Effects of Lake Sediment on Soil Properties, Crop Growth, and the phoD-Harboring Microbial Community
Abstract
:1. Introduction
2. Materials and Methods
2.1. Site Description
2.2. Experimental Design
2.3. Sampling and Measurements
2.4. Microbial Analysis
2.5. Statistical Analysis
3. Results and Discussion
3.1. Soil Properties
3.2. Crop Yield and Growth-Related Traits
3.3. phoD-Harboring Microbial Abundance Analysis
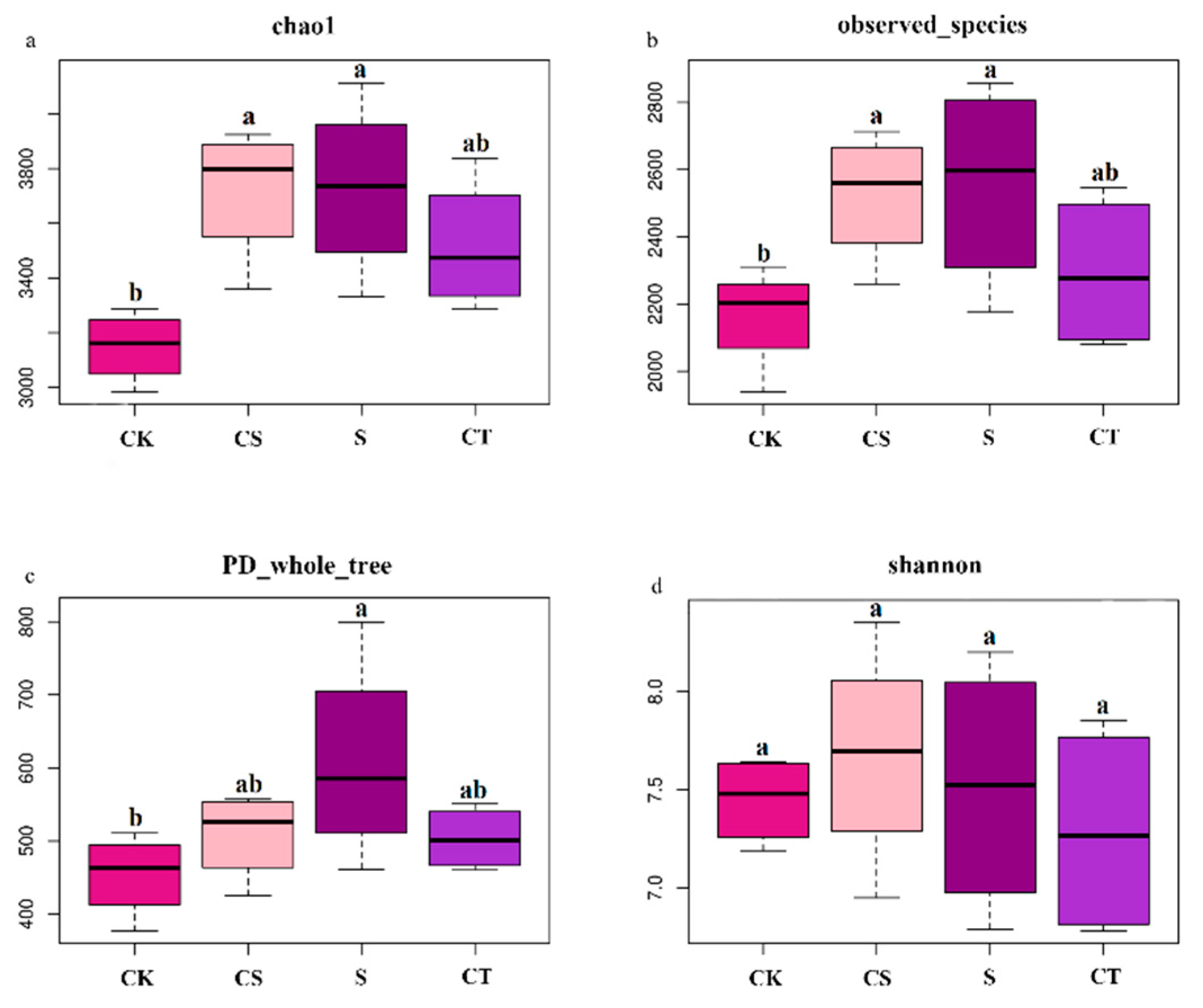
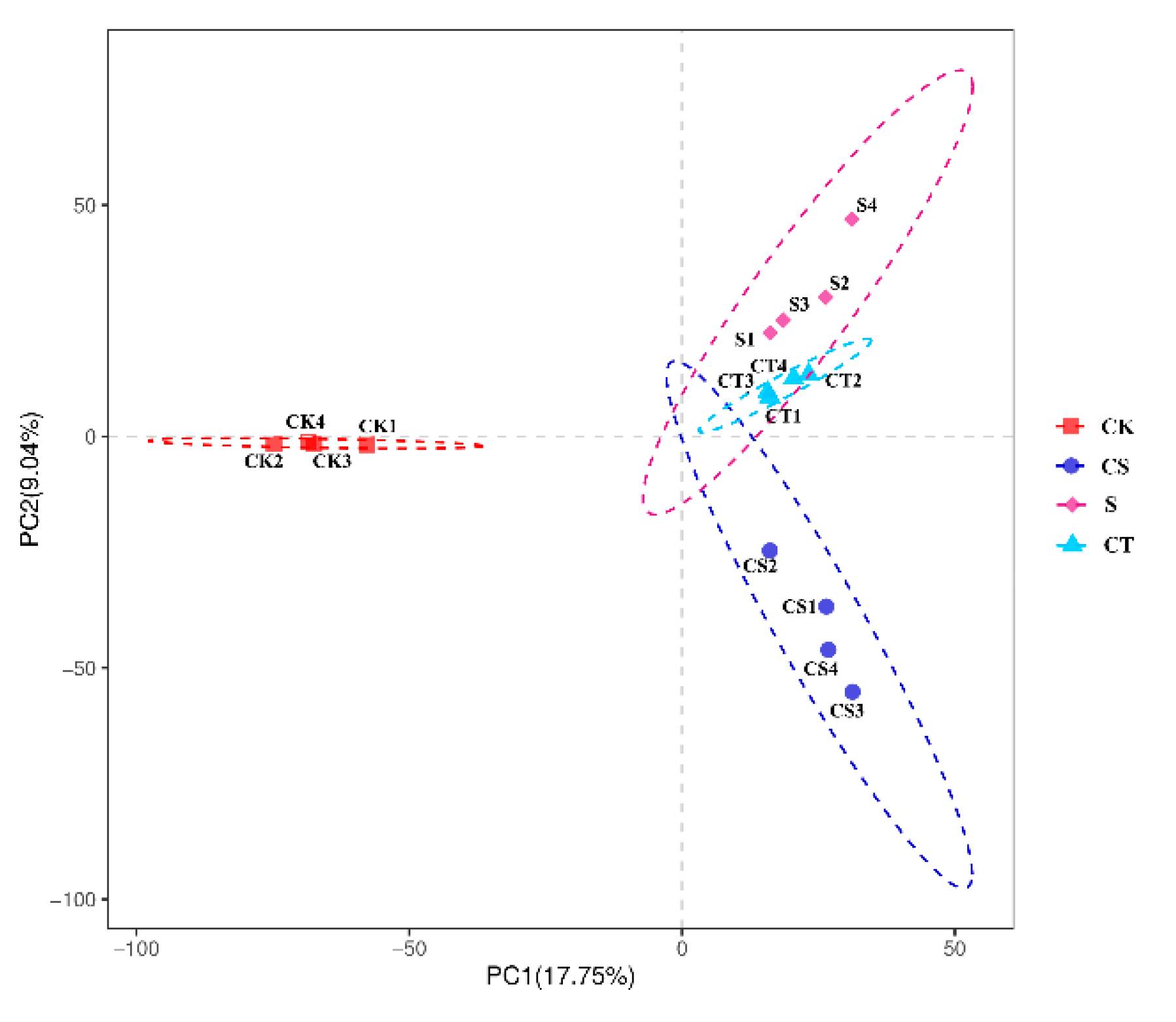
3.4. phoD Microbial Community α and β Diversity Analysis
3.5. Comparison of phoD Microbial Community Composition among the Different Treatments
3.6. Comparison of phoD Microbial Community Composition among the Different Treatments
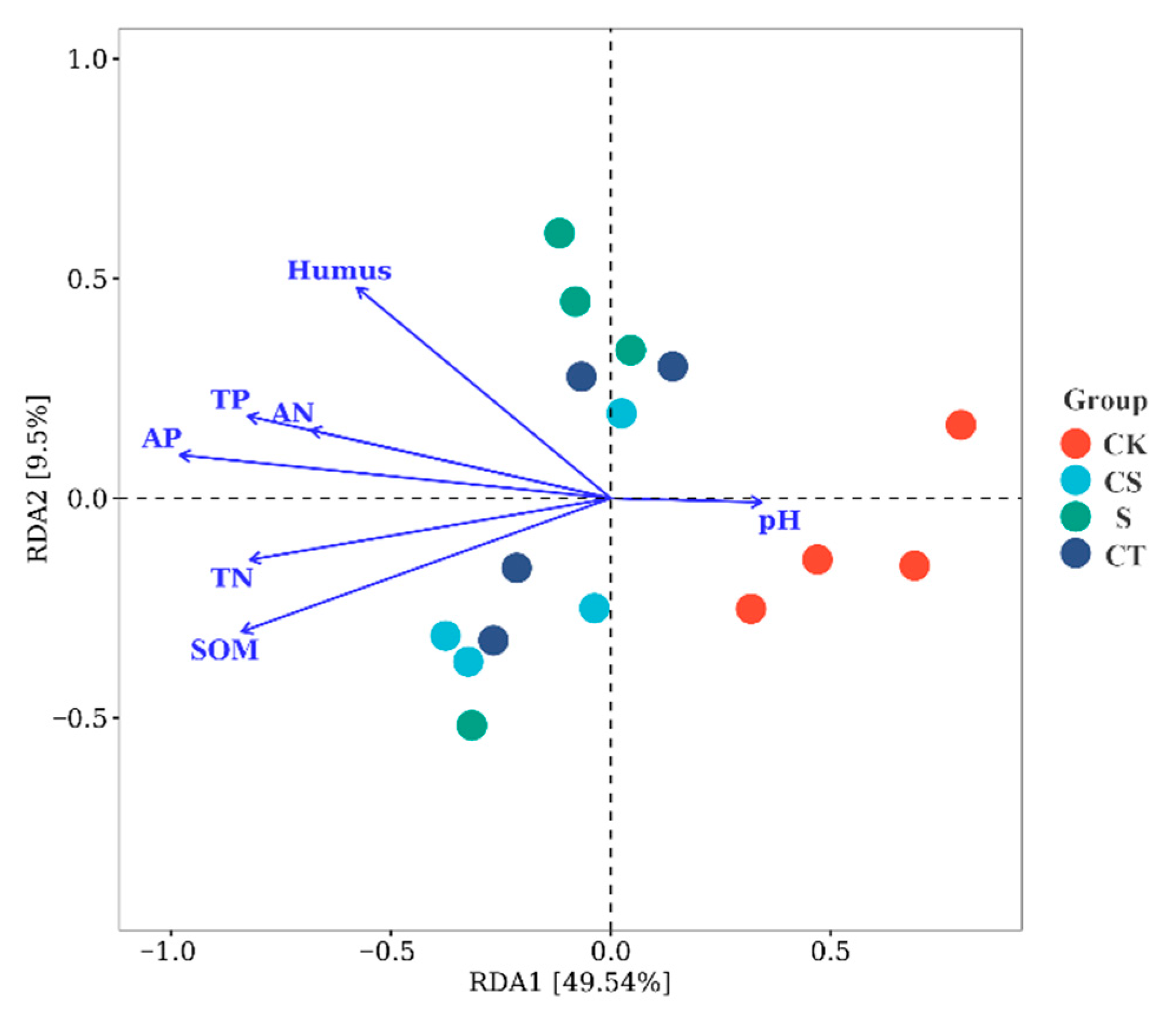
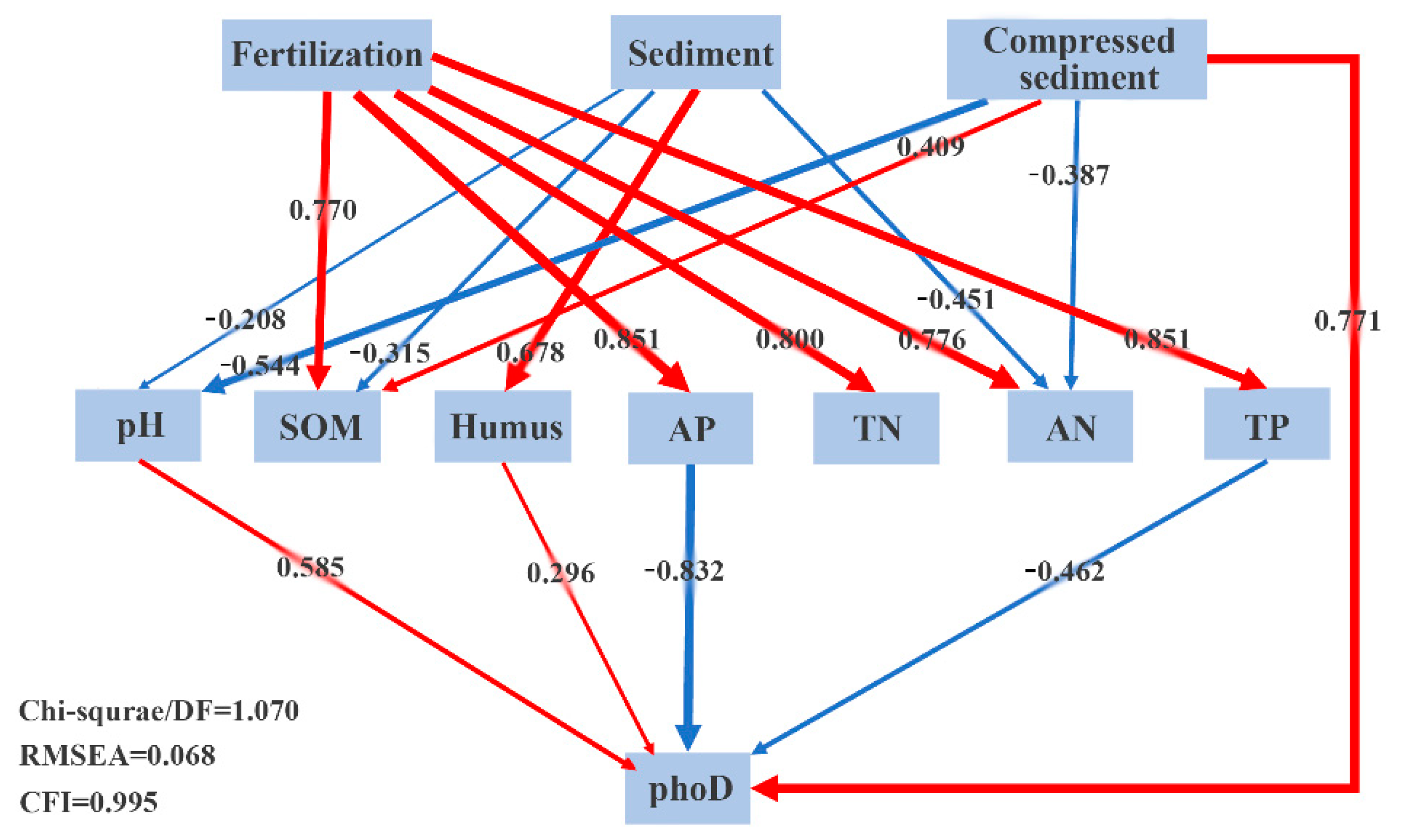
3.7. Structural Equation Model Analysis of the phoD Microbial Communities
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Tiessen, H. Phosphorus in the global environment. In The Ecophysiology of Plant-Phosphorus Interactions; Springer: Berlin/Heidelberg, Germany; Dordrecht, The Netherlands, 2008; pp. 1–7. [Google Scholar]
- Guang-Can, T.A.O.; Shu-Jun, T.I.A.N.; Miao-Ying, C.A.I.; Guang-Hui, X.I.E. Phosphate-solubilizing and-mineralizing abilities of bacteria isolated from soils. Pedosphere 2008, 18, 515–523. [Google Scholar]
- Turner, B.L.; Frossard, E.; Baldwin, D.S. (Eds.) Organic Phosphorus in the Environment; CABI Pub.: Wallingford, UK, 2005; pp. 269–294. [Google Scholar]
- Rawat, P.; Das, S.; Shankhdhar, D.; Shankhdhar, S.C. Phosphate-solubilizing microorganisms: Mechanism and their role in phosphate solubilization and uptake. J. Soil Sci. Plant Nutr. 2021, 21, 49–68. [Google Scholar] [CrossRef]
- Ahmed, S.; Iqbal, N.; Tang, X.; Ahmad, R.; Irshad, M.; Irshad, U. Organic amendment plus inoculum drivers: Who drives more P nutrition for wheat plant fitness in small duration soil experiment. PLoS ONE 2022, 17, e0266279. [Google Scholar] [CrossRef] [PubMed]
- Fraser, T.; Lynch, D.H.; Entz, M.H.; Dunfield, K.E. Linking alkaline phosphatase activity with bacterial phoD gene abundance in soil from a long-term management trial. Geoderma 2015, 257, 115–122. [Google Scholar] [CrossRef]
- Neal, A.L.; Blackwell, M.; Akkari, E.; Guyomar, C.; Clark, I.; Hirsch, P.R. Phylogenetic distribution, biogeography and the effects of land management upon bacterial non-specific Acid phosphatase Gene diversity and abundance. Plant Soil 2018, 427, 175–189. [Google Scholar] [CrossRef] [Green Version]
- Sarr, P.S.; Tibiri, E.B.; Fukuda, M.; Zongo, A.N.; Compaore, E.; Nakamura, S. Phosphate-solubilizing fungi and alkaline phosphatase trigger the P solubilization during the co-composting of sorghum straw residues with Burkina Faso phosphate rock. Front. Environ. Sci. 2020, 8, 559195. [Google Scholar] [CrossRef]
- Luo, G.; Ling, N.; Nannipieri, P.; Chen, H.; Raza, W.; Wang, M.; Guo, S.; Shen, Q. Long-term fertilisation regimes affect the composition of the alkaline phosphomonoesterase encoding microbial community of a vertisol and its derivative soil fractions. Biol. Fertil. Soils 2017, 53, 375–388. [Google Scholar] [CrossRef]
- Ragot, S.A.; Huguenin-Elie, O.; Kertesz, M.A.; Frossard, E.; Bünemann, E.K. Total and active microbial communities and phoD as affected by phosphate depletion and pH in soil. Plant Soil 2016, 408, 15–30. [Google Scholar] [CrossRef]
- Hu, M.; Peñuelas, J.; Sardans, J.; Tong, C.; Chang, C.T.; Cao, W. Dynamics of phosphorus speciation and the phoD phosphatase gene community in the rhizosphere and bulk soil along an estuarine freshwater-oligohaline gradient. Geoderma 2020, 365, 114236. [Google Scholar] [CrossRef]
- Neal, A.L.; Rossmann, M.; Brearley, C.; Akkari, E.; Guyomar, C.; Clark, I.M.; Allen, E.; Hirsch, P.R. Land-use influences phosphatase gene microdiversity in soils. Environ. Microbiol. 2017, 19, 2740–2753. [Google Scholar] [CrossRef] [Green Version]
- Wang, F.; Zhang, Y.; Xia, Y.; Cui, Z.; Cao, C. Soil Microbial Community Succession Based on phoD and Gcd Genes along a Chronosequence of Sand-Fixation Forest. Forests 2021, 12, 1707. [Google Scholar] [CrossRef]
- Itelima, J.U.; Bang, W.J.; Onyimba, I.A.; Sila, M.D.; Egbere, O.J. Bio-fertilizers as key player in enhancing soil fertility and crop productivity: A review. Direct Res. J. Agric. Food Sci. 2018, 6, 73–83. [Google Scholar]
- Qin, B.; Gao, G.; Zhu, G.; Zhang, Y.; Song, Y.; Tang, X.; Xu, H.; Deng, J. Lake eutrophication and its ecosystem response. Chin. Sci. Bull. 2013, 58, 961–970. [Google Scholar] [CrossRef] [Green Version]
- Kiani, M.; Raave, H.; Simojoki, A.; Tammeorg, O.; Tammeorg, P. Recycling lake sediment to agriculture: Effects on plant growth, nutrient availability, and leaching. Sci. Total Environ. 2021, 753, 141984. [Google Scholar] [CrossRef]
- Huser, B.J.; Futter, M.; Lee, J.T.; Perniel, M. In-lake measures for phosphorus control: The most feasible and cost-effective solution for long-term management of water quality in urban lakes. Water Res. 2016, 97, 142–152. [Google Scholar] [CrossRef] [PubMed]
- Nixon, S.W. Replacing the Nile: Are anthropogenic nutrients providing the fertility once brought to the Mediterranean by a great river? AMBIO A J. Hum. Environ. 2003, 32, 30–39. [Google Scholar] [CrossRef]
- Canet, R.; Chaves, C.; Pomares, F.; Albiach, R. Agricultural use of sediments from the Albufera Lake (eastern Spain). Agric. Ecosyst. Environ. 2003, 95, 29–36. [Google Scholar] [CrossRef]
- Kazberuk, W.; Szulc, W.; Rutkowska, B. Use bottom sediment to agriculture—Effect on plant and heavy metal content in soil. Agronomy 2021, 11, 1077. [Google Scholar] [CrossRef]
- Zhang, H.; Sun, H.; Zhou, S.; Bai, N.; Zheng, X.; Li, S.; Zhang, J.; Lv, W. Effect of Straw and Straw Biochar on the Community Structure and Diversity of Ammonia-oxidizing Bacteria and Archaea in Rice-wheat Rotation Ecosystems. Sci. Rep. 2019, 9, 9367. [Google Scholar] [CrossRef] [Green Version]
- Dupont, A.Ö.C.; Griffiths, R.I.; Bell, T.; Bass, D. Differences in soil micro-eukaryotic communities over soil pH gradients are strongly driven by parasites and saprotrophs. Environ. Microbiol. 2016, 18, 2010–2024. [Google Scholar] [CrossRef] [Green Version]
- Khan, S.A.; Mulvaney, R.L.; Mulvaney, C.S. Accelerated diffusion methods for inorganic-nitrogen analysis of soil extracts and water. Soil Sci. Soc. Am. J. 1997, 61, 936–942. [Google Scholar] [CrossRef]
- Song, H.; Li, Z.; Du, B.; Wang, G.; Ding, Y. Bacterial communities in sediments of the shallow Lake Dongping in China. J. Appl. Microbiol. 2012, 112, 79–89. [Google Scholar] [CrossRef] [PubMed]
- Tan, Y.; Cui, Y.; Li, H.; Kuang, A.; Li, X.; Wei, Y.; Ji, X. Rhizospheric soil and root endogenous fungal diversity and composition in response to continuous Panax notoginseng cropping practices. Microbiol. Res. 2017, 194, 10–19. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Wei, Z.; Zhu, Z.; Zhao, Y.; Jia, L.; Lv, P. Humus formation driven by ammonia-oxidizing bacteria during mixed materials composting. Bioresour. Technol. 2020, 311, 123500. [Google Scholar] [CrossRef] [PubMed]
- Huang, B.; Yan, D.; Ouyang, C.; Zhang, D.; Zhu, J.; Liu, J.; Li, Y.; Wang, Q.; Han, Q.; Cao, A. Chloropicrin fumigation alters the soil phosphorus and the composition of the encoding alkaline phosphatase PhoD gene microbial community. Sci. Total Environ. 2020, 711, 135080. [Google Scholar] [CrossRef] [PubMed]
- Lagos, L.M.; Acuña, J.J.; Maruyama, F.; Ogram, A.; de la Luz Mora, M.; Jorquera, M.A. Effect of phosphorus addition on total and alkaline phosphomonoesterase-harboring bacterial populations in ryegrass rhizosphere microsites. Biol. Fertil. Soils 2016, 52, 1007–1019. [Google Scholar] [CrossRef]
- Chen, X.; Jiang, N.; Condron, L.M.; Dunfield, K.E.; Chen, Z.; Wang, J.; Chen, L. Impact of long-term phosphorus fertilizer inputs on bacterial phoD gene community in a maize field, Northeast China. Sci. Total Environ. 2019, 669, 1011–1018. [Google Scholar] [CrossRef]
- Nannipieri, P.; Giagnoni, L.; Renella, G.; Puglisi, E.; Ceccanti, B.; Masciandaro, G.; Fornasier, F.L.A.V.I.O.; Moscatelli, M.C.; Marinari, S.A.R.A. Soil enzymology: Classical and molecular approaches. Biol. Fertil. Soils 2012, 48, 743–762. [Google Scholar] [CrossRef]
- Tan, H.; Barret, M.; Rice, O.; Dowling, D.N.; Burke, J.; Morrissey, J.P.; O’Gara, F. Long-term agrichemical use leads to alterations in bacterial community diversity. Plant Soil Environ. 2012, 58, 452–458. [Google Scholar] [CrossRef] [Green Version]
- Liu, W.; Ling, N.; Luo, G.; Guo, J.; Zhu, C.; Xu, Q.; Liu, M.; Shen, Q.; Guo, S. Active phoD-harboring bacteria are enriched by long-term organic fertilization. Soil Biol. Biochem. 2021, 152, 108071. [Google Scholar] [CrossRef]
- Chen, X.; Jiang, N.; Chen, Z.; Tian, J.; Sun, N.; Xu, M.; Chen, L. Response of soil phoD phosphatase gene to long-term combined applications of chemical fertilizers and organic materials. Appl. Soil Ecol. 2017, 119, 197–204. [Google Scholar] [CrossRef]
- Coelho, J.J.; Hennessy, A.; Casey, I.; Bragança, C.R.S.; Woodcock, T.; Kennedy, N. Biofertilisation with anaerobic digestates: A field study of effects on soil microbial abundance and diversity. Appl. Soil Ecol. 2020, 147, 103403. [Google Scholar] [CrossRef]
- Sapp, M.; Harrison, M.; Hany, U.; Charlton, A.; Thwaites, R. Comparing the effect of digestate and chemical fertiliser on soil bacteria. Appl. Soil Ecol. 2015, 86, 1–9. [Google Scholar] [CrossRef]
- Huang, B.; Yan, D.; Wang, Q.; Fang, W.; Song, Z.; Cheng, H.; Li, Y.; Ouyang, C.; Han, Q.; Jin, X.; et al. Effects of dazomet fumigation on soil phosphorus and the composition of phoD-harboring microbial communities. J. Agric. Food Chem. 2020, 68, 5049–5058. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cheng, J.; Zhang, Y.; Wang, H.; Cui, Z.; Cao, C. Sand-fixation plantation type affects soil phosphorus transformation microbial community in a revegetation area of Horqin Sandy Land, Northeast China. Ecol. Eng. 2022, 180, 106644. [Google Scholar] [CrossRef]
- Liu, J.; Ma, Q.; Hui, X.; Ran, J.; Ma, Q.; Wang, X.; Wang, Z. Long-term high-P fertilizer input decreased the total bacterial diversity but not phoD-harboring bacteria in wheat rhizosphere soil with available-P deficiency. Soil Biol. Biochem. 2020, 149, 107918. [Google Scholar] [CrossRef]
- Wang, J.; Liu, S.; Li, S. Effect of long-term plastic film mulching and fertilization on inorganic N distribution and organic N mineralization in brown earth. J. Soil Water Conserv. 2006, 20, 107–110. [Google Scholar]


| Items | pH | SOM (g·kg−1) | TN (g·kg−1) | TP (g·kg−1) | Humus (g·kg−1) | AN (mg·kg−1) | AP (mg·kg−1) |
|---|---|---|---|---|---|---|---|
| CK | 8.44 ± 0.06ab | 30.36 ± 0.88c | 1.33 ± 0.04b | 1.19 ± 0.21b | 3.41 ± 0.25b | 57.6 ± 2.92c | 24.64 ± 1.97b |
| CS | 7.77 ± 0.04c | 49.26 ± 2.89a | 1.84 ± 0.19a | 1.87 ± 0.22a | 4.06 ± 0.16ab | 77.89 ± 6.64b | 46.30 ± 2.45a |
| S | 8.23 ± 0.22b | 37.68 ± 3.25b | 1.64 ± 0.06a | 1.90 ± 0.03a | 4.48 ± 0.46a | 74.55 ± 6.13b | 44.66 ± 2.16a |
| CT | 8.52 ± 0.04a | 40.95 ± 5.18b | 1.73 ± 0.08a | 1.83 ± 0.06a | 3.71 ± 0.18b | 98.03 ± 1.6a | 45.11 ± 1.34a |
| Items | Yield (kg·667m−2) | Plant Height (cm) | Chlorophyll (mg·kg−1) | p-TN (g·kg−1) | p-TP (g·kg−1) |
|---|---|---|---|---|---|
| CK | 1266.55 ± 45.13c | 11.3 ± 0.88c | 28.85 ± 0.91c | 1.19 ± 0.08b | 0.20 ± 0.01c |
| CS | 3992.67 ± 85.27a | 23.38 ± 0.56a | 34.93 ± 0.47ab | 2.04 ± 0.06a | 0.34 ± 0.01a |
| S | 2835.2 ± 108.69b | 23.74 ± 1.04a | 36.27 ± 1.32a | 1.98 ± 0.16a | 0.35 ± 0.00a |
| CT | 2847.95 ± 105.48b | 19.46 ± 1.38b | 33.18 ± 1.57b | 2.06 ± 0.12a | 0.30 ± 0.01b |
| phoD | pH | SOM | TN | AN | TP | AP | Humus | Chao1 | Shannon | |
|---|---|---|---|---|---|---|---|---|---|---|
| PhoD | 1 | |||||||||
| pH | 0.193 | 1 | ||||||||
| SOM | −0.534 * | −0.662 ** | 1 | |||||||
| TN | −0.683 * | −0.500 * | 0.772 ** | 1 | ||||||
| AN | −0.712 * | 0.045 | 0.496 | 0.670 ** | 1 | |||||
| TP | −0.888 ** | −0.369 | 0.630 ** | 0.700 ** | 0.599 * | 1 | ||||
| AP | −0.885 ** | −0.398 | 0.718 ** | 0.835 ** | 0.689 ** | 0.869 ** | 1 | |||
| Humus | −0.471 | −0.493 | 0.375 | 0.371 | 0.110 | 0.599 * | 0.592 * | 1 | ||
| Chao1 | −0.628 ** | −0.379 | 0.431 | 0.399 | 0.371 | 0.561 * | 0.682 ** | 0.422 | 1 | |
| Shannon | −0.084 | −0.221 | 0.073 | −0.083 | −0.056 | −0.057 | 0.033 | −0.146 | 0.628 ** | 1 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhong, F.; Bai, N.; Chu, X.; He, Y.; Zhang, H.; Li, H. Effects of Lake Sediment on Soil Properties, Crop Growth, and the phoD-Harboring Microbial Community. Agriculture 2022, 12, 2065. https://doi.org/10.3390/agriculture12122065
Zhong F, Bai N, Chu X, He Y, Zhang H, Li H. Effects of Lake Sediment on Soil Properties, Crop Growth, and the phoD-Harboring Microbial Community. Agriculture. 2022; 12(12):2065. https://doi.org/10.3390/agriculture12122065
Chicago/Turabian StyleZhong, Feng, Naling Bai, Xiangqian Chu, Yu He, Hanlin Zhang, and Haibo Li. 2022. "Effects of Lake Sediment on Soil Properties, Crop Growth, and the phoD-Harboring Microbial Community" Agriculture 12, no. 12: 2065. https://doi.org/10.3390/agriculture12122065
APA StyleZhong, F., Bai, N., Chu, X., He, Y., Zhang, H., & Li, H. (2022). Effects of Lake Sediment on Soil Properties, Crop Growth, and the phoD-Harboring Microbial Community. Agriculture, 12(12), 2065. https://doi.org/10.3390/agriculture12122065





