Analysis of Phylogeny and Genetic Diversity of Endangered Romanian Grey Steppe Cattle Breed, a Reservoir of Valuable Genes to Preserve Biodiversity
Abstract
1. Introduction
2. Materials and Methods
2.1. The Biological Material Studied
2.2. Blood Samples
2.3. Extraction and Quantification of Total DNA from Blood Samples
2.4. Primer, Amplification, Sequencing
2.5. Data Analysis
3. Results
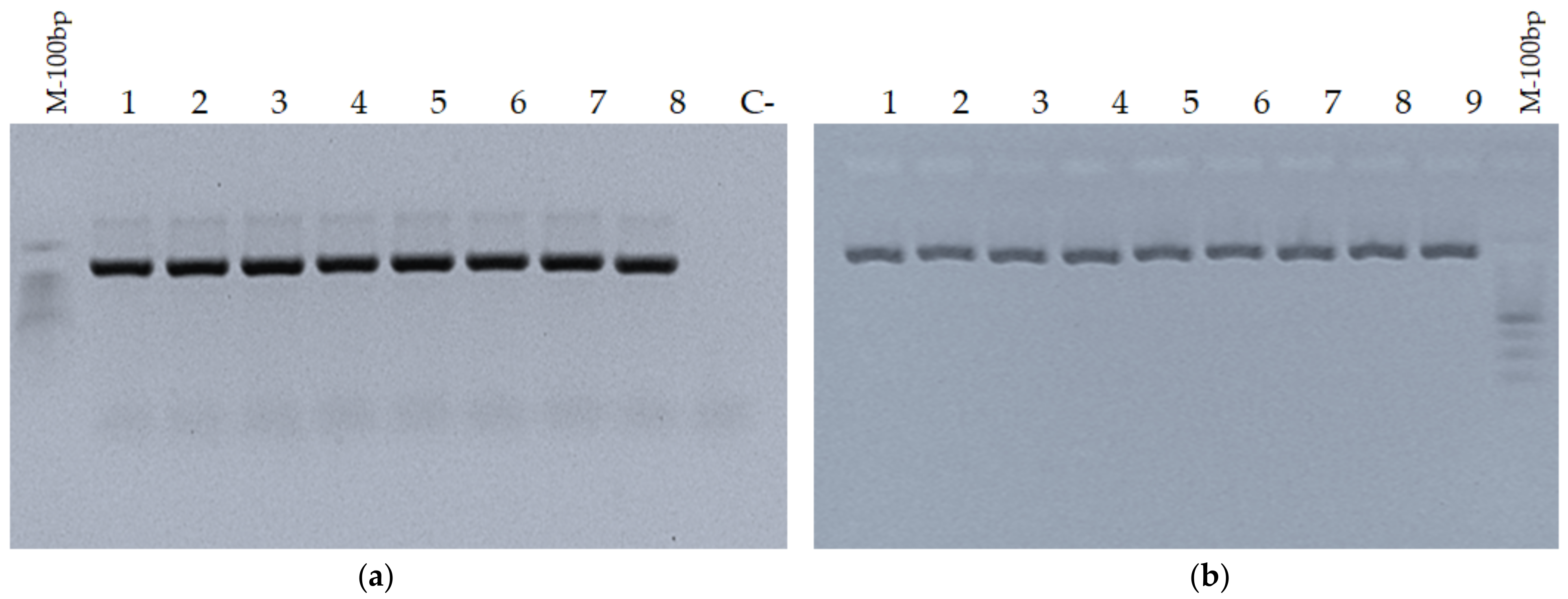
3.1. Validation of Amplification of PCR Products in Agarose Gel Electrophoresis
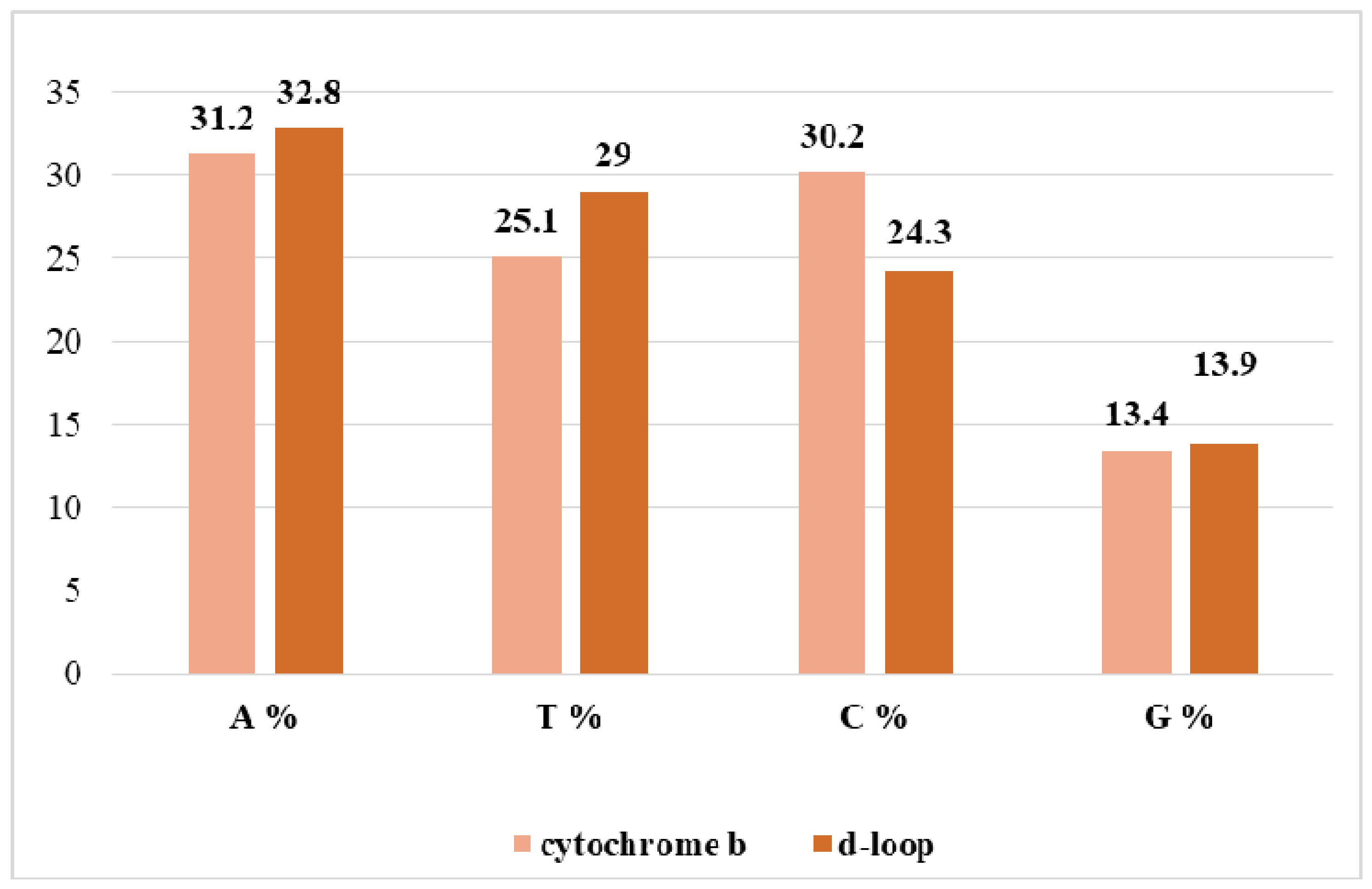
3.2. The Proportion of Nitrogenous Bases in the Nucleotide Sequences of Grey Steppe for Cytochrome b and d-Loop Mitochondrial Markers
3.3. The Specificity Coefficient
3.4. Dynamics of the Rate Evolution of Cytochrome b and d-Loop Mitochondrial Markers
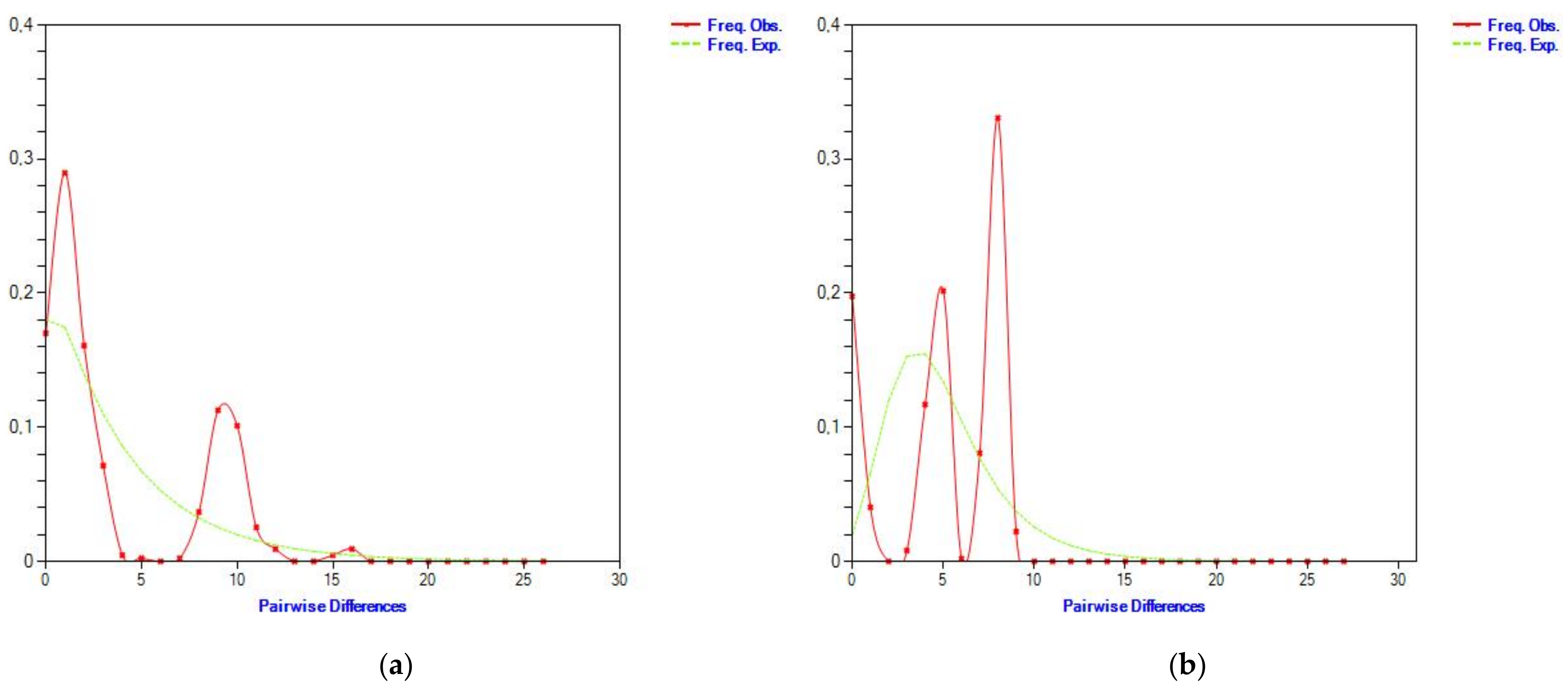
3.5. Haplotype Frequency Assessment by Analysis of Cytochrome b Gene Sequences and Mitochondrial d-Loop Control Region
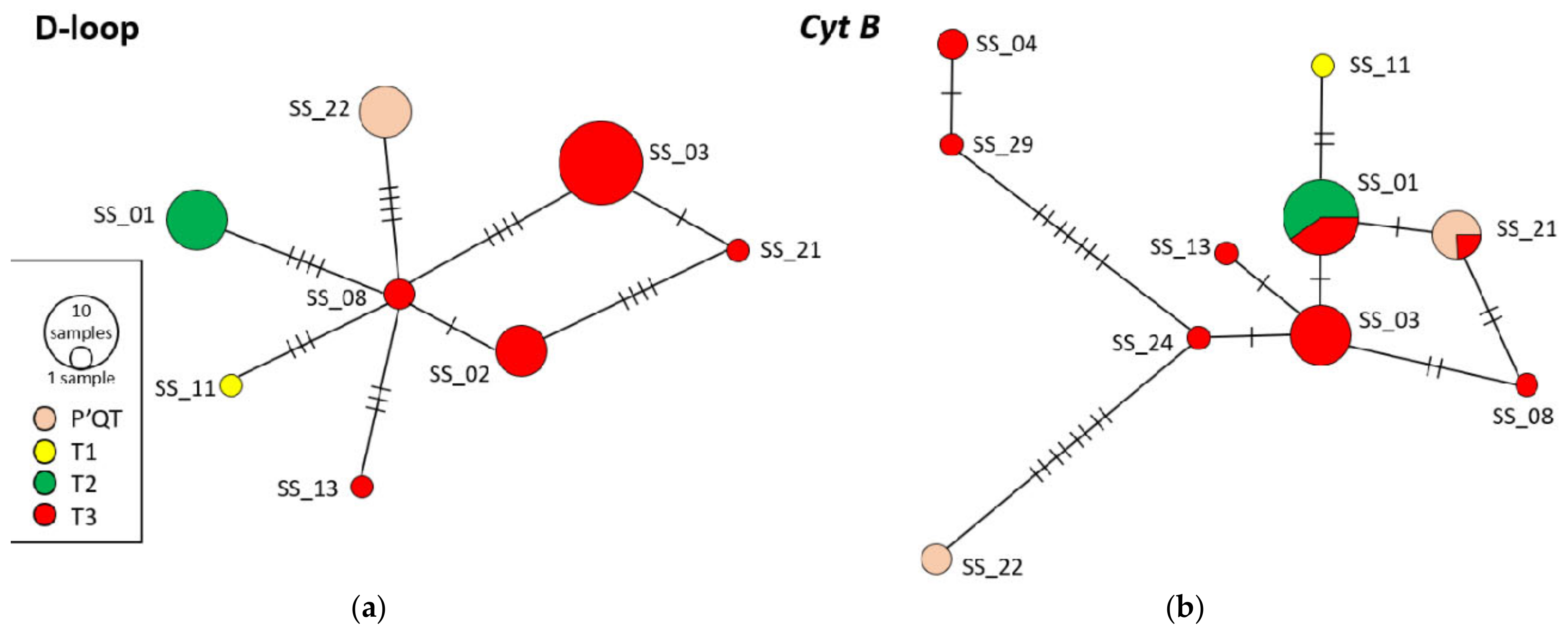
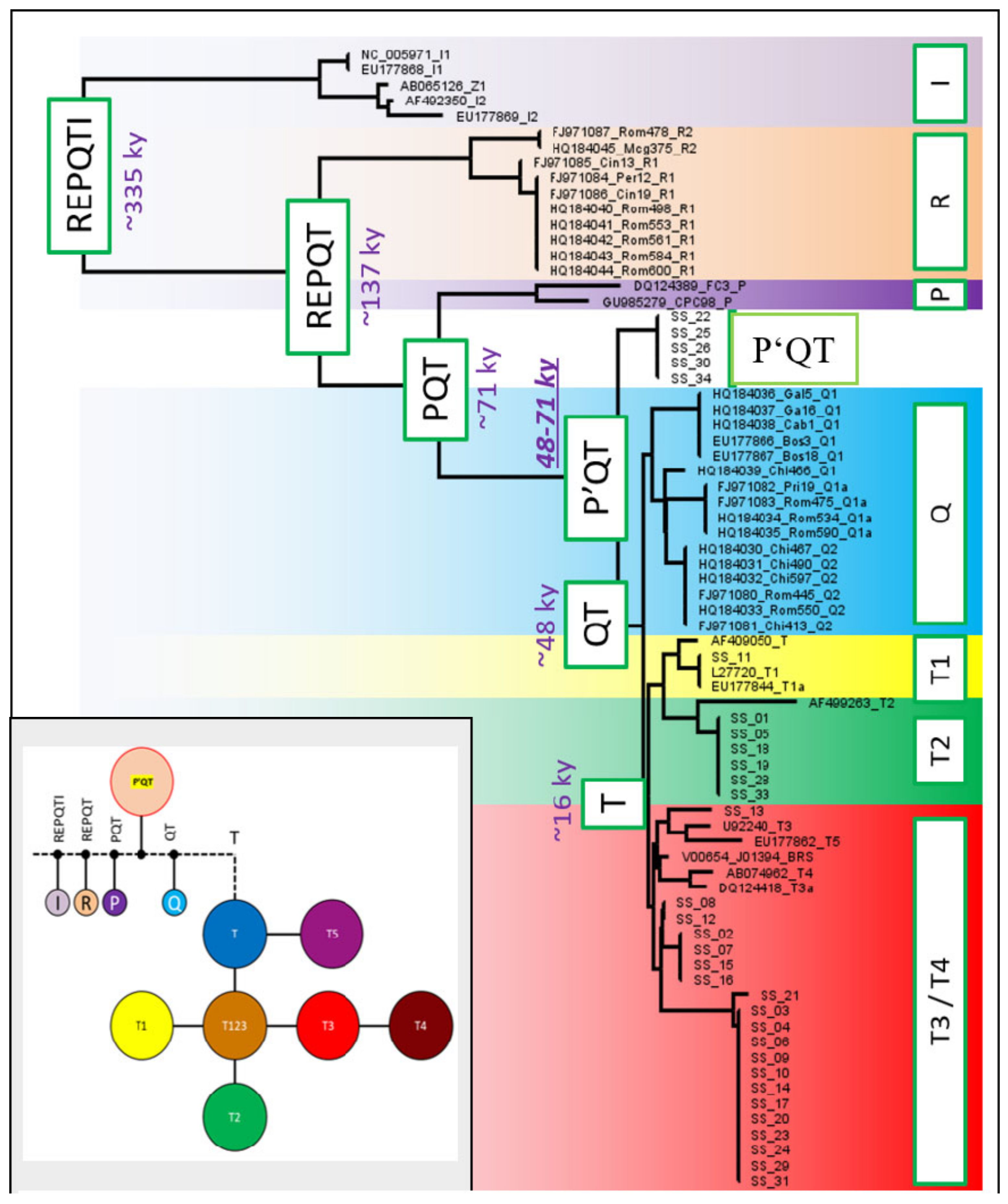
3.6. Haplotype Network Analysis and Phylogenetic Tree Construction
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Groeneveld, L.F.; Lenstra, J.A.; Eding, H.; Toro, M.A.; Scherf, B.; Pilling, D. Genetic diversity in farm animals—A review. Anim. Genet. 2010, 11, 6–31. [Google Scholar] [CrossRef]
- Pariset, L.; Mariotti, M.; Nardone, A.; Soysal, M.I.; Ozkan, E.; Williams, J.L.; Dunner, S.; Leveziel, H.; Maroti-Agots, A.; Bodo, I.; et al. Relationships between Podolic cattle breeds assessed by single nucleotide polymorphisms (SNPs) genotyping. J. Anim. Breed. Genet. 2010, 127, 481–488. [Google Scholar] [CrossRef]
- Ilie, D.E.; Cean, A.; Cziszter, L.T.; Gavojdian, D.; Ivan, A.; Kusza, S. Microsatellite and Mitochondrial DNA study of native Eastern European cattle populations: The case of the Romanian Grey. PLoS ONE 2015, 10, e0138736. [Google Scholar] [CrossRef]
- Scherf, B.D. World Watch List for Domestic Animal Diversity, 3rd ed.; FAO: Rome, Italy, 2000; Available online: http://www.fao.org/docrep/009/x8750e/x8750e00.htm (accessed on 23 May 2022).
- Creangă, Ș.; Dascălu, D.L.; Ariton, A.M. Conservation of Grey Steppe Cattle Breed from Romania in Order to Ensure the Biodiversity of the Genetic Resources of the Animal Populations; Pim: Iași, Romania, 2018; pp. 15–23. ISBN 20978-606-13-4638-7. [Google Scholar]
- Teneva, A.; Todorovska, E.; Tyufekchiev, N.; Kozelov, L.; Atanassov, A.; Foteva, S.; Ralcheva, S.; Zlatarev, S. Molecular characterization of Bulgarian livestock genetic resources: Genetic diversity in Bulgarian grey cattle as revealed by microsatellite markers. Biotechnol. Anim. Husb. 2005, 21, 35–42. [Google Scholar] [CrossRef]
- Creangă, Ş.; Maciuc, V.; Leonte, C. Genetical conservation for Romanian Grey Steppe cattle breed from SCDCB Dancu, Iasi county. Sci. Pap. Vet. Med. Ser. 2008, 51, 702–706. [Google Scholar]
- Georgescu, S.E.; Manea, M.A.; Zaulet, M.; Costache, M. Genetic diversity among Romanian cattle breeds with a special focus on the Romanian Grey Steppe Breed. Rom. Biotechnol. Lett. 2008, 14, 4194–4200. [Google Scholar]
- Creangă, Ș.; Chelmu, S.S.; Maciuc, V.; Bâlteanu, V.A. Molecular characterization of Grey Steppe cattle breed for its genetic biopreservation. Rom. Biotechnol. Lett. 2013, 18, 8893–8900. [Google Scholar]
- Maciuc, V.; Radu-Rusu, R.M. Assessment of Grey Steppe cattle genetic and phenotypic traits as valuable resources in preserving biodiversity. Environ. Eng. Manag. J. 2018, 17, 2741–2748. [Google Scholar]
- Maciuc, V. Cattle Breeding Management; Alfa: Iași, Romania, 2006; pp. 76–78. ISBN 973-8953-18-9. [Google Scholar]
- Creangă, Ș.; Maciuc, V. Grey Steppe Cattle Breed from Romania; Alfa: Iași, Romania, 2010; pp. 47–58. ISBN 978-606-540-034-4. [Google Scholar]
- Dascălu, D.L.; Creangă, Ș.; Borș, S.I.; Doliș, M.G.; Simeanu, D.; Donosă, R. The Romanian Grey Steppe characterisation—The breed evolution in the last century from the numerical and morphological point of view. Danub. Anim. Genet. Resour. 2017, 2, 70–75. [Google Scholar]
- Davidescu, M.A.; Ciorpac, M.; Madescu, B.M.; Porosnicu, I.; Creangă, Ș. Analysis of the Genetic diversity of endangered cattle breeds based on studies of genetic markers. Sci. Pap. Anim. Sci. Biotechnol. 2021, 54, 60–63. [Google Scholar]
- Davidescu, M.A.; Grădinaru, A.C.; Creangă, Ș. Endangered romanian cattle breeds–between traditional breeding and genetic conservation. Sci. Pap.-Anim. Sci. Ser. Sci. Pap.-Anim. Husb. Ser. 2021, 75, 66–75. [Google Scholar]
- Grădinaru, A.C.; Petrescu-Mag, I.V.; Oroian, F.C.; Balint, C.; Oltean, I. Milk Protein Polymorphism Characterization: A Modern Tool for Sustainable Conservation of Endangered Romanian Cattle Breeds in the Context of Traditional Breeding. Sustainability 2018, 10, 534. [Google Scholar] [CrossRef]
- Bollongino, R.; Edwards, C.J.; Alt, K.W.; Burger, J.; Bradley, D.G. Early history of European domestic cattle as revealed by ancient DNA. Biol. Lett. 2006, 2, 155–159. [Google Scholar] [CrossRef] [PubMed]
- Bollongino, R.; Elsner, J.; Vigne, J.D.; Burger, J. Y-SNPs do not indicate hybridisation between European aurochs and domestic cattle. PLoS ONE 2008, 3, e3418. [Google Scholar] [CrossRef]
- Achilli, A.; Bonfiglio, S.; Olivieri, A.; Malusà, A.; Pala, M. The multifaceted origin of taurine cattle reflected by the mitochondrial genome. PLoS ONE 2009, 4, e5753. [Google Scholar] [CrossRef] [PubMed]
- Sinding, M.H.S.; Gilbert, M.T.P. The draft genome of extinct European Aurochs and its implications for de-extinction. Open Quat. 2016, 2, 7. [Google Scholar] [CrossRef]
- Senczuk, G.; Mastrangelo, S.; Ajmone-Marsan, P.; Becskei, Z.; Colangelo, P.; Colli, L.; Ferretti, L.; Karsli, T.; Lancioni, H.; Lasagna, E.; et al. On the origin and diversification of Podolian cattle breeds: Testing scenarios of European colonization using genome-wide SNP data. Genet. Sel. 2021, 53, 48. [Google Scholar] [CrossRef]
- Dascălu, D.L. Conservation of the Romanian Grey Steppe Breed. Ph.D. Thesis, Iasi University of Life Sciences, Iași, Romania, 2014. [Google Scholar]
- PCR Master Mix—Promega Corporation. PCR Master Mix. Available online: https://www.promega.ro/products/pcr/taq-polymerase/master-mix-pcr/?catNum=M7502 (accessed on 12 May 2021).
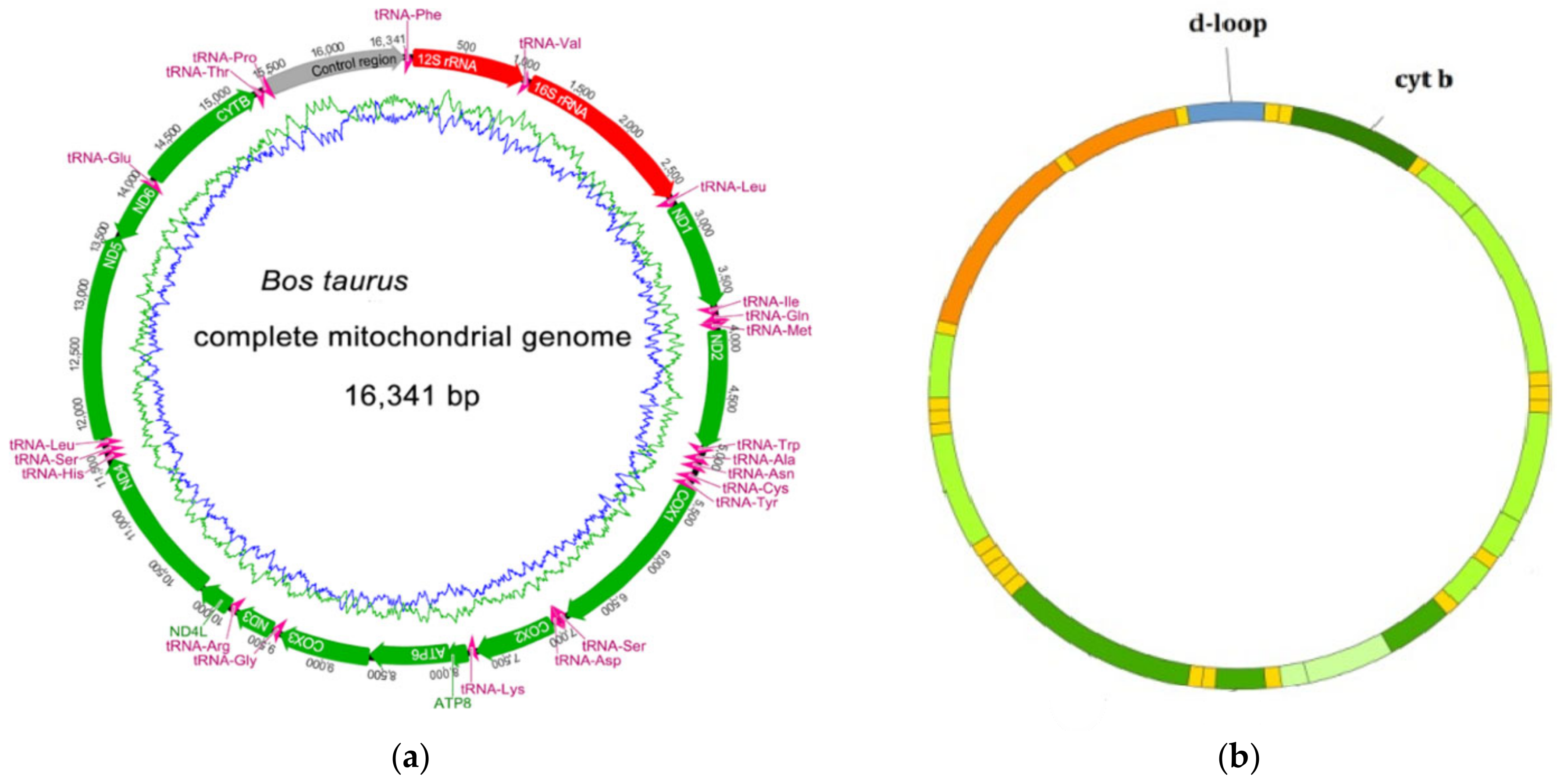
- GenBank Overview-NCBI. Bos taurus Complete Mitochondrial Genome. Available online: https://www.ncbi.nlm.nih.gov/nuccore/V00654.1 (accessed on 15 September 2021).
- Seroussi, E.; Yakobson, E. Bovine mtDNA D-loop haplotypes exceed mutations in number despite reduced recombination: An effective alternative for identity control. Animal 2010, 4, 1818–1822. [Google Scholar] [CrossRef]
- Wang, X.; Zhang, Y.Q.; He, D.C.; Yang, X.M.; Li, B.; Wang, D.C.; Guang, J.; Xu, F.; Li, J.Y.; Gao, X.; et al. The complete mitochondrial genome of Bos taurus coreanae (Korean native cattle). Mitochondrial DNA A DNA Mapp. Seq. Anal. 2016, 27, 120–121. [Google Scholar] [CrossRef]
- Liu, S.J.; Lv, J.Z.; Tan, Z.Y.; Ge, X.Y. The complete mitochondrial genome of Uruguayan native cattle (Bos taurus). Mitochondrial DNA B Resour. 2020, 5, 443–444. [Google Scholar] [CrossRef]
- Arbizu, C.I.; Ferro-Mauricio, R.D.; Chávez-Galarza, J.C.; Vásquez, H.V.; Maicelo, J.L.; Poemape, C.; Gonzales, J.; Quilcate, C.; Corredor, F.-A. The Complete Mitochondrial Genome of a Neglected Breed, the Peruvian Creole Cattle (Bos taurus), and Its Phylogenetic Analysis. Data 2022, 7, 76. [Google Scholar] [CrossRef]
- Guo, X.; Pei, J.; Xiong, L.; Bao, P.; Zhu, Y.; Wangdui, B.; Wu, X.; Chu, M.; Yan, P.; Ding, X. The complete mitochondrial genome of Shigaste humped cattle (Bos taurus). Conserv. Genet. Resour. 2017, 10, 789–790. [Google Scholar] [CrossRef]
- Thompson, J.D.; Higgins, D.G.; Gibson, T.J. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994, 22, 4673–4680. [Google Scholar] [CrossRef] [PubMed]
- Rozas, J.; Sánchez-DelBarrio, J.C.; Messeguer, X.; Rozas, R. DnaSP, DNA polymorphism analyses by the coalescent and other methods. Bioinformatics 2003, 19, 2496–2497. [Google Scholar] [CrossRef] [PubMed]
- Tarekegn, G.M.; Ji, X.Y.; Bai, X.; Liu, B.; Zhang, W.; Birungi, J.; Djikeng, A.; Tesfaye, K. Variations in mitochondrial cytochrome b region among Ethiopian indigenous cattle populations assert Bos taurus maternal origin and historical dynamics. Asian-Australas. J. Anim. Sci. 2018, 31, 1393–1400. [Google Scholar] [CrossRef]
- Yang, S.L.; Lin, R.Y.; Xu, L.X.; Cheng, L. Analysis of polymorphisms of mitochondrial DNA D-loop and Mc1R gene in Chinese Wuchuan Black cattle. J. Appl. Anim. Res. 2014, 42, 487–491. [Google Scholar] [CrossRef]
- Darriba, D.; Taboada, G.L.; Doallo, R.; Posada, D. jModelTest 2: More models, new heuristics and parallel computing. Nature Methods 2012, 9, 772. [Google Scholar] [CrossRef]
- Akaike, H. Anew look at the statistical model identification. IEEE Trans. Autom. Control 1974, 19, 716–723. [Google Scholar] [CrossRef]
- Achilli, A.; Olivieri, A.; Pellecchia, M.; Uboldi, C.; Colli, L. Mitochondrial genomes of extinct aurochs survive in domestic cattle. Curr. Biol. 2008, 18, 157–158. [Google Scholar] [CrossRef]
- Bonfiglio, S.; Ginja, C.; De Gaetano, A.; Achilli, A.; Olivieri, A.; Colli, L.; Tesfaye, K.; Agha, S.H.; Gama, L.T.; Cattonaro, F.; et al. Origin and spread of Bos taurus: New clues from mitochondrial genomes belonging to haplogroup T1. PLoS ONE 2012, 7, e38601. [Google Scholar] [CrossRef]
- Di Lorenzo, P.; Lancioni, H.; Ceccobelli, S.; Colli, L.; Cardinali, I.; Karsli, T.; Capodiferro, M.R.; Sahin, E.; Ferretti, L.; Marsan, P.A.; et al. Mitochondrial DNA variants of Podolian cattle breeds testify for a dual maternal origin. PLoS ONE 2018, 13, e0192567. [Google Scholar] [CrossRef]
- Olivieri, A.; Gandini, F.; Achilli, A.; Fichera, A.; Rizzi, E.; Bonfiglio, S.; Battaglia, V.; Brandini, S.; De-Gaetano, A.; El-Beltagi, A.; et al. Mitogenomes fromEgyptian Cattle Breeds: New Clues on the Origin of Haplogroup Q and the Early Spread of Bos taurus from the Near East. PLoS ONE 2015, 10, e0141170. [Google Scholar] [CrossRef] [PubMed]
- Park, S.D.E.; Magee, D.A.; McGettigan, P.A.; Teasdale, M.D.; Edwards, C.J.; Lohan, A.J.; Murphy, A.; Braud, M.; Donoghue, M.T.; Liu, Y.; et al. Genome sequencing of the extinct Eurasian wild aurochs, Bos primigenius, illuminates the phylogeography and evolution of cattle. Genome Biol. 2015, 16, 234. [Google Scholar] [CrossRef]
- Cubric-Curik, V.; Novosel, D.; Brajkovic, V.; Rota Stabelli, O.; Krebs, S.; Sölkner, J.; Šalamon, D.; Ristov, S.; Berger, B.; Trivizaki, S.; et al. Large-scale mitogenome sequencing reveals consecutive expansions of domestic taurine cattle and supports sporadic aurochs introgression. Evol. Appl. 2022, 15, 663–678. [Google Scholar] [CrossRef] [PubMed]
- Dorji, J.; Vander-Jagt, C.; Chamberlain, A.; Cocks, B.; MacLeod, I.; Daetwyler, H. Cattle Maternal Diversity Inferred from 1,883 Taurine and Indicine Mitogenomes. Res. Sq. 2021. [Google Scholar] [CrossRef]
- Dorji, J.; Vander Jagt, C.J.; Chamberlain, A.J.; Cocks, B.G.; MacLeod, I.M.; Daetwyler, H.D. Recovery of mitogenomes from whole genome sequences to infer maternal diversity in 1883 modern taurine and indicine cattle. Sci. Rep. 2022, 12, 5582. [Google Scholar] [CrossRef]
- Liu, H.; Zhai, J.; Wu, H.; Wang, J.; Zhang, S.; Li, J.; Niu, Z.; Shen, C.; Zhang, K.; Liu, Z.; et al. Diversity of Mitochondrial DNA Haplogroups and Their Association with Bovine Antral Follicle Count. Animals 2022, 12, 2350. [Google Scholar] [CrossRef]
- Edwards, J.; Bollongino, R.; Scheu, A.; Chamberlain, A.; Tresset, A.; Vigne, J.D.; Baird, J.F.; Larson, G.; Ho, S.Y.; Heupink, T.H. Mitochondrial DNA analysis shows a Near Eastern Neolithic origin for domestic cattle and no indication of domestication of European aurochs. Proc. Biol. Sci. 2007, 274, 1377–1385. [Google Scholar] [CrossRef]
- Stock, F.; Edwards, C.J.; Bollongino, R.; Finlay, E.K.; Burger, J.; Bradley, D.G. Cytochrome b sequences of ancient cattle and wild ox support phylogenetic complexity in the ancient and modern bovine populations. Anim. Genet. 2009, 40, 694–700. [Google Scholar] [CrossRef]
- Magee, D.A.; Hugh, D.E.; Ceiridwen, J.E. Interrogation of modern and ancient genomes reveals the complex domestic history of cattle. Anim. Front. 2014, 4, 7–22. [Google Scholar] [CrossRef]
- Maretto, F.; Ramljak, J.; Sbarra, F.; Penasa, M.; Mantovani, R.; Ivankovic, A.; Bittantea, G. Genetic relationship among Italian and Croatian Podolian cattle breeds assessed by microsatellite markers. Livest. Sci. 2012, 150, 256–264. [Google Scholar] [CrossRef]
- Felius, M.; Beerling, M.-L.; Buchanan, D.S.; Theunissen, B.; Koolmees, P.A.; Lenstra, J.A. On the History of Cattle Genetic Resources. Diversity 2014, 6, 705–750. [Google Scholar] [CrossRef]
- Pitt, D.; Sevane, N.; Nicolazzi, E.L.; Hugh, D.E.; Park, S.D.E.; Colli, L.; Martinez, R.; Bruford, M.W.; Orozco-terWenge, P. Domestication of cattle: Two or three events? Evol. Appl. 2019, 12, 123–136. [Google Scholar] [CrossRef] [PubMed]
- Verdugo, M.P.; Mullin, V.E.; Scheu, A.; Mattiangeli, V.; Daly, K.G.; Maisano Delser, P.; Hare, A.J.; Burger, J.; Collins, M.J.; Kehati, R.; et al. Ancient cattle genomics, origins, and rapid turnover in the Fertile Crescent. Science 2019, 365, 173–176. [Google Scholar] [CrossRef]
- Loftus, R.T.; MacHugh, D.E.; Bradley, D.G.; Sharp, P.M.; Cunningham, E.P. Evidence for two independent domestications of cattle. Proc. Natl. Acad. Sci. USA 1994, 91, 2757–2761. [Google Scholar] [CrossRef]
- Upadhyay, M.R.; Chen, W.; Lenstra, J.A.; Goderie, C.R.; MacHugh, D.E.; Park, S.D.; Magee, D.A.; Matassino, D.; Ciani, F.; Megens, H.J.; et al. European Cattle Genetic Diversity Consortium & RPMA Crooijmans Genetic origin, admixture and population history of aurochs (Bos primigenius) and primitive European cattle. Heredity 2017, 118, 169–176. [Google Scholar] [CrossRef]
- Ivankovic, A.; Paprika, S.; Ramljak, J.; Dovc, P.; Konjacic, M. Mitochondrial DNA-based genetic evaluation of autochthonous cattle breeds in Croatia. Czech J. Anim. Sci. 2014, 59, 519–528. [Google Scholar] [CrossRef]
- Hristov, P.; Teofanova, D.; Neov, B.; Radoslavov, G. Haplotype diversity in autochthonous Balkan cattle breeds. Anim. Genet. 2014, 46, 92–94. [Google Scholar] [CrossRef]
- Kantanen, J.; Edwards, C.J.; Bradley, D.G.; Viinalass, H.; Thessler, S.; Ivanova, Z.; Kiselyova, T.; Ćinkulov, M.; Popov, R.; Stojanović, S.; et al. Maternal and paternal genealogy of Eurasian taurine cattle (Bos taurus). Heredity 2009, 103, 404–415. [Google Scholar] [CrossRef]

| Current No. | Cattle Identification No. | Age of Cattle (Months) | Sex | Blood Sample Identification Code 1 |
|---|---|---|---|---|
| 1. | RO242000109988 | 288 | female | blood-ss01 |
| 2. | RO242000110002 | 242 | female | blood-ss02 |
| 3. | RO245000109998 | 228 | female | blood-ss03 |
| 4. | RO241000109812 | 202 | female | blood-ss04 |
| 5. | RO247000109811 | 193 | female | blood-ss05 |
| 6. | RO243000109786 | 187 | female | blood-ss06 |
| 7. | RO243000109800 | 186 | female | blood-ss07 |
| 8. | RO242000110205 | 185 | female | blood-ss08 |
| 9. | RO243000109723 | 185 | female | blood-ss09 |
| 10. | RO241000110120 | 184 | female | blood-ss10 |
| 11. | RO241000110274 | 178 | female | blood-ss11 |
| 12. | RO242000120579 | 175 | female | blood-ss12 |
| 13. | RO243000120587 | 175 | female | blood-ss13 |
| 14. | RO242000120656 | 171 | female | blood-ss14 |
| 15. | RO243000120678 | 171 | female | blood-ss15 |
| 16. | RO243000174164 | 152 | female | blood-ss16 |
| 17. | RO242000218328 | 144 | female | blood-ss17 |
| 18. | RO243000218587 | 136 | female | blood-ss18 |
| 19. | RO143000108432 | 128 | female | blood-ss19 |
| 20. | RO248001058693 | 111 | female | blood-ss20 |
| 21. | RO507000112314 | 109 | female | blood-ss21 |
| 22. | RO506001166289 | 97 | female | blood-ss22 |
| 23. | RO504001166308 | 96 | female | blood-ss23 |
| 24. | RO508003831422 | 69 | female | blood-ss24 |
| 25. | RO502003831482 | 68 | female | blood-ss25 |
| 26. | RO504005500784 | 61 | female | blood-ss26 |
| 27. | RO501007300790 | 39 | female | blood-ss27 |
| 28. | RO508007300858 | 37 | female | blood-ss28 |
| 29. | RO507007405358 | 29 | female | blood-ss29 |
| 30. | RO504007405388 | 28 | female | blood-ss30 |
| 31. | RO507007448267 | 27 | female | blood-ss31 |
| 32. | RO500007448406 | 22 | female | blood-ss32 |
| Primer Set | Primer Specificity | Sequenced (5′-3′) | Content G 1 + C 2 (%) | GenBank Accession no./Position in Genome [23] | Length of PCR Product (bp) |
|---|---|---|---|---|---|
| BCYT | cytochrome b | Forward: TTCTTACATGGAATCTAACCATGA | 33.3 | V00654.1 14,443–14,466 | 1140 cytochrome b |
| cytochrome b | Reverse: GGGAGGTTAGTTGTTCTCCTTCTC | 50.0 | V00654.1 473–497 | ||
| BRS | d-loop | Forward: CCTAAGACTCAAGGAAGAAACTGC | 45.8 | V00654.1 15,718–15,741 | 910 d-loop |
| d-loop | Reverse: CAGTGAGAATGCCCTCTAGGTT | 50.0 | V00654.1 496–517 |
| Stages * | Cytochrome b Amplification Conditions | d-Loop Amplification Conditions | ||
|---|---|---|---|---|
| Temperature | Time | Temperature | Time | |
| 1. Denaturing | 94 °C | 2 min | 94 °C | 2 min |
| 2. Annealing | 62 °C | 30 s | 60 °C | 30 s |
| 3. Extending | 72 °C | 5 min | 72 °C | 5 min |
| Data Analysis | Program Used |
|---|---|
Amplicon sequencing
|  |
Alignment of chromatograms and correction of raw sequences
|  |
Sequence alignment
|  |
Calculation of the optimal substitution model
|  |
Haplotype network construction
|  |
Construction of phylogenetic trees
|  |
Nucleotide sequence diversity analysis
|  |
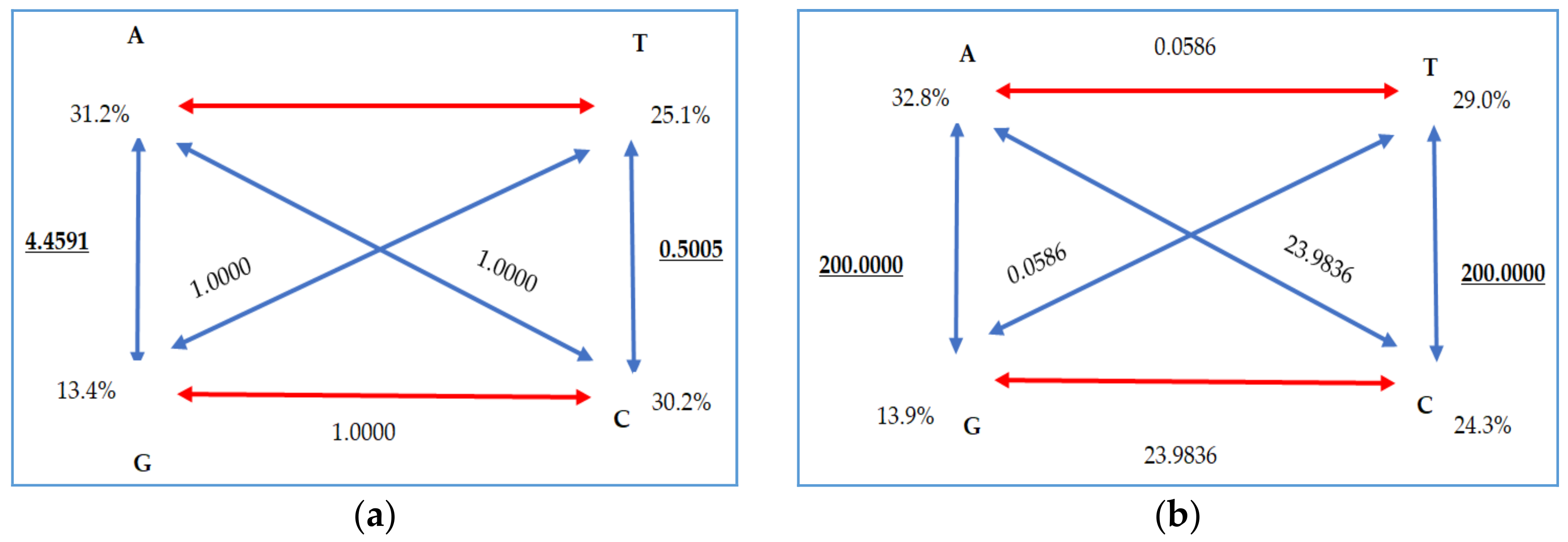
| Cytochrome b | d-Loop | |||||
|---|---|---|---|---|---|---|
| Model | TrN+I 6 | TPM3uf | ||||
| Partition | 010020 | 012012 | ||||
| -lnL 1 | 1708.1951 | 1354.6687 | ||||
| K 2 | 64 | 67 | ||||
| freqA 3 | 0.3125 | R(a) | 1.0000 | 0.3278 | R(a) | 23.9836 |
| freqC 3 | 0.3021 | R(b) | 4.4591 | 0.2440 | R(b) | 200.0000 |
| freqG 3 | 0.1352 | R(c) | 1.0000 | 0.1379 | R(c) | 0.0586 |
| cytochrome b | d-loop | |||||
| freqT 3 | 0.2501 | R(d) | 1.0000 | 0.2902 | R(d) | 23.9836 |
| ti/tv 4 | - | R(e) | 0.5005 | - | R(e) | 200.0000 |
| - | R(f) | 1.0000 | - | R(f) | 0.0586 | |
| p-inv 5 | 0.8680 | gamma | - | gamma | - | |
| Sequence | No. of Informative Sites | % | No. of Variable Sites | % | Ti/Tv 1 |
|---|---|---|---|---|---|
| cytochrome b | 18 | 1.6 | 22 | 1.9 | 0.755 |
| d-loop | 13 | 1.4 | 18 | 2.0 | 5.107 |
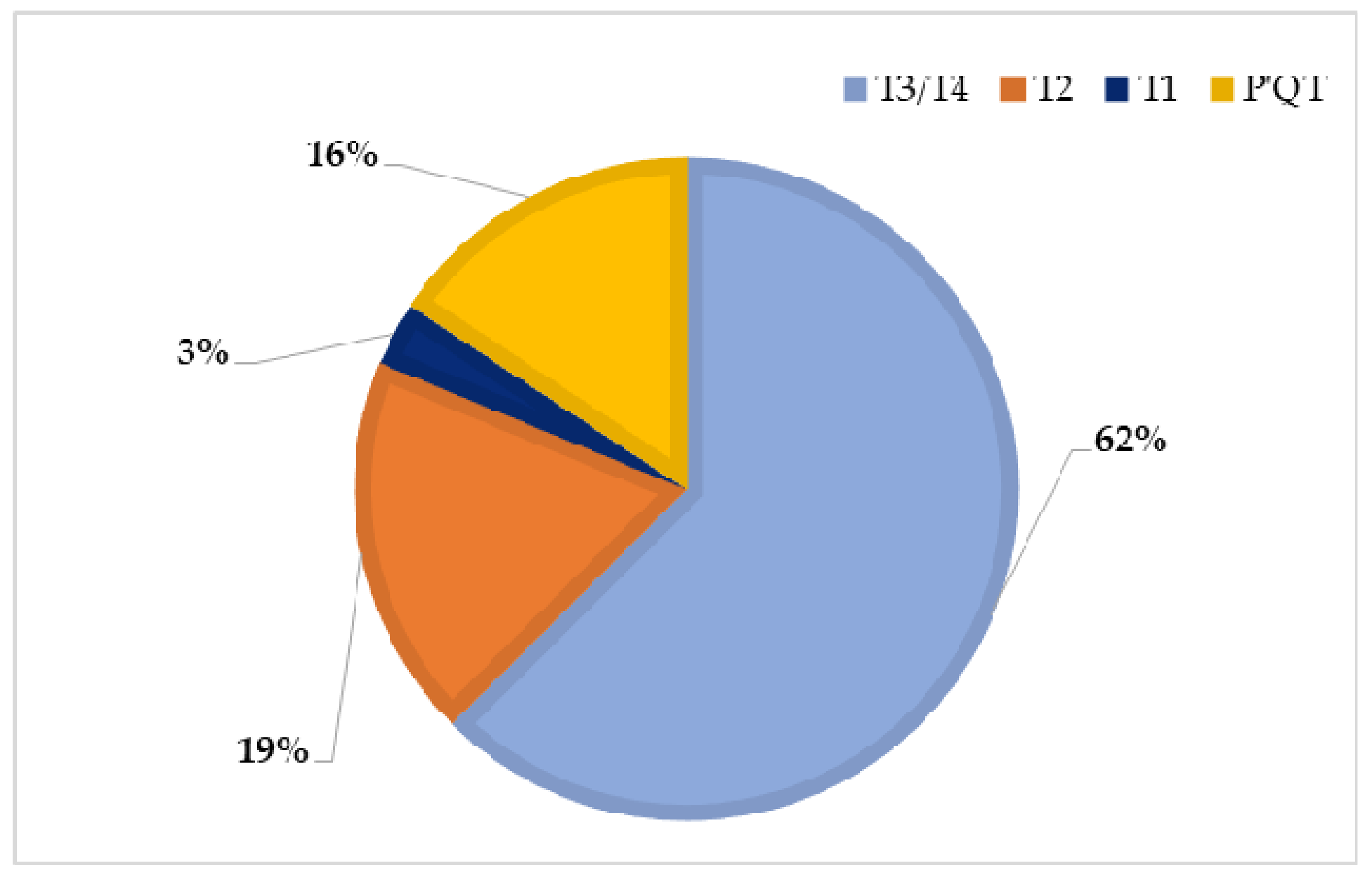
| Haplotypes Identified | Representative Individuals | Total Individuals/Haplotype |
|---|---|---|
| T3/T4 | SS_02; SS_03; SS_04; SS_06; SS_07; SS_08; SS_09; SS_10; SS_12; SS_13; SS_14; SS_15; SS_16; SS_17; SS_20; SS_21; SS_23; SS_24; SS_29; SS_31 | 20 |
| T2 | SS_01; SS_05; SS_18; SS_19; SS_28; SS_33 | 6 |
| T1 | SS_11 | 1 |
| P′QT | SS_22; SS_25; SS_26; SS_30; SS_34 | 5 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Davidescu, M.-A.; Simeanu, D.; Gorgan, D.-L.; Ciorpac, M.; Creanga, S. Analysis of Phylogeny and Genetic Diversity of Endangered Romanian Grey Steppe Cattle Breed, a Reservoir of Valuable Genes to Preserve Biodiversity. Agriculture 2022, 12, 2059. https://doi.org/10.3390/agriculture12122059
Davidescu M-A, Simeanu D, Gorgan D-L, Ciorpac M, Creanga S. Analysis of Phylogeny and Genetic Diversity of Endangered Romanian Grey Steppe Cattle Breed, a Reservoir of Valuable Genes to Preserve Biodiversity. Agriculture. 2022; 12(12):2059. https://doi.org/10.3390/agriculture12122059
Chicago/Turabian StyleDavidescu, Madalina-Alexandra, Daniel Simeanu, Dragos-Lucian Gorgan, Mitica Ciorpac, and Steofil Creanga. 2022. "Analysis of Phylogeny and Genetic Diversity of Endangered Romanian Grey Steppe Cattle Breed, a Reservoir of Valuable Genes to Preserve Biodiversity" Agriculture 12, no. 12: 2059. https://doi.org/10.3390/agriculture12122059
APA StyleDavidescu, M.-A., Simeanu, D., Gorgan, D.-L., Ciorpac, M., & Creanga, S. (2022). Analysis of Phylogeny and Genetic Diversity of Endangered Romanian Grey Steppe Cattle Breed, a Reservoir of Valuable Genes to Preserve Biodiversity. Agriculture, 12(12), 2059. https://doi.org/10.3390/agriculture12122059







