Constitutive and Inducible Expression of Genes Related to Salicylic Acid and Ethylene Pathways in a Moderately Resistant Tomato Cultivar Leads to Delayed Development of Meloidogyne javanica
Abstract
:1. Introduction
2. Material and Methods
2.1. Nematode Inoculum
2.2. Identification of RKN Species
2.3. Plant Materials
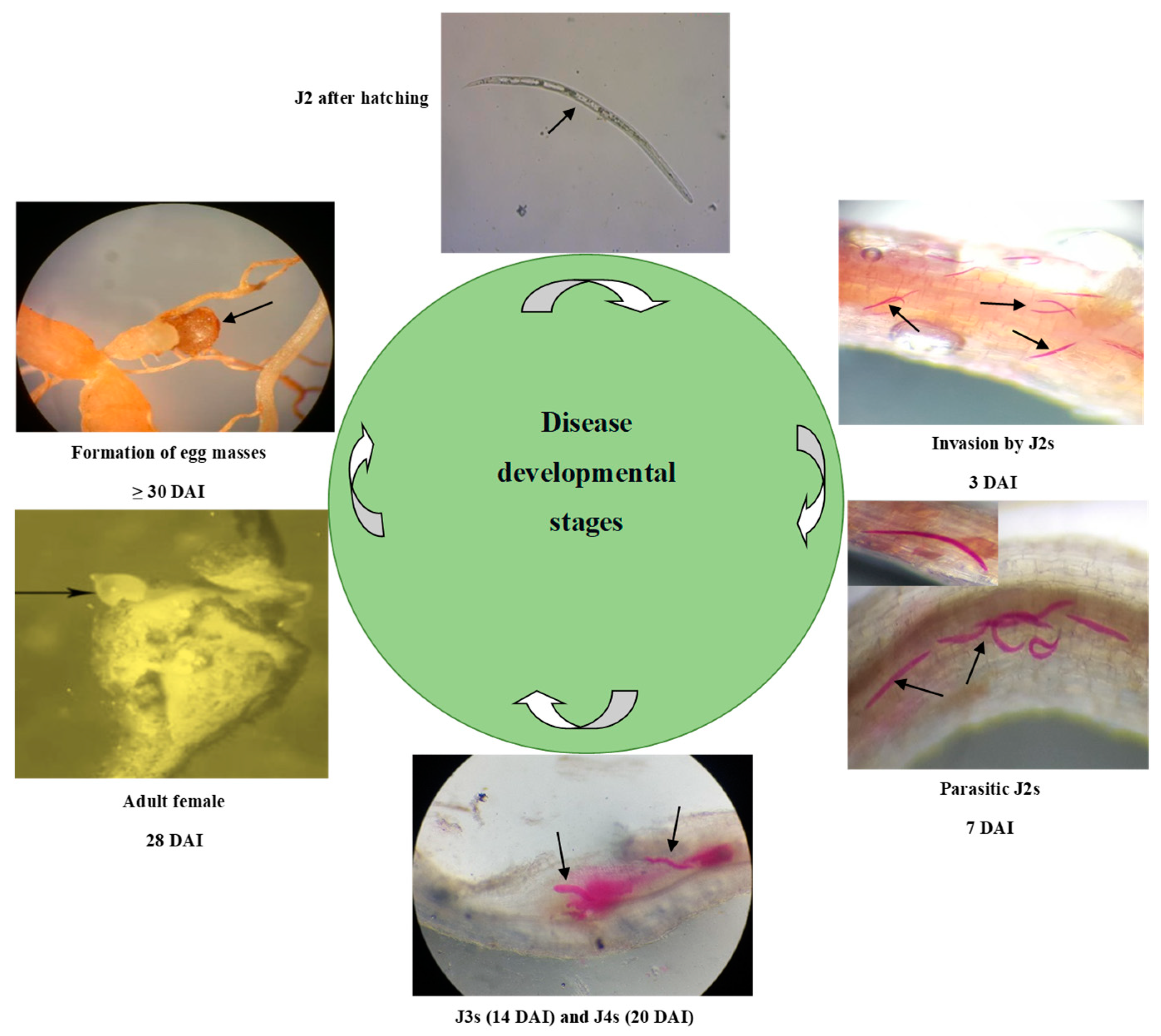
2.4. Disease Development Stages
2.5. Expression Analysis of Genes Related to JA, SA, and ET Pathways 14 Days following RKN Attack in Highly Susceptible and Moderately Resistant Tomato Cultivars
2.5.1. RNA Extraction and cDNA Synthesis
2.5.2. Quantitative Real-Time PCR (qRT-PCR)
2.6. Statistical Analysis
3. Results
3.1. Comparison of M. javanica Invasion, Development, and Reproduction in Highly Susceptible and Moderately Resistant Tomato Cultivars
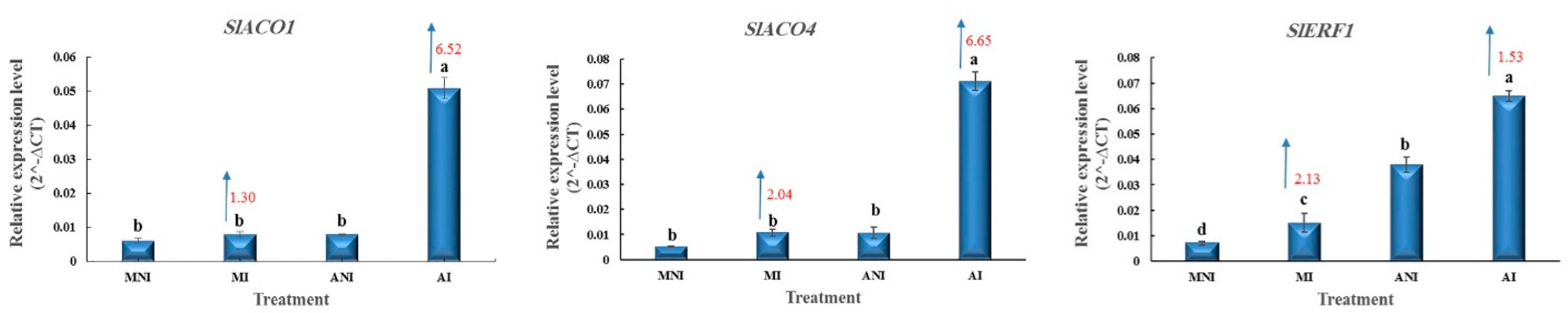
3.2. ET-Related Responses in the Roots of ALYSTE F-1 and Dutch Mobil at 14 DAI with M. javanica (Gene Bank Accession Number: OM281060)
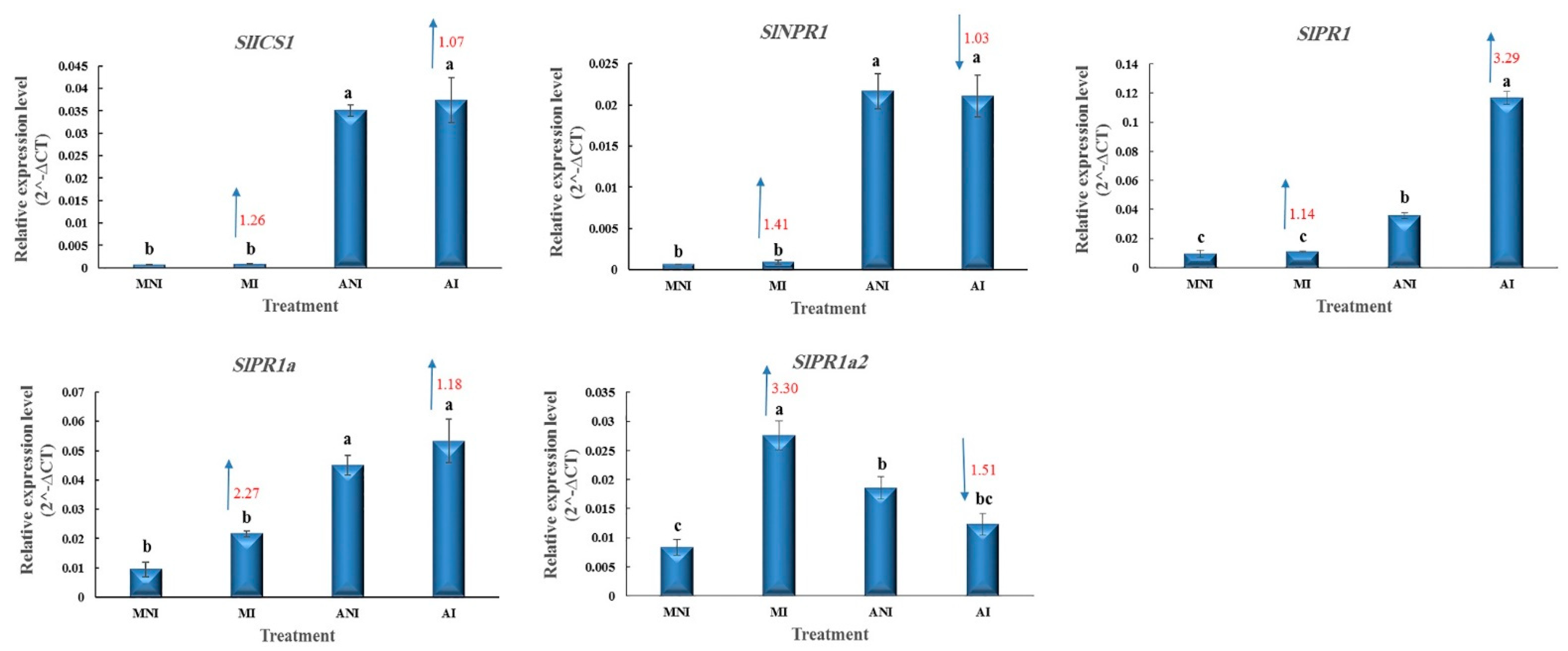
3.3. SA-Related Responses in the Roots of ALYSTE F-1 and Dutch Mobil at 14 DAI with M. javanica
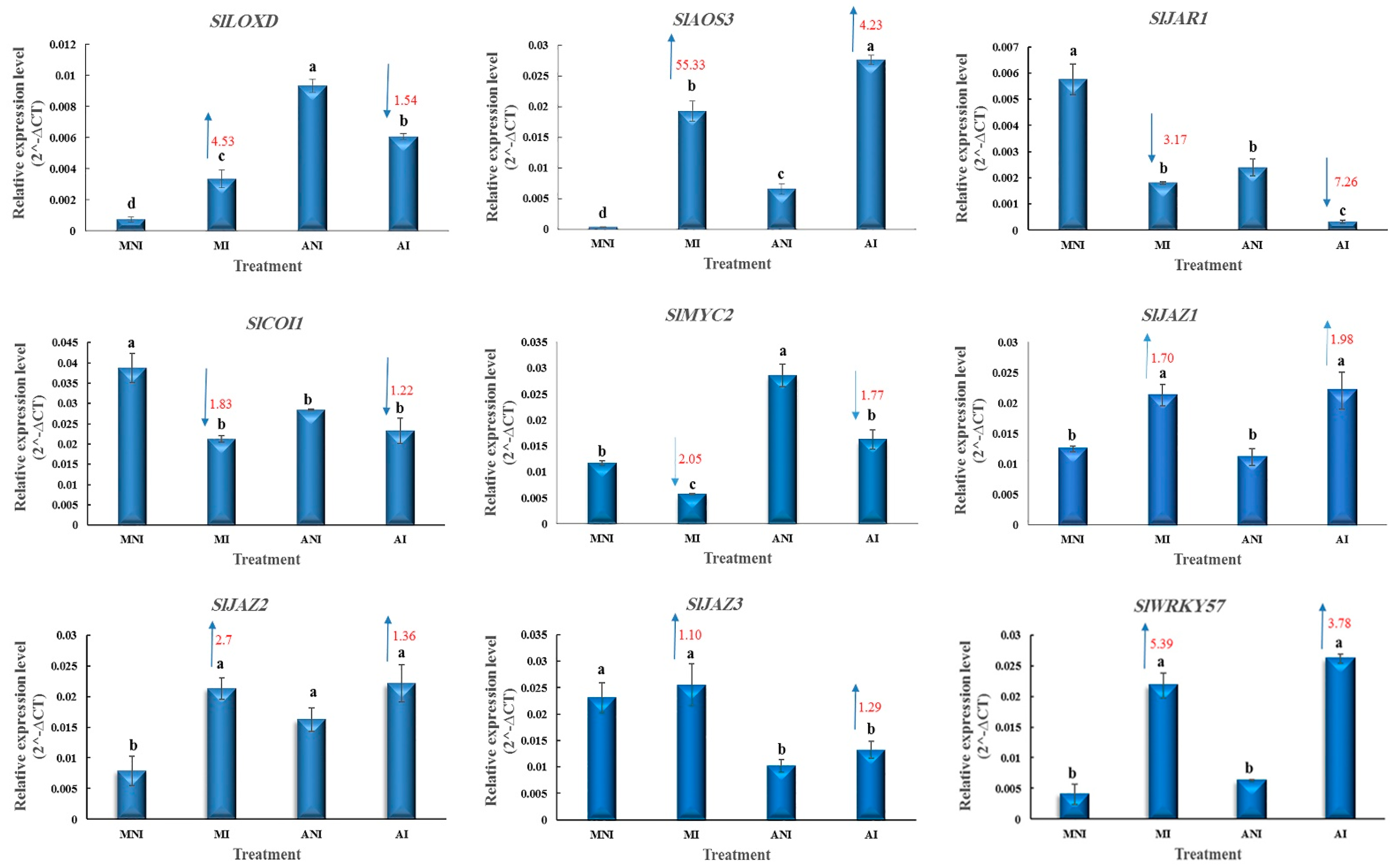
3.4. JA-Related Responses in the Roots of ALYSTE F-1 and Dutch Mobil at 14 DAI with M. javanica
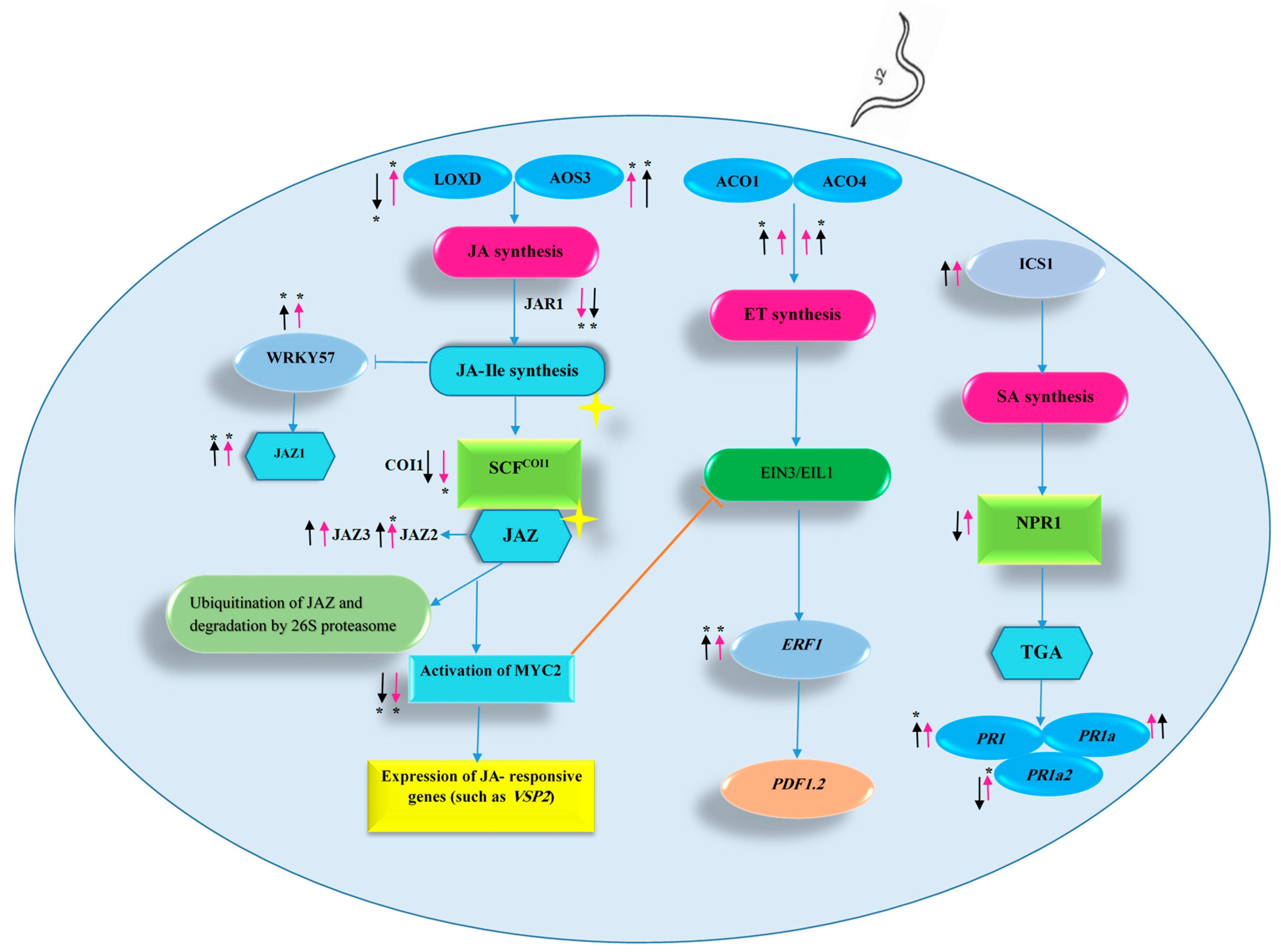
4. Discussion
4.1. Disease Development Stages
4.2. ET-Related Responses in the Roots of ALYSTE F-1 and Dutch Mobil at 14 DAI
4.3. SA-Related Responses in the Roots of ALYSTE F-1 and Dutch Mobil at 14 DAI
4.4. JA-Related Responses in the Roots of ALYSTE F-1 and Dutch Mobil at 14 DAI
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Janati, S.; Houari, A.; Wifaya, A.; Essarioui, A.; Mimouni, A.; Hormatallah, A.; Sbaghi, M.; Dababat, A.; Mokrini, F. Occurrence of the root-knot nematode species in vegetable crops in Souss region of Morocco. Plant Pathol. J. 2018, 34, 308. [Google Scholar] [CrossRef] [PubMed]
- Abad, P.; Williamson, V.M. Plant nematode interaction. A sophisticated dialogue. In Advances in Botanical Research; Kader, J.C., Delseny, M., Eds.; Elsevier: Philadelphia, PA, USA, 2010; pp. 147–192. [Google Scholar]
- Mitchum, M.G.; Hussey, R.S.; Baum, T.J.; Wang, X.; Elling, A.A.; Wubben, M.; Davis, E.L. Nematode effector proteins: An emerging paradigm of parasitism. New Phytol. 2013, 199, 879–894. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shukla, N.; Kaur, P.; Kumar, A. Molecular aspects of plant-nematode interactions. Indian J. Plant Physiol. 2016, 21, 477–488. [Google Scholar] [CrossRef]
- Shukla, N.; Yadav, R.; Kaur, P.; Rasmussen, S.; Goel, S.; Agarwal, M.; Jagannath, A.; Gupta, R.; Kumar, A. Transcriptome analysis of root-knot nematode (Meloidogyne incognita)-infected tomato (Solanum lycopersicum) roots reveals complex gene expression profiles and metabolic networks of both host and nematode during susceptible and resistance responses. Mol. Plant Pathol. 2018, 19, 615–633. [Google Scholar] [CrossRef] [Green Version]
- Huang, C.S. Formation, anatomy and physiology of giant cells induced by root-knot nematodes. In An Advanced Treatise on Meloidogyne; Sasser, J.N., Carter, C.C., Eds.; NCSU and USAID Cooperative Publication: Raleigh, NC, USA, 1985; pp. 155–164. [Google Scholar]
- Jatala, P.; Russel, C.C. Nature of sweet potato resistance to Meloidogyne incognita and the effects of temperature on parasitism. J. Nematol. 1972, 4, 1. [Google Scholar]
- Giebel, J. Mechanism of resistance to plant nematodes. Ann. Rev. Phytopathol. 1982, 20, 257–279. [Google Scholar] [CrossRef]
- Anwar, S.A.; Mc Kenry, M.V. Penetration, development and reproduction of Meloidogyne arenaria on two new resistant Vitis spp. Nematropica 2000, 30, 9–17. [Google Scholar]
- Leon-Reyes, A.; Spoel, S.H.; De Lange, E.S.; Abe, H.; Kobayashi, M.; Tsuda, S.; Millenaar, F.F.; Welschen, R.A.; Ritsema, T.; Pieterse, C.M. Ethylene modulates the role of NONEXPRESSOR OF PATHOGENESIS-RELATED GENES1 in cross talk between salicylate and jasmonate signaling. Plant Physiol. 2009, 149, 1797–1809. [Google Scholar] [CrossRef] [Green Version]
- Ithal, N.; Recknor, J.; Nettleton, D.; Maier, T.; Baum, T.J.; Mitchum, M.G. Developmental transcript profiling of cyst nematode feeding cells in soybean roots. Mol. Plant Microbe Interact. 2007, 20, 510–525. [Google Scholar] [CrossRef] [Green Version]
- Swiecicka, M.; Filipecki, M.; Lont, D.; Van Vliet, J.; Qin, L.; Goverse, A.; Bakker, J.; Helder, J. Dynamics in the tomato root transcriptome on infection with the potato cyst nematode Globodera rostochiensis. Mol. Plant Pathol. 2009, 10, 487–500. [Google Scholar] [CrossRef]
- Cabrera, J.; Barcala, M.; Fenoll, C.; Escobar, C. Transcriptomic signatures of transfer cells in early developing nematode feeding cells of Arabidopsis focused on auxin and ethylene signaling. Front. Plant Sci. 2014, 5, 107. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bhattarai, K.K.; Xie, Q.G.; Mantelin, S.; Bishnoi, U.; Girke, T.; Navarre, D.A.; Kaloshian, I. Tomato susceptibility to root-knot nematodes requires an intact jasmonic acid signaling pathway. Mol. Plant Microbe Interact. 2008, 21, 1205–1214. [Google Scholar] [CrossRef] [PubMed]
- Uehara, T.; Sugiyama, S.; Matsuura, H.; Arie, T.; Masuta, C. Resistant and susceptible responses in tomato to cyst nematode are differentially regulated by salicylic acid. Plant Cell Physiol. 2010, 51, 1524–1536. [Google Scholar] [CrossRef]
- Kyndt, T.; Denil, S.; Haegeman, A.; Trooskens, G.; Bauters, L.; Van Criekinge, W.; De Meyer, T.; Gheysen, G. Transcriptional reprogramming by root knot and migratory nematode infection in rice. New Phytol. 2012, 196, 887–900. [Google Scholar] [CrossRef] [PubMed]
- Nahar, K.; Kyndt, T.; Nzogela, Y.B.; Gheysen, G. Abscisic acid interacts antagonistically with classical defense pathways in rice–migratory nematode interaction. New Phytol. 2012, 196, 901–913. [Google Scholar] [CrossRef]
- Matthews, B.F.; Beard, H.; MacDonald, M.H.; Kabir, S.; Youssef, R.M.; Hosseini, P.; Brewer, E. Engineered resistance and hypersusceptibility through functional metabolic studies of 100 genes in soybean to its major pathogen, the soybean cyst nematode. Planta 2013, 237, 1337–1357. [Google Scholar] [CrossRef] [Green Version]
- Li, R.; Rashotte, A.M.; Singh, N.K.; Weaver, D.B.; Lawrence, K.S.; Locy, R.D. Integrated signaling networks in plant responses to sedentary endoparasitic nematodes: A perspective. Plant Cell Rep. 2015, 34, 5–22. [Google Scholar] [CrossRef]
- Wubben, M.J.E., II; Su, H.; Rodermel, S.R.; Baum, T.J. Susceptibility to the sugar beet cyst nematode is modulated by ethylene signal transduction in Arabidopsis thaliana. Mol. Plant Microbe Interact. 2001, 14, 1206–1212. [Google Scholar] [CrossRef] [Green Version]
- Wubben, M.J.E., II; Rodermel, S.R.; Baum, T.J. Mutation of a UDP-glucose-4-epimerase alters nematode susceptibility and ethylene responses in Arabidopsis roots. Plant J. 2004, 40, 712–724. [Google Scholar] [CrossRef]
- Fudali, S.L.; Wang, C.; Williamson, V.M. Ethylene signaling pathway modulates attractiveness of host roots to the root-knot nematode Meloidogyne hapla. Mol. Plant Microbe Interact. 2013, 26, 75–86. [Google Scholar] [CrossRef] [Green Version]
- Kammerhofer, N.; Radakovic, Z.; Regis, J.M.; Dobrev, P.; Vankova, R.; Grundler, F.M.; Siddique, S.; Hofmann, J.; Wieczorek, K. Role of stress-related hormones in plant defense during early infection of the cyst nematode Heterodera schachtii in Arabidopsis. New Phytol. 2015, 207, 778–789. [Google Scholar] [CrossRef] [Green Version]
- Hu, Y.; You, J.; Li, C.; Williamson, V.M.; Wang, C. Ethylene response pathway modulates attractiveness of plant roots to soybean cyst nematode Heterodera glycines. Sci. Rep. 2017, 7, 41282. [Google Scholar] [CrossRef] [PubMed]
- Huang, W.K.; Ji, H.L.; Gheysen, G.; Debode, J.; Kyndt, T. Biochar-amended potting medium reduces the susceptibility of rice to root-knot nematode infections. BMC Plant Biol. 2015, 15, 267. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Goode, K.; Mitchum, M.G. Pattern-triggered immunity against root-knot nematode infection: A minireview. Physiol. Plant. 2022, 174, e13680. [Google Scholar] [CrossRef] [PubMed]
- Goverse, A.; Overmars, H.; Engelbertink, J.; Schots, A.; Bakker, J.; Helder, J. Both induction and morphogenesis of cyst nematode feeding cells are mediated by auxin. Mol. Plant Microbe Interact. 2000, 13, 1121–1129. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gheysen, G.; Mitchum, M.G. Phytoparasitic nematode control of plant hormone pathways. Plant Physiol. 2019, 179, 1212–1226. [Google Scholar] [CrossRef] [Green Version]
- Houben, M.; Van de Poel, B. 1-Aminocyclopropane-1-carboxylic acid oxidase (ACO): The enzyme that makes the plant hormone ethylene. Front. Plant Sci. 2019, 695. [Google Scholar] [CrossRef] [Green Version]
- Ju, C.; Chang, C. Mechanistic insights in ethylene perception and signal transduction. Plant Physiol. 2015, 169, 85–95. [Google Scholar] [CrossRef] [Green Version]
- Alonso, J.M.; Hirayama, T.; Roman, G.; Nourizadeh, S.; Ecker, J.R. EIN2, a bifunctional transducer of ethylene and stress responses in Arabidopsis. Science 1999, 284, 2148–2152. [Google Scholar] [CrossRef]
- Ju, C.; Yoon, G.M.; Shemansky, J.M.; Lin, D.Y.; Ying, Z.I.; Chang, J.; Garrett, W.M.; Kessenbrock, M.; Groth, G.; Tucker, M.L.; et al. CTR1 phosphorylates the central regulator EIN2 to control ethylene hormone signaling from the ER membrane to the nucleus in Arabidopsis. Proc. Natl. Acad. Sci. USA 2012, 109, 19486–19491. [Google Scholar] [CrossRef] [Green Version]
- Qiao, H.; Shen, Z.; Huang, S.S.C.; Schmitz, R.J.; Urich, M.A.; Briggs, S.P.; Ecker, J.R. Processing and subcellular trafficking of ER-tethered EIN2 control response to ethylene gas. Science 2012, 338, 390–393. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Solano, R.; Stepanova, A.; Chao, Q.; Ecker, J.R. Nuclear events in ethylene signaling: A transcriptional cascade mediated by ETHYLENE-INSENSITIVE3 and ETHYLENE-RESPONSE-FACTOR1. Genes Dev. 1998, 12, 3703–3714. [Google Scholar] [CrossRef] [PubMed]
- Pré, M.; Atallah, M.; Champion, A.; De Vos, M.; Pieterse, C.M.; Memelink, J. The AP2/ERF domain transcription factor ORA59 integrates jasmonic acid and ethylene signals in plant defense. Plant Physiol. 2008, 147, 1347–1357. [Google Scholar] [CrossRef] [Green Version]
- Li, N.; Han, X.; Feng, D.; Yuan, D.; Huang, L.J. Signaling crosstalk between salicylic acid and ethylene/jasmonate in plant defense: Do we understand what they are whispering? Int. J. Mol. Sci. 2019, 20, 671. [Google Scholar] [CrossRef] [Green Version]
- Wubben, M.J.E., II; Jin, J.; Baum, T.J. Cyst nematode parasitism of Arabidopsis thaliana is inhibited by salicylic acid (SA) and elicits uncoupled SA-independent pathogenesis-related gene expression in roots. Mol. Plant Microbe Interact. 2008, 21, 424–432. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nahar, K.; Kyndt, T.; De Vleesschauwer, D.; Höfte, M.; Gheysen, G. The jasmonate pathway is a key player in systemically induced defense against root knot nematodes in rice. Plant Physiol. 2011, 157, 305–316. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Molinari, S.; Fanelli, E.; Leonetti, P. Expression of tomato salicylic acid (SA)-responsive pathogenesis-related genes in Mi-1-mediated and SA-induced resistance to root-knot nematodes. Mol. Plant Pathol. 2014, 15, 255–264. [Google Scholar] [CrossRef] [PubMed]
- Barcala, M.; García, A.; Cabrera, J.; Casson, S.; Lindsey, K.; Favery, B.; García-Casado, G.; Solano, R.; Fenoll, C.; Escobar, C. Early transcriptomic events in microdissected Arabidopsis nematode-induced giant cells. Plant J. 2010, 61, 698–712. [Google Scholar] [CrossRef]
- El-Shafeey, E.S.I.; Ghareeb, R.Y.; Abd-Elhady, M.A.; Abd-Elhady, S.H.; Salim, M.S. Defense-related genes induced by application of silver nanoparticles, ascorbic acid and salicylic acid for enhancing the immune response system of eggplant against invasion of root–knot nematode, Meloidogyne Javanica. Biotechnol. Biotechnol. Equip. 2021, 35, 917–933. [Google Scholar] [CrossRef]
- Sahebani, N.; Gholamrezaee, N. The ability of Meloidogyne javanica to suppress salicylic acid-induced plant defense responses. Nematology 2022, 24, 499–508. [Google Scholar] [CrossRef]
- Priya, D.B.; Somasekhar, N.; Prasad, J.S.; Kirti, P.B. Transgenic tobacco plants constitutively expressing Arabidopsis NPR1 show enhanced resistance to root-knot nematode, Meloidogyne incognita. BMC Res. Notes 2011, 4, 231. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lin, J.; Mazarei, M.; Zhao, N.; Zhu, J.J.; Zhuang, X.; Liu, W.; Pantalone, V.R.; Arelli, P.R.; Stewart, C.N., Jr.; Chen, F. Overexpression of a soybean salicylic acid methyltransferase gene confers resistance to soybean cyst nematode. Plant Biotechnol. J. 2013, 11, 1135–1145. [Google Scholar] [CrossRef] [PubMed]
- Youssef, R.M.; MacDonald, M.H.; Brewer, E.P.; Bauchan, G.R.; Kim, K.H.; Matthews, B.F. Ectopic expression of AtPAD4 broadens resistance of soybean to soybean cyst and root-knot nematodes. BMC Plant Biol. 2013, 13, 67. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ojeda-Rivera, J.O.; Ulloa, M.; Roberts, P.A.; Kottapalli, P.; Wang, C.; Nájera-González, H.R.; Payton, P.; Lopez-Arredondo, D.; Herrera-Estrella, L. Root-knot nematode resistance in Gossypium hirsutum determined by a constitutive defense-response transcriptional program avoiding a fitness penalty. Front. Plant Sci. 2022, 13, 858313. [Google Scholar] [CrossRef] [PubMed]
- Dewdney, J.; Reuber, T.L.; Wildermuth, M.C.; Devoto, A.; Cui, J.; Stutius, L.M.; Drummond, E.P.; Ausubel, F.M. Three unique mutants of Arabidopsis identify eds loci required for limiting growth of a biotrophic fungal pathogen. Plant J. 2000, 24, 205–218. [Google Scholar] [CrossRef] [Green Version]
- Wildermuth, M.C.; Dewdney, J.; Wu, G.; Ausubel, F.M. Isochorismate synthase is required to synthesize salicylic acid for plant defense. Nature 2001, 414, 562–565. [Google Scholar] [CrossRef]
- Zhang, X.; Chen, S.; Mou, Z. Nuclear localization of NPR1 is required for regulation of salicylate tolerance, isochorismate synthase 1 expression and salicylate accumulation in Arabidopsis. J. Plant Physiol. 2010, 167, 144–148. [Google Scholar] [CrossRef]
- Thomma, B.P.; Eggermont, K.; Penninckx, I.A.; Mauch-Mani, B.; Vogelsang, R.; Cammue, B.P.; Broekaert, W.F. Separate jasmonate-dependent and salicylate-dependent defense-response pathways in Arabidopsis are essential for resistance to distinct microbial pathogens. Proc. Natl. Acad. Sci. USA 1998, 95, 15107–15111. [Google Scholar] [CrossRef] [Green Version]
- Huang, H.; Zhao, W.; Qiao, H.; Li, C.; Sun, L.; Yang, R.; Ma, X.; Ma, J.; Song, S.; Wang, S. SlWRKY45 interacts with jasmonate-ZIM domain proteins to negatively regulate defense against the root-knot nematode Meloidogyne incognita in tomato. Hortic. Res. 2022, 9, uhac197. [Google Scholar] [CrossRef]
- Cooper, W.R.; Jia, L.; Goggin, L. Effects of jasmonate-induced defenses on root-knot nematode infection of resistant and susceptible tomato cultivars. J. Chem. Ecol. 2005, 31, 1953–1967. [Google Scholar] [CrossRef]
- Fujimoto, T.; Tomitaka, Y.; Abe, H.; Tsuda, S.; Futai, K.; Mizukubo, T. Expression profile of jasmonic acid-induced genes and the induced resistance against the root-knot nematode (Meloidogyne incognita) in tomato plants (Solanum lycopersicum) after foliar treatment with methyl jasmonate. J. Plant Physiol. 2011, 168, 1084–1097. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Cheng, J.; Yang, R.; Sun, Z.; Wu, C.; Wang, S. Effects of JA synthesis-related genes Spr2 and LePrs on the resistance to root-knot nematodes in tomato. Sci. Agric. Sin. 2011, 44, 4022–4028. [Google Scholar]
- Zhou, J.; Jia, F.; Shao, S.; Zhang, H.; Li, G.; Xia, X.; Zhou, Y.; Yu, J.; Shi, K. Involvement of nitric oxide in the jasmonate-dependent basal defense against root-knot nematode in tomato plants. Front. Plant Sci. 2015, 6, 193. [Google Scholar] [CrossRef]
- Kyndt, T.; Nahar, K.; Haeck, A.; Verbeek, R.; Demeestere, K.; Gheysen, G. Interplay between carotenoids, abscisic acid and jasmonate guides the compatible rice-Meloidogyne graminicola interaction. Front. Plant Sci. 2017, 8, 951. [Google Scholar] [CrossRef] [Green Version]
- Ruan, J.; Zhou, Y.; Zhou, M.; Yan, J.; Khurshid, M.; Weng, W.; Zhang, K. Jasmonic acid signaling pathway in plants. Int. J. Mol. Sci. 2019, 20, 2479. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baker, A.; Graham, I.A.; Holdsworth, M.; Smith, S.M.; Theodoulou, F.L. Chewing the fat: β-oxidation in signaling and development. Trends Plant Sci. 2006, 11, 124–132. [Google Scholar] [CrossRef]
- Fonseca, S.; Chini, A.; Hamberg, M.; Adie, B.; Porzel, A.; Kramell, R.; Miersch, O.; Wasternack, C.; Solano, R. (+)-7-iso-Jasmonoyl-L-isoleucine is the endogenous bioactive jasmonate. Nat. Chem. Biol. 2009, 5, 344–350. [Google Scholar] [CrossRef]
- Liu, H.; Timko, M.P. Jasmonic acid signaling and molecular crosstalk with other phytohormones. Int. J. Mol. Sci. 2021, 22, 2914. [Google Scholar] [CrossRef]
- Thines, B.; Katsir, L.; Melotto, M.; Niu, Y.; Mandaokar, A.; Liu, G.; Nomura, K.; He, S.Y.; Howe, G.A.; Browse, J. JAZ repressor proteins are targets of the SCFCOI1 complex during jasmonate signaling. Nature 2007, 448, 661–665. [Google Scholar] [CrossRef]
- Xie, D.X.; Feys, B.F.; James, S.; Nieto-Rostro, M.; Turner, J.G. COI1: An Arabidopsis gene required for jasmonate-regulated defense and fertility. Science 1998, 280, 1091–1094. [Google Scholar] [CrossRef]
- Zhai, Q.; Zhang, X.; Wu, F.; Feng, H.; Deng, L.; Xu, L.; Zhang, M.; Wang, Q.; Li, C. Transcriptional mechanism of jasmonate receptor COI1-mediated delay of flowering time in Arabidopsis. Plant Cell. 2015, 27, 2814–2828. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiang, Y.; Yu, D. The WRKY57 transcription factor affects the expression of jasmonate ZIM-domain genes transcriptionally to compromise Botrytis cinerea resistance. Plant Physiol. 2016, 171, 2771–2782. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Williamson, V.M.; Roberts, P.A. Mechanisms and genetics of resistance. In Root-Knot Nematodes; Perry, R.N., Moens, M., Starr, J.L., Eds.; CAB International: Wallingford, UK, 2009; pp. 301–319. [Google Scholar]
- Hussey, R.S.; Barker, K.R. A comparison of methods of collecting inocula of Meloidogyne spp. including a new technique. Plant Dis. Rep. 1973, 57, 1025–1028. [Google Scholar]
- Stirling, G.; Nicol, J.; Reay, F. Advisory Services for Nematode Pests (Operational Guidelines); Rural Industries Research Development Corporation: Barton, ACT, Australia, 2002; pp. 22–32. [Google Scholar]
- Jepson, S.B. Identification of Root-Knot Nematodes Meloidogyne Species; CABI Publishing: Wallingford, UK, 1987; pp. 1–265. [Google Scholar]
- Zijlstra, C.; Donkers-Venne, D.; Fargette, M. Identification of Meloidogyne incognita, M. javanica, and M. arenaria using sequence characterized amplified region (SCAR) based PCR assay. Nematology 2000, 2, 847–853. [Google Scholar]
- Asadi-Sardari, A.; Mahdikhani-Moghadam, E.; Zaki-Aghl, M. The biochemical changes in two moderately resistant and highly susceptible tomato cultivars at the later stages of Meloidogyne javanica infection. Nematology 2022, 24, 1085–1103. [Google Scholar] [CrossRef]
- Daykin, M.E.; Hussey, R.S. Staining and histopathological techniques in nematology. In An Advance Treatise on Meloidogyne; Barker, K.R., Carter, C.C., Sasser, J.N., Eds.; North Carolina State University Graphics: Raleigh, NC, USA, 1985; pp. 39–48. [Google Scholar]
- Kamali, S.; Javadmanesh, A.; Stelinski, L.L.; Kyndt, T.; Seifi, A.; Cheniany, M.; Zaki-Aghl, M.; Hosseini, M.; Heydarpour, M.; Asili, J.; et al. Beneficial worm allies warn plants of parasite attack below-ground and reduce above-ground herbivore preference and performance. Mol. Ecol. 2022, 31, 691–712. [Google Scholar] [CrossRef]
- Lamovšek, J.; Stare, B.G.; Pleško, I.M.; Širca, S.; Urek, G. Agrobacteria enhance plant defense against root-knot nematodes on tomato. Phytopathology 2017, 107, 681–691. [Google Scholar] [CrossRef] [Green Version]
- Rubio, M.B.; Quijada, N.M.; Pérez, E.; Domínguez, S.; Monte, E.; Hermosa, R. Identifying beneficial qualities of Trichoderma parareesei for plants. Appl. Environ. Microbiol. 2014, 80, 1864–1873. [Google Scholar] [CrossRef] [Green Version]
- Song, W.; Ma, X.; Tan, H.; Zhou, J. Abscisic acid enhances resistance to Alternaria solani in tomato seedlings. Plant Physiol. Biochem. 2011, 49, 693–700. [Google Scholar] [CrossRef]
- Ludwig, A.; Schulte, A.; Schnack, C.; Hundhausen, C.; Reiss, K.; Brodway, N.; Held-Feindt, J.; Mentlein, R. Enhanced expression and shedding of the transmembrane chemokine CXCL16 by reactive astrocytes and glioma cells. J. Neurochem. 2005, 93, 1293–1303. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2DDCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Goossens, J.; Fernández-Calvo, P.; Schweizer, F.; Goossens, A. Jasmonates: Signal transduction components and their roles in environmental stress responses. Plant Mol. Biol. 2016, 91, 673–689. [Google Scholar] [CrossRef]
- Oka, Y.; Cohen, Y.; Spiegel, Y. Local and systemic induced resistance to the root-knot nematode in tomato by DL-β-amino-n-butyric acid. Phytopathology 1999, 89, 1138–1143. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kumari, C.; Dutta, T.K.; Banakar, P.; Rao, U. Comparing the defense-related gene expression changes upon root-knot nematode attack in susceptible versus resistant cultivars of rice. Sci. Rep. 2016, 6, 22846. [Google Scholar] [CrossRef] [Green Version]
- Kyndt, T.; Nahar, K.; Haegeman, A.; De Vleesschauwer, D.; Höfte, M.; Gheysen, G. Comparing systemic defense-related gene expression changes upon migratory and sedentary nematode attack in rice. Plant Biol. 2012, 14, 73–82. [Google Scholar] [CrossRef] [PubMed]
- Mantelin, S.; Bhattarai, K.K.; Jhaveri, T.Z.; Kaloshian, I. Mi-1-mediated resistance to Meloidogyne incognita in tomato may not rely on ethylene but hormone perception through ETR3 participates in limiting nematode infection in a susceptible host. PLoS ONE 2013, 8, e63281. [Google Scholar] [CrossRef] [Green Version]
- Dyer, S.; Weir, R.; Cox, D.; Cheseto, X.; Torto, B.; Dalzell, J.J. Ethylene Response Factor (ERF) genes modulate plant root exudate composition and the attraction of plant parasitic nematodes. Int. J. Parasitol. 2019, 49, 999–1003. [Google Scholar] [CrossRef] [PubMed]
- Iberkleid, I.; Ozalvo, R.; Feldman, L.; Elbaz, M.; Patricia, B.; Horowitz, S.B. Responses of tomato genotypes to avirulent and Mi-virulent Meloidogyne javanica isolates occurring in Israel. Phytopathology 2014, 104, 484–496. [Google Scholar] [CrossRef] [Green Version]
- Zhan, L.P.; Peng, D.L.; Wang, X.L.; Kong, L.A.; Peng, H.; Liu, S.M.; Liu, Y.; Huang, W.K. Priming effect of root-applied silicon on the enhancement of induced resistance to the root-knot nematode Meloidogyne graminicola in rice. BMC Plant Biol. 2018, 18, 50. [Google Scholar] [CrossRef] [Green Version]
- Agrawal, G.K.; Rakwal, R.; Jwa, N.S. Rice (Oryza sativa L.) OsPR1b gene is phytohormonally regulated in close interaction with light signals. Biochem. Biophys. Res. Commun. 2000, 278, 290–298. [Google Scholar] [CrossRef]
- Mitsuhara, I.; Iwai, T.; Seo, S.; Yanagawa, Y.; Kawahigasi, H.; Hirose, S.; Ohkava, Y.; Ohashi, Y. Characteristic expression of twelve rice PR1 family genes in response to pathogen infection, wounding, and defense-related signal compounds (121/180). Mol. Genet. Genom. Med. 2008, 279, 415–427. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mei, C.; Qi, M.; Sheng, G.; Yang, Y. Inducible overexpression of a rice allene oxide synthase gene increases the endogenous jasmonic acid level, PR gene expression, and host resistance to fungal infection. Mol. Plant Microbe Interact. 2006, 19, 1127–1137. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tirumalaraju, S.V.; Jain, M.; Gallo, M. Differential gene expression in roots of nematode-resistant and-susceptible peanut (Arachis hypogaea) cultivars in response to early stages of peanut root-knot nematode (Meloidogyne arenaria) parasitization. J. Plant Physiol. 2011, 168, 481–492. [Google Scholar] [CrossRef]
- Qtu, J.; Hallmann, J.; Kokalis-Burelle, N.; Weaver, D.B.; Rodríguez-Kábana, R.; Tuzun, S. Activity and differential induction of chitinase isozymes in soybean cultivars resistant or susceptible to root-knot nematodes. J. Nematol. 1997, 29, 523. [Google Scholar] [PubMed]
- Hamamouch, N.; Li, C.; Seo, P.J.; Park, C.M.; Davis, E.L. Expression of Arabidopsis pathogenesis-related genes during nematode infection. Mol. Plant Pathol. 2011, 12, 355–364. [Google Scholar] [CrossRef]
- Guimaraes, P.M.; Guimaraes, L.A.; Morgante, C.V.; Silva, O.B., Jr.; Araujo, A.C.G.; Martins, A.C.; Saraiva, M.A.P.; Oliveira, T.N.; Togawa, R.C.; Leal-Bertioli, S.C.M.; et al. Root transcriptome analysis of wild peanut reveals candidate genes for nematode resistance. PLoS ONE 2015, 10, e0140937. [Google Scholar] [CrossRef]
- Lee, I.H.; Shim, D.; Jeong, J.C.; Sung, Y.W.; Nam, K.J.; Yang, J.W.; Ha, J.; Lee, J.J.; Kim, Y.H. Transcriptome analysis of root-knot nematode (Meloidogyne incognita)-resistant and susceptible sweet potato cultivars. Planta 2019, 249, 431–444. [Google Scholar] [CrossRef]
- Fan, J.W.; Hu, C.L.; Zhang, L.N.; Li, Z.L.; Zhao, F.K.; Wang, S.H. Jasmonic acid mediates tomato’s response to root knot nematodes. J. Plant Growth Regul. 2015, 34, 196–205. [Google Scholar] [CrossRef]
- Ibrahim, H.M.; Hosseini, P.; Alkharouf, N.W.; Hussein, E.H.; Abd El Kader, Y.; Aly, M.A.; Matthews, B.F. Analysis of gene expression in soybean (Glycine max) roots in response to the root knot nematode Meloidogyne incognita using microarrays and KEGG pathways. BMC Genom. 2011, 12, 220. [Google Scholar] [CrossRef] [Green Version]
- Clevenger, J.; Chu, Y.; Guimaraes, L.A.; Maia, T.; Bertioli, D.; Leal-Bertioli, S.; Timper, P.; Holbrook, C.C.; Ozias-Akins, P. Gene expression profiling describes the genetic regulation of Meloidogyne arenaria resistance in Arachis hypogaea and reveals a candidate gene for resistance. Sci. Rep. 2017, 7, 1317. [Google Scholar] [CrossRef] [Green Version]
- Song, H.; Lin, B.; Huang, Q.; Sun, T.; Wang, W.; Liao, J.; Zhuo, K. The Meloidogyne javanica effector Mj2G02 interferes with jasmonic acid signaling to suppress cell death and promote parasitism in Arabidopsis. Mol. Plant Pathol. 2021, 22, 1288–1301. [Google Scholar] [CrossRef]
- Dave, A.; Graham, I.A. Oxylipin signaling: A distinct role for the jasmonic acid precursor cis-(+)-12-oxo-phytodienoic acid (cis-OPDA). Front. Plant Sci. 2012, 3, 42. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Naor, N.; Gurung, F.B.; Ozalvo, R.; Bucki, P.; Sanadhya, P.; Miyara, S.B. Tight regulation of allene oxide synthase (AOS) and allene oxide cyclase-3 (AOC3) promote Arabidopsis susceptibility to the root-knot nematode Meloidogyne javanica. Eur. J. Plant Pathol. 2018, 150, 149–165. [Google Scholar] [CrossRef]
- Gleason, C.; Leelarasamee, N.; Meldau, D.; Feussner, I. OPDA has key role in regulating plant susceptibility to the root-knot nematode Meloidogyne hapla in Arabidopsis. Front. Plant Sci. 2016, 7, 1565. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kolomiets, M.V.; Chen, H.; Gladon, R.J.; Braun, E.J.; Hannapel, D.J. A leaf lipoxygenase of potato induced specifically by pathogen infection. Plant Physiol. 2000, 124, 1121–1130. [Google Scholar] [CrossRef]
- Göbel, C.; Feussner, I.; Schmidt, A.; Scheel, D.; Sanchez-Serrano, J.; Hamberg, M.; Rosahl, S. Oxylipin profiling reveals the preferential stimulation of the 9-lipoxygenase pathway in elicitor-treated potato cells. J. Biol. Chem. 2001, 276, 6267–6273. [Google Scholar] [CrossRef] [Green Version]
- Prost, I.; Dhondt, S.; Rothe, G.; Vicente, J.; Rodriguez, M.J.; Kift, N.; Carbonne, F.; Griffiths, G.; Esquerré-Tugayé, M.T.; Rosahl, S.; et al. Evaluation of the antimicrobial activities of plant oxylipins supports their involvement in defense against pathogens. Plant Physiol. 2005, 139, 1902–1913. [Google Scholar] [CrossRef] [Green Version]
- Vellosillo, T.; Martínez, M.; López, M.A.; Vicente, J.; Cascón, T.; Dolan, L.; Hamberg, M.; Castresana, C. Oxylipins produced by the 9-lipoxygenase pathway in Arabidopsis regulate lateral root development and defense responses through a specific signaling cascade. Plant Cell. 2007, 19, 831–846. [Google Scholar] [CrossRef] [Green Version]
- Hwang, I.S.; Hwang, B.K. The pepper 9-lipoxygenase gene CaLOX1 functions in defense and cell death responses to microbial pathogens. Plant Physiol. 2010, 152, 948–967. [Google Scholar] [CrossRef] [Green Version]
- Christensen, S.A.; Huffaker, A.; Kaplan, F.; Sims, J.; Ziemann, S.; Doehlemann, G.; Ji, L.; Schmitz, R.J.; Kolomiets, M.V.; Alborn, H.T.; et al. Maize death acids, 9-lipoxygenase–derived cyclopente (a) nones, display activity as cytotoxic phytoalexins and transcriptional mediators. Proc. Natl. Acad. Sci. USA 2015, 112, 11407–11412. [Google Scholar] [CrossRef] [Green Version]
- Ozalvo, R.; Cabrera, J.; Escobar, C.; Christensen, S.A.; Borrego, E.J.; Kolomiets, M.V.; Castresana, C.; Iberkleid, I.; Brown Horowitz, S. Two closely related members of Arabidopsis 13-lipoxygenases (13-LOXs), LOX3 and LOX4, reveal distinct functions in response to plant-parasitic nematode infection. Mol. Plant Pathol. 2014, 15, 319–332. [Google Scholar] [CrossRef]
- Macharia, T.N.; Bellieny-Rabelo, D.; Moleleki, L.N. Transcriptome profiling of potato (Solanum tuberosum L.) responses to root-knot nematode (Meloidogyne javanica) infestation during a compatible interaction. Microorganisms 2020, 8, 1443. [Google Scholar] [CrossRef]
- Ghadamgahi, F.; Tarighi, S.; Taheri, P.; Saripella, G.V.; Anzalone, A.; Kalyandurg, P.B.; Catara, V.; Ortiz, R.; Vetukuri, R.R. Plant growth-promoting activity of Pseudomonas aeruginosa FG106 and its ability to act as a biocontrol agent against potato, tomato and Taro pathogens. Biology 2022, 11, 140. [Google Scholar] [CrossRef] [PubMed]
- Song, S.; Huang, H.; Gao, H.; Wang, J.; Wu, D.; Liu, X.; Yang, S.; Zhai, Q.; Li, C.; Qi, T.; et al. Interaction between MYC2 and ETHYLENE INSENSITIVE3 modulates antagonism between jasmonate and ethylene signaling in Arabidopsis. Plant Cell 2014, 26, 263–279. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, X.; Zhu, Z.; An, F.; Hao, D.; Li, P.; Song, J.; Yi, C.; Guo, H. Jasmonate-activated MYC2 represses ETHYLENE INSENSITIVE3 activity to antagonize ethylene-promoted apical hook formation in Arabidopsis. Plant Cell 2014, 26, 1105–1117. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Y.; Lan, Y.; Shi, T.; Zhu, Z. Diverse contributions of MYC 2 and EIN 3 in the regulation of Arabidopsis jasmonate-responsive gene expression. Plant Direct. 2017, 1, e00015. [Google Scholar] [CrossRef] [PubMed]

| Gene | Forward Primer (5′to 3′) | Reverse Primer (5′to 3′) | Locus ID (solgenomics.net) 8 May 2022 | Amplicon Length (bp) | Reference |
|---|---|---|---|---|---|
| SlCOI1 a | GGGTACAAGGATACAGGGCAT | GGCAAGAGAATAGTAGGCAAGT | Solyc05g052620.2 | 173 | This study |
| SlMYC2 a | ATGCTTCCAAATCTATGCCGTT | TAATAACCATCTCCCCAACCCA | Solyc08g076930.1 | 163 | This study |
| SlWRKY57 a | GGACTTATCAATCACGAAGCAT | CATCTGGTTGACTTGTTTCTGG | Solyc05g012500.2 | 190 | This study |
| SlJAZ1 a | GTGATTCATCGTCGTCATCGTC | TCATTTGTGCCTTCTCTGGTTG | Solyc12g009220.1 | 147 | This study |
| SlJAZ2 a | TCAGAGTTCATTTGGGACTTTC | CTGGCTTAATCTGGAGGTGTT | Solyc03g122190.2 | 134 | This study |
| SlJAZ3 a | GGAATGAAGGCTGAGTCGGAAC | GAAACTCGGAACCACCAAATCG | Solyc07g042170.2 | 186 | This study |
| SlJAR1 a | GCCATTTATAAGAAAGGAGGGA | CAGCATCTTTAGTCAACACCT | Solyc10g011660.2 | 109 | This study |
| SlAOS3 a | CACTTTCCCTCTACCTTACATCCT | AACCGCCATACGAATTGAATCC | Solyc10g007960.1 | 170 | [72] |
| SlLOXD a | ATCCCTGACGAGAACGATCC | TCCAAGTAGACGGTTGCTGT | Solyc03g122340.2 | 178 | This study |
| SlERF1 a | GGGTCCTTGGTCTCTACTCA | TCTCTTGTGCTTGACTCTTCTA | Solyc05g051200.1 | 142 | This study |
| SlACO1 a | TGAGTTGGTGAACCATGGAA | AGTAGGAAGATGGCGCAAGA | Solyc07g049530.2 | 190 | [73] |
| SlACO4 a | CGCAGGAGGCATCATACTTC | CCGAGTCCCATCTGTTTGTG | Solyc07g049550.2 | 196 | [72] |
| SlICS1 a | GTTCCTCTCCAAGAAATGTCC | TCCTTCAAGCTCATCAAACTC | Solyc06g071030.2 | 142 | [74] |
| SlNPR1 a | TACCAAGTCTACAGAGGAAGGA | CAAATCATCGCCTGCCATAG | Solyc07g040690.2 | 133 | This study |
| SlPR 1a a | GCTGTGAAGATGTGGGTTGATG | CGTTGTCCTCTCCAGTTACCT | Solyc01g106620.2 | 200 | [72] |
| SlPR1a2 a | TTGGGATGCCGACTTGGAAT | CCGCTAACACATTCATTCGTATCG | Solyc09g007020.1 | 192 | [72] |
| SlPR1 a | TGTCCGAGAGGCCAAGCTATAAC | AATGAACCACCATCCGTTGTTGC | Solyc00g174340.1 | 143 | [75] |
| SlTAC b | CTCACGCATTGACCACAAGT | CAGCACCAACCTCCTCATAATC | Solyc08g006890.2 | 149 | [72] |
| Invasion by J2s | Parasitic J2s | J3s | J4s | Females | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Days post inoculation | Dutch Mobil | ALYSTE F-1 | Dutch Mobil | ALYSTE F-1 | Dutch Mobil | ALYSTE F-1 | Dutch Mobil | ALYSTE F-1 | Dutch Mobil | ALYSTE F-1 |
| 1 | 35.67 ± 8.09 e | 1.67 ± 0.88 e | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 2 | 269 ± 16.26 c | 9.33 ± 1.20 d | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 3 | 338.33 ± 12.73 a | 22.67 ± 2.90 b | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 4 | 296 ± 7.77 b | 29 ± 0.58a | 42.33 ± 15.39 fg | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 5 | 163 ± 18.90 d | 29.67 ± 1.33 a | 157.35 ± 17.57 d | 0.5 ± 0.34 j | 0 | 0 | 0 | 0 | 0 | 0 |
| 6 | 22.33 ± 1.45 ef | 19.67 ± 1.20 b | 293 ± 9.81 b | 10.33 ± 1.20 ghi | 0 | 0 | 0 | 0 | 0 | 0 |
| 7 | 0.5 ± 0.34 f | 15.33 ± 1.77 c | 367.52 ± 21.36 a | 14 ± 2.08 efgh | 0 | 0 | 0 | 0 | 0 | 0 |
| 8 | 0 | 9 ± 0.58 d | 354.40 ± 17.68 a | 19 ± 1.15 d | 0 | 0 | 0 | 0 | 0 | 0 |
| 9 | 0 | 0 | 289.57 ± 12.98 b | 30 ± 0.58 a | 10.57 ± 3.48 jk | 0 | 0 | 0 | 0 | 0 |
| 10 | 0 | 0.33 ± 0.33 e | 242.40 ± 23.82 c | 25 ± 2.52 bc | 56 ± 7.81 gh | 0 | 0 | 0 | 0 | 0 |
| 11 | 0 | 0 | 252 ± 16.59 c | 27 ± 2.52 abc | 48.62 ± 8.64 hi | 0 | 0 | 0 | 0 | 0 |
| 12 | 0 | 0 | 153.65 ± 24.51 d | 24.63 ± 3.38 bc | 138.43 ± 22 e | 2 ± 1.15 h | 0 | 0 | 0 | 0 |
| 13 | 0 | 0 | 87 ± 19.05 e | 26 ± 0.58 abc | 234.70 ± 28.91 c | 2.57 ± 0.83 gh | 0 | 0 | 0 | 0 |
| 14 | 0 | 0 | 54.48 ± 10.68 ef | 23.54 ± 2.40 c | 310.35 ± 8.40 a | 4.63 ± 2.03 fg | 0 | 0 | 0 | 0 |
| 15 | 0 | 0 | 27 ± 2 fgh | 29 ± 1.15 ab | 280.39 ± 11.83 ab | 7.33 ± 1.20 ef | 24.63 ± 1.45 ghi | 0 | 0 | 0 |
| 16 | 0 | 0 | 14.40 ± 3.84 gh | 26.52 ± 2.18 abc | 245.67 ± 19.27 c | 9 ± 1.52 cde | 39.34 ± 7.53 ghi | 0 | 0 | 0 |
| 17 | 0 | 0 | 0.5 ± 0.34 h | 18.37 ± 0.88 de | 259 ± 19.08 bc | 12.33 ± 1.20 abc | 74.33 ± 13.96 f | 0 | 0 | 0 |
| 18 | 0 | 0 | 0 | 16.75 ± 0.33 def | 198.89 ± 8.95 d | 14.33 ± 1.33 ab | 119.70 ± 16.19 e | 0 | 0 | 0 |
| 19 | 0 | 0 | 0 | 14.63 ± 1.45 defg | 110 ± 7.5 ef | 16 ± 2.51 a | 232.33 ± 3.93 bc | 0 | 0 | 0 |
| 20 | 0 | 0 | 0 | 14 ± 1.73 efgh | 80 ± 3.76 fg | 12 ± 2.31 abc | 303.78 ± 6.96 a | 4.73 ± 1.86 bcde | 0 | 0 |
| 21 | 0 | 0 | 0 | 12 ± 1.52 ghi | 34 ± 4.05 hig | 9.33 ± 1.45 cde | 299.30 ± 11.29 a | 8.67 ± 1.20 bcde | 12 ± 1.15 f | 0 |
| 22 | 0 | 0 | 0 | 12.66 ± 2.33 fghi | 24 ± 3 ijk | 11.67 ± 1.53 bcd | 255 ± 15.40 b | 5.76 ± 1.45 e | 32.34 ± 4.84 ef | 0 |
| 23 | 0 | 0 | 0 | 11.62 ± 2.03 ghi | 11.83 ± 2.73 jk | 12 ± 1.45 abc | 230 ± 11.54 bc | 6 ± 0.60 de | 55 ± 4.16 e | 0 |
| 24 | 0 | 0 | 0 | 9.36 ± 0.37 i | 1.65 ± 0.88 k | 7.38 ± 2.35 ef | 214.37 ± 1.76 c | 12 ± 2.30 abc | 88.67 ± 6.98 d | 0 |
| 25 | 0 | 0 | 0 | 11 ± 0.58 ghi | 0 | 5.34 ± 0.32 efg | 167 ± 9.07 d | 14 ± 0.6 a | 139.70 ± 18.52 c | 0 |
| 26 | 0 | 0 | 0 | 9 ± 0.52 i | 0.5 ± 0.22 k | 7 ± 2.31 ef | 78 ± 9.29 f | 13.70 ± 2.73 a | 231 ± 4.93 b | 0 |
| 27 | 0 | 0 | 0 | 9.63 ± 1.45 hi | 0 | 6 ± 1 efg | 47.34 ± 8.87 g | 12.36 ± 2.97 ab | 245.33 ± 7.96 b | 1.67 ± 0.89 b |
| 28 | 0 | 0 | 0 | 9 ± 1.15 i | 0 | 5.67 ± 1.76 efg | 7 ± 1.15 i | 11 ± 1.53 abc | 326.67 ± 12.91 a | 1.67 ± 0.5 ab |
| 29 | 0 | 0 | 0 | 10.33 ± 0.88 ghi | 0 | 7.69 ± 0.88 def | 12 ± 4.04 i | 10 ± 1.53 abcd | 319 ± 9.85 a | 0 |
| 30 | 0 | 0 | 0 | 9.42 ± 1.20 i | 0 | 7.33 ± 0.88 ef | 13.67 ± 3.84 hi | 8.40 ± 1.20 cde | 307.58 ± 10.48 a | 3.40 ± 1.20 a |
| Number of egg masses 30 days after inoculation | ||||||||||
| Dutch Mobil | ALYSTE F-1 | |||||||||
| 34 ± 1.45 | 0 | |||||||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Asadi-Sardari, A.; Mahdikhani-Moghadam, E.; Zaki-Aghl, M.; Vetukuri, R.R. Constitutive and Inducible Expression of Genes Related to Salicylic Acid and Ethylene Pathways in a Moderately Resistant Tomato Cultivar Leads to Delayed Development of Meloidogyne javanica. Agriculture 2022, 12, 2122. https://doi.org/10.3390/agriculture12122122
Asadi-Sardari A, Mahdikhani-Moghadam E, Zaki-Aghl M, Vetukuri RR. Constitutive and Inducible Expression of Genes Related to Salicylic Acid and Ethylene Pathways in a Moderately Resistant Tomato Cultivar Leads to Delayed Development of Meloidogyne javanica. Agriculture. 2022; 12(12):2122. https://doi.org/10.3390/agriculture12122122
Chicago/Turabian StyleAsadi-Sardari, Ameneh, Esmat Mahdikhani-Moghadam, Mohammad Zaki-Aghl, and Ramesh Raju Vetukuri. 2022. "Constitutive and Inducible Expression of Genes Related to Salicylic Acid and Ethylene Pathways in a Moderately Resistant Tomato Cultivar Leads to Delayed Development of Meloidogyne javanica" Agriculture 12, no. 12: 2122. https://doi.org/10.3390/agriculture12122122
APA StyleAsadi-Sardari, A., Mahdikhani-Moghadam, E., Zaki-Aghl, M., & Vetukuri, R. R. (2022). Constitutive and Inducible Expression of Genes Related to Salicylic Acid and Ethylene Pathways in a Moderately Resistant Tomato Cultivar Leads to Delayed Development of Meloidogyne javanica. Agriculture, 12(12), 2122. https://doi.org/10.3390/agriculture12122122






