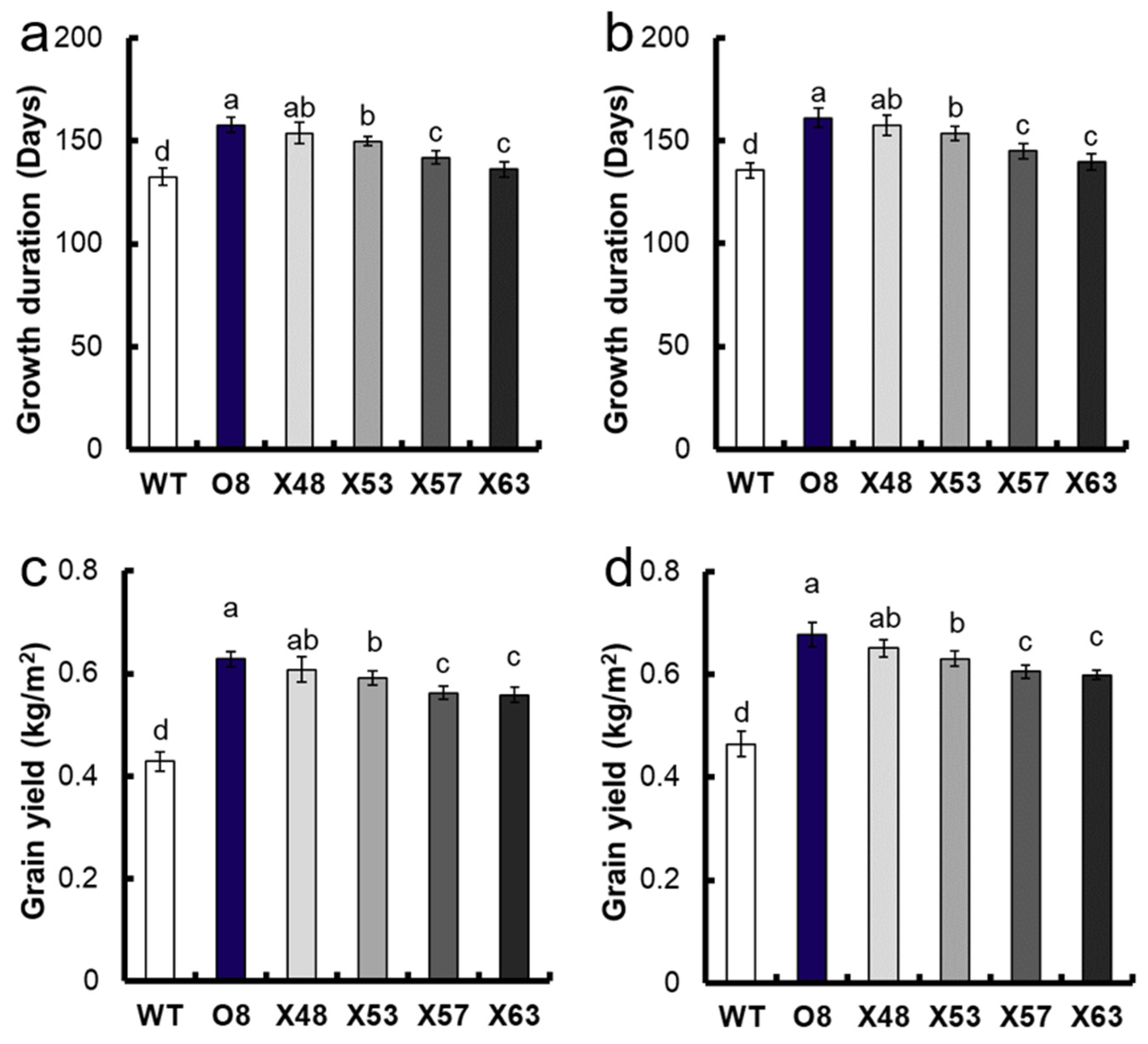
3.1. Analysis of Growth Duration of Mutants
3.2. Analysis of Yield and Agronomic Characteristics of Mutants
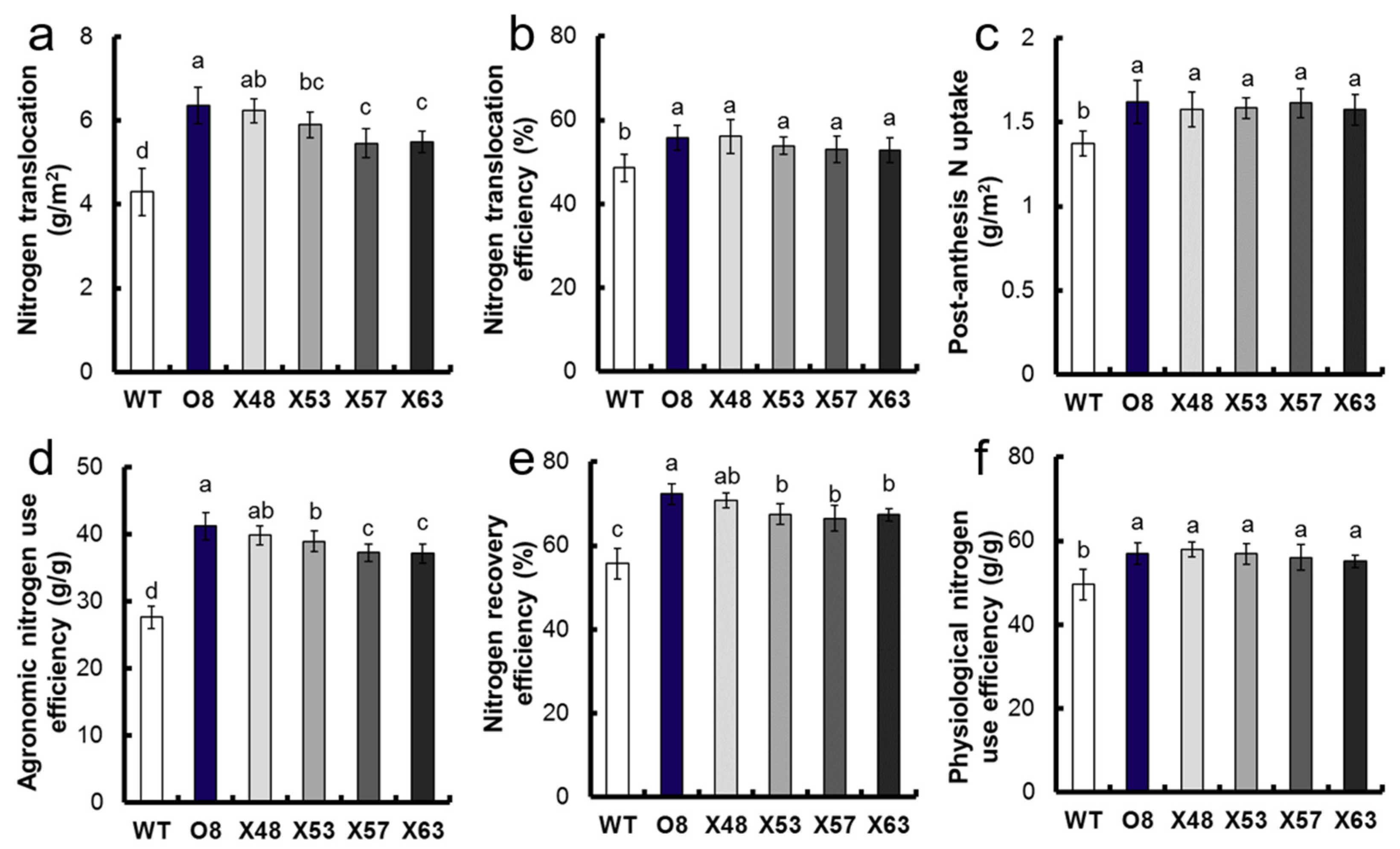
3.3. NUE Analysis of Mutants
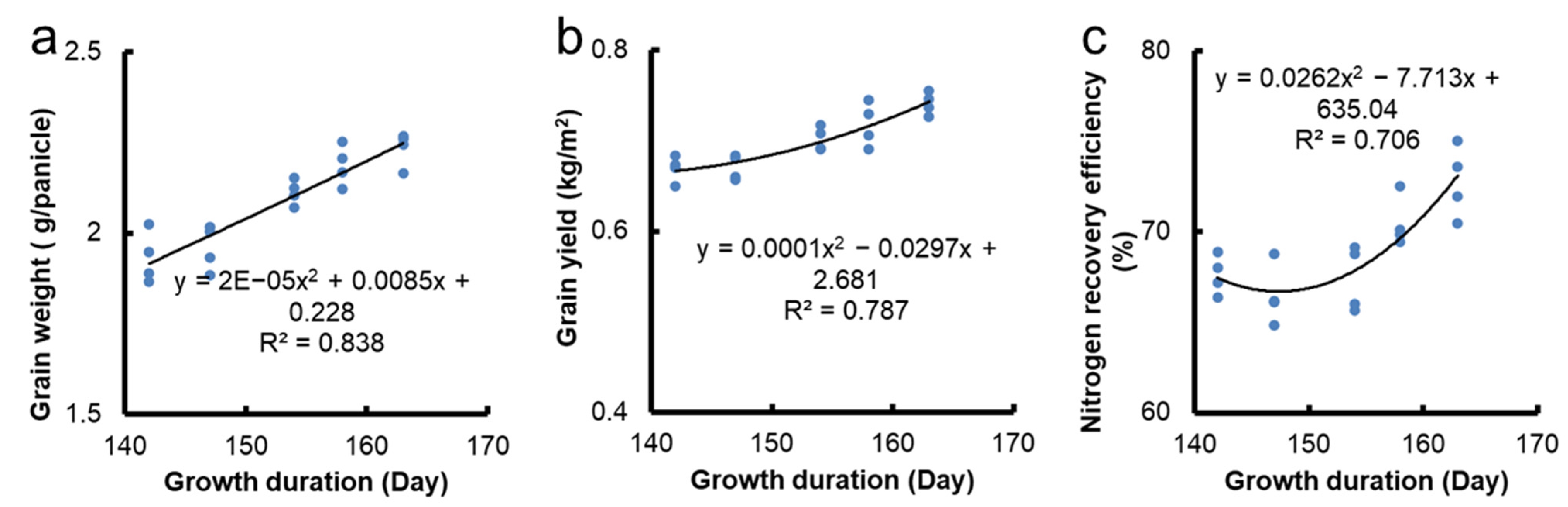
3.4. Regression Analysis of Growth Duration and Grain Yield
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Xue, W.; Xing, Y.; Weng, X.; Zhao, Y.; Tang, W.; Wang, L.; Zhou, H.; Yu, S.; Xu, C.; Li, X.; et al. Natural variation in Ghd7 is an important regulator of heading date and yield potential in rice. Nat. Genet. 2008, 40, 761–767. [Google Scholar] [CrossRef]
- Weng, X.; Wang, L.; Wang, J.; Hu, Y.; Du, H.; Xu, C.; Xing, Y.; Li, X.; Xiao, J.; Zhang, Q. Grain number, plant height, and heading date7 is a central regulator of growth, development, and stress response. Plant Physiol. 2014, 164, 735–747. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, J.; Zhang, H.; Ying, J.; Ma, L.; Zhuang, J. Natural variation at qHd1 affects heading date acceleration at high temperatures with pleiotropism for yield traits in rice. BMC Plant Biol. 2018, 18, 112. [Google Scholar]
- Hou, M.; Yu, M.; Li, Z.; Ai, Z.; Chen, J. Molecular Regulatory Networks for Improving Nitrogen Use Efficiency in Rice. Int. J. Mol. Sci. 2021, 22, 9040. [Google Scholar] [CrossRef] [PubMed]
- Hori, K.; Matsubara, K.; Yano, M. Genetic control of flowering time in rice: Integration of Mendelian genetics and genomics. Theor. Appl. Genet. 2016, 129, 2241–2252. [Google Scholar] [CrossRef] [PubMed]
- Wickland, D.P.; Hanzawa, Y. The FLOWERING LOCUS T/TERMINAL FLOWER 1 gene family: Functional evolution and molecular mechanisms. Mol. Plant 2015, 8, 983–997. [Google Scholar] [CrossRef] [Green Version]
- Hayama, R.; Yokoi, S.; Tamaki, S.; Yano, M.; Shimamoto, K. Adaptation of photoperiodic control pathways produces short-day flowering in rice. Nature 2003, 422, 719–722. [Google Scholar] [CrossRef]
- Komiya, R.; Yokoi, S.; Shimamoto, K. A gene network for longday flowering activates RFT1 encoding a mobile flowering signal in rice. Development 2009, 136, 3443–3450. [Google Scholar] [CrossRef] [Green Version]
- Du, A.; Tian, W.; Wei, M.; Yan, W.; He, H.; Zhou, D.; Huang, X.; Li, S.; Ouyang, X. The DTH8-Hd1 module mediates Day-Length-Dependent regulation of rice flowering. Mol. Plant 2017, 10, 948–961. [Google Scholar] [CrossRef] [Green Version]
- Tamaki, S.; Matsuo, S.; Wong, H.L.; Yokoi, S.; Shimamoto, K. Hd3a protein is a mobile flowering signal in rice. Science 2007, 316, 1033–1036. [Google Scholar] [CrossRef]
- Wu, W.; Zheng, X.M.; Lu, G.; Zhong, Z.; Gao, H.; Chen, L.; Wu, C.; Wang, H.J.; Wang, Q.; Zhou, K.; et al. Association of functional nucleotide polymorphisms at DTH2 with the northward expansion of rice cultivation in Asia. Proc. Natl. Acad. Sci. USA 2013, 110, 2775–2780. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pasriga, R.; Yoon, J.; Cho, L.H.; An, G. Overexpression of RICE FLOWERING LOCUS T 1 (RFT1) induces extremely early flowering in rice. Mol. Cells 2019, 42, 406–417. [Google Scholar]
- Cho, L.H.; Yoon, J.; An, G. The control of flowering time by environmental factors. Plant J. 2017, 90, 708–719. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Zhang, Y.; Li, K.; Yan, M.; Zhang, J.; Yu, M.; Tang, S.; Wang, L.; Qu, H.; Luo, L.; et al. Nitrogen Mediates Flowering Time and Nitrogen Use Efficiency via Floral Regulators in Rice. Curr. Biol. 2021, 31, 671–683. [Google Scholar] [CrossRef] [PubMed]
- Wada, K.C.; Takeno, K. Stress-induced flowering. Plant Signal. Behav. 2010, 5, 944–947. [Google Scholar] [CrossRef] [PubMed]
- Tienderen, P.H.V.; Hammad, I.; Zwaal, F.C. Pleiotropic Effects of Flowering Time Genes in the Annual Crucifer Arabidopsis thaliana (Brassicaceae). Am. J. Bot. 1996, 83, 169–174. [Google Scholar] [CrossRef] [Green Version]
- Kolár, J.; Senková, J. Reduction of mineral nutrient availability accelerates flowering of Arabidopsis thaliana. J. Plant Physiol. 2008, 165, 1601–1609. [Google Scholar] [CrossRef]
- Dielen, V.; Lecouvet, V.; Dupont, S.; Kinet, J.M. In vitro control of floral transition in tomato (Lycopersicon esculentum Mill.), the model for autonomously flowering plants, using the late flowering uniflora mutant. J. Exp. Bot. 2001, 52, 715–723. [Google Scholar] [CrossRef] [Green Version]
- Castro, M.I.; Loef, I.; Bartetzko, L.; Searle, I.; Coupland, G.; Stitt, M.; Osuna, D. Nitrate regulates floral induction in Arabidopsis, acting independently of light, gibberellin and autonomous pathways. Planta 2011, 233, 539–552. [Google Scholar] [CrossRef] [Green Version]
- Li, H.; Hu, B.; Chu, C. Nitrogen use efficiency in crops: Lessons from Arabidopsis and rice. J. Exp. Bot. 2017, 68, 2477–2488. [Google Scholar] [CrossRef]
- Liu, T.; Li, Y.; Ren, J.; Qian, Y.; Yang, X.; Duan, W.; Hou, X. Nitrate or NaCl regulates floral induction in Arabidopsis thaliana. Biologia 2013, 68, 215–222. [Google Scholar] [CrossRef]
- Wang, W.; Hu, B.; Yuan, D.; Liu, Y.; Che, R.; Hu, Y.; Ou, S.; Liu, Y.; Zhang, Z.; Wang, H.; et al. Expression of the nitrate transporter gene OsNRT1.1A/OsNPF6.3 confers high yield and early maturation in rice. Plant Cell. 2018, 30, 638–651. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sanagi, M.; Aoyama, S.; Kubo, A.; Lu, Y.; Sato, Y.; Ito, S.; Abe, M.; Mitsuda, N.; Ohme-Takagi, M.; Kiba, T.; et al. Low nitrogen conditions accelerate flowering by modulating the phosphorylation state of FLOWERING BHLH 4 in Arabidopsis. Proc. Natl. Acad. Sci. USA 2021, 118, e2022942118. [Google Scholar] [CrossRef]
- Song, Y.H.; Shim, J.S.; Kinmonth-Schultz, H.A.; Imaizumi, T. Photoperiodic flowering: Time measurement mechanisms in leaves. Annu. Rev. Plant Biol. 2015, 66, 441–464. [Google Scholar] [CrossRef] [Green Version]
- Fan, X.; Tang, Z.; Tan, Y.; Zhang, Y.; Luo, B.; Yang, M.; Lian, X.; Shen, Q.; Miller, A.J.; Xu, G. Overexpression of a pH-sensitive nitrate transporter in rice increases crop yields. Proc. Natl. Acad. Sci. USA 2016, 113, 7118–7123. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Feng, H.; Li, B.; Zhi, Y.; Chen, J.; Li, R.; Xia, X.; Xu, G.; Fan, X. Overexpression of the nitrate transporter, OsNRT2.3b, improves rice phosphorus uptake and translocation. Plant Cell Rep. 2017, 36, 1287–1296. [Google Scholar] [CrossRef]
- Chen, J.; Fan, X.; Qian, K.; Zhang, Y.; Song, M.; Liu, Y.; Xu, G.; Fan, X. pOsNAR2.1:OsNAR2.1 expression enhances nitrogen uptake efficiency and grain yield in transgenic rice plants. Plant Biotechnol. J. 2017, 15, 1273–1283. [Google Scholar] [CrossRef] [Green Version]
- Chen, J.; Qi, T.; Hu, Z.; Fan, X.; Zhu, L.; Iqbal, M.F.; Yin, X.; Xu, G.; Fan, X. OsNAR2.1 positively regulates drought tolerance and grain yield under drought stress conditions in rice. Front. Plant Sci. 2019, 10, 197. [Google Scholar] [CrossRef] [Green Version]
- Chen, J.; Liu, X.; Liu, S.; Fan, X.; Zhao, L.; Song, M.; Fan, X.; Xu, G. Co-Overexpression of OsNAR2.1 and OsNRT2.3a Increased Agronomic Nitrogen Use Efficiency in Transgenic Rice Plants. Front. Plant Sci. 2020, 11, 1245. [Google Scholar] [CrossRef]
- Xu, G.; Fan, X.; Miller, A.J. Plant nitrogen assimilation and use efficiency. Annu. Rev. Plant Biol. 2012, 63, 153–182. [Google Scholar] [CrossRef] [Green Version]
- Cockram, J.; Jones, H.; Leigh, F.J.; O’Sullivan, D.; Powell, W.; Laurie, D.A.; Greenland, A.J. Control of flowering time in temperate cereals: Genes, domestication, and sustainable productivity. J. Exp. Bot. 2007, 58, 1231–1244. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Liu, H.; Qi, F.; Zhang, Z.; Li, Q.; Han, Z.; Xing, Y. Genetic interactions among Ghd7, Ghd8, OsPRR37 and Hd1 contribute to large variation in heading date in rice. Rice 2019, 12, 48. [Google Scholar] [CrossRef] [Green Version]
- Roux, F.; Touzet, P.; Cuguen, J.; Le Corre, V. How to be early flowering: An evolutionary perspective. Trends Plant Sci. 2006, 11, 375–381. [Google Scholar] [CrossRef] [PubMed]
- Feys, K.; Demuynck, K.; De Block, J.; Bisht, A.; De Vliegher, A.; Inzé, D.; Nelissen, H. Growth rate rather than growth duration drives growth heterosis in maize B104 hybrids. Plant Cell Environ. 2018, 41, 374–382. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, H.; Gu, F.; Dong, S.; Liu, W.; Wang, H.; Chen, Z.; Wang, J. CONSTANS-like 9 (COL9) delays the flowering time in Oryza sativa by repressing the Ehd1 pathway. Biochem. Biophys. Res. Commun. 2016, 479, 173–178. [Google Scholar] [CrossRef] [Green Version]
- Zheng, T.; Sun, J.; Zhou, S.; Chen, S.; Lu, J.; Cui, S.; Tian, Y.; Zhang, H.; Cai, M.; Zhu, S.; et al. Post-transcriptional regulation of Ghd7 protein stability by phytochrome and OsGI in photoperiodic control of flowering in rice. New Phytol. 2019, 224, 306–320. [Google Scholar] [CrossRef] [PubMed]
- Wu, W.; Zheng, X.M.; Chen, D.; Zhang, Y.; Ma, W.; Zhang, H.; Sun, L.; Yang, Z.; Zhao, C.; Zhan, X.; et al. OsCOL16, encoding a CONSTANS-like protein, represses flowering by up-regulating Ghd7 expression in rice. Plant Sci. 2017, 260, 60–69. [Google Scholar] [CrossRef]


| Genotype | WT | O8 | X48 | X53 | X57 | X63 |
|---|---|---|---|---|---|---|
| Total tiller number per plant | 28.03 ± 1.41 a | 23.06 ± 2.01 b | 24.62 ± 1.66 b | 23.00 ± 2.31 b | 23.60 ± 1.60 b | 24.07 ± 1.65 b |
| Effective tiller number per plant | 20.77 ± 1.05 a | 19.85 ± 1.61 a | 21.01 ± 1.54 a | 19.50 ± 2.04 a | 20.80 ± 1.35 a | 20.96 ± 1.34 a |
| Panicle length (cm) | 20.80 ± 1.17 d | 29.49 ± 1.13 a | 28.75 ± 1.39 ab | 27.77 ± 0.61 b | 26.46 ± 0.59 c | 26.25 ± 0.41 c |
| Grain number per panicle | 77.69 ± 6.70 d | 113.58 ± 3.56 a | 110.39 ± 5.32 ab | 106.75 ± 3.24 b | 100.31 ± 3.16 c | 98.89 ± 4.05 c |
| Seed setting rate (%) | 80.63 ± 6.36 a | 85.46 ± 3.72 a | 83.97 ± 3.73 a | 81.67 ± 3.02 a | 83.87 ± 5.37 a | 80.91 ± 5.89 a |
| Grain weight (g/panicle) | 1.55 ± 0.10 d | 2.24 ± 0.08 a | 2.19 ± 0.11 ab | 2.11 ± 0.06 b | 1.96 ± 0.07 c | 1.93 ± 0.09 c |
| Grain width (mm) | 3.11 ± 0.06 a | 2.75 ± 0.07 b | 2.64 ± 0.05 b | 2.72 ± 0.11 b | 2.69 ± 0.18 b | 2.69 ± 0.11 b |
| Grain length (mm) | 7.28 ± 013 b | 8.67 ± 0.09 a | 8.73 ± 0.21 a | 8.63 ± 0.24 a | 8.67 ± 0.22 a | 8.66 ± 0.20 a |
| Grain thickness (mm) | 2.25 ± 0.05 a | 2.28 ± 0.06 a | 2.21 ± 0.04 a | 2.29 ± 0.05 a | 2.28 ± 0.06 a | 2.24 ± 0.04 a |
| 1000-grain weight (g) | 23.18 ± 0.12 a | 23.63 ± 0.06 a | 22.04 ± 0.10 a | 22.96 ± 0.08 a | 22.16 ± 0.19 a | 22.65 ± 0.21 a |
| Grain yield (kg/m2) | 0.50 ± 0.03 d | 0.74 ± 0.02 a | 0.72 ± 0.02 ab | 0.70 ± 0.02 b | 0.67 ± 0.01 c | 0.67 ± 0.01 c |
| Dry weight (kg/m2) | 1.03 ± 0.06 d | 1.43 ± 0.02 a | 1.40 ± 0.05 ab | 1.37 ± 0.03 b | 1.32 ± 0.02 c | 1.32 ± 0.02 c |
| Grain–straw ratio | 0.93 ± 0.05 b | 1.07 ± 0.07 a | 1.05 ± 0.07 a | 1.05 ± 0.03 a | 1.03 ± 0.05 a | 1.02 ± 0.04 a |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, J.; Wang, F.; Lei, B.; Qian, K.; Wei, J.; Fan, X. Modulation of Growth Duration, Grain Yield and Nitrogen Recovery Efficiency by EMS Mutagenesis under OsNRT2.3b Overexpression Background in Rice. Agriculture 2022, 12, 799. https://doi.org/10.3390/agriculture12060799
Chen J, Wang F, Lei B, Qian K, Wei J, Fan X. Modulation of Growth Duration, Grain Yield and Nitrogen Recovery Efficiency by EMS Mutagenesis under OsNRT2.3b Overexpression Background in Rice. Agriculture. 2022; 12(6):799. https://doi.org/10.3390/agriculture12060799
Chicago/Turabian StyleChen, Jingguang, Fan Wang, Biqi Lei, Kaiyun Qian, Jia Wei, and Xiaorong Fan. 2022. "Modulation of Growth Duration, Grain Yield and Nitrogen Recovery Efficiency by EMS Mutagenesis under OsNRT2.3b Overexpression Background in Rice" Agriculture 12, no. 6: 799. https://doi.org/10.3390/agriculture12060799
APA StyleChen, J., Wang, F., Lei, B., Qian, K., Wei, J., & Fan, X. (2022). Modulation of Growth Duration, Grain Yield and Nitrogen Recovery Efficiency by EMS Mutagenesis under OsNRT2.3b Overexpression Background in Rice. Agriculture, 12(6), 799. https://doi.org/10.3390/agriculture12060799







