Evaluating Data Augmentation Effects on the Recognition of Sugarcane Leaf Spot
Abstract
1. Introduction
2. Materials and Methods
2.1. Research Roadmap
2.2. Dataset
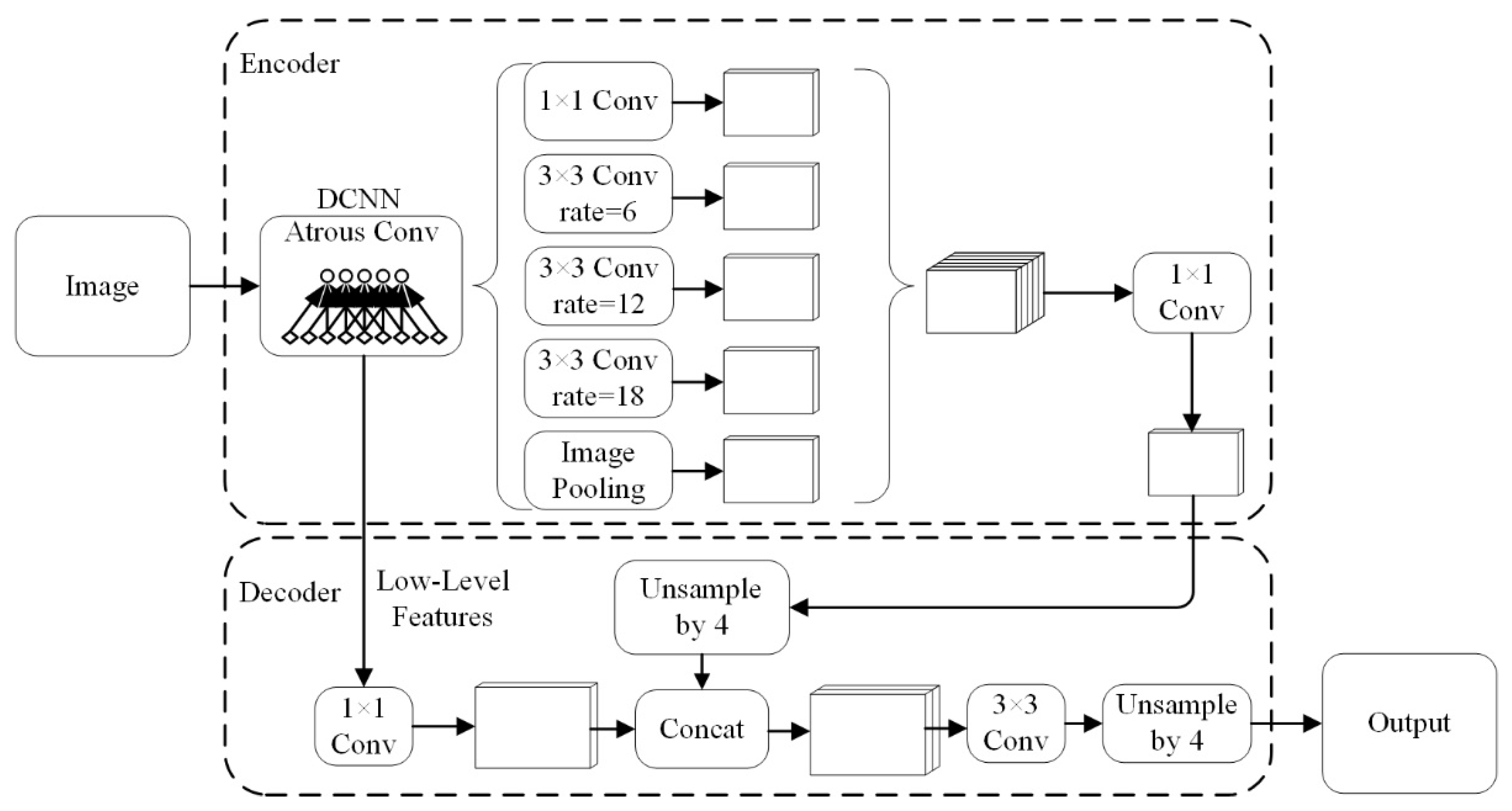
2.3. DeepLabV3+
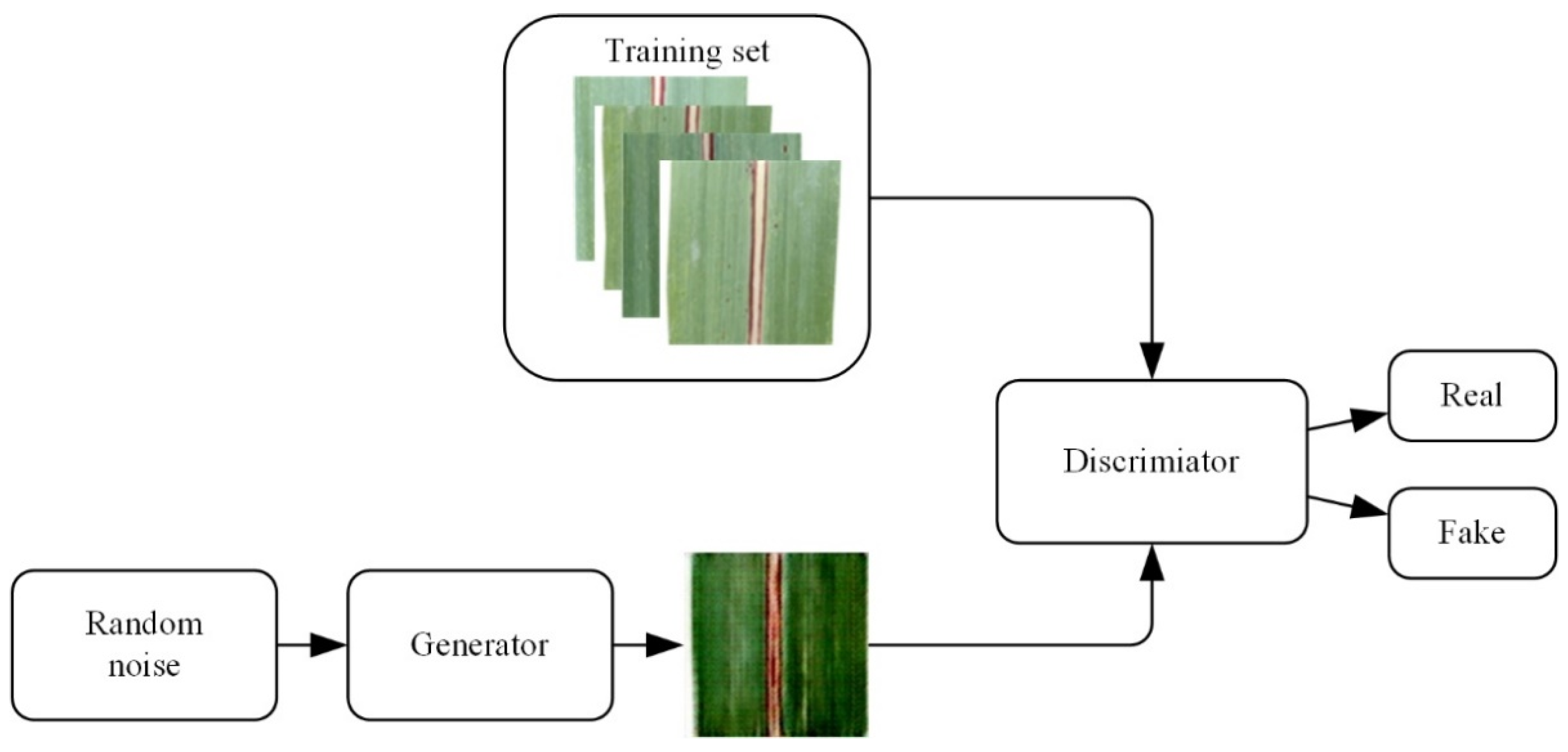
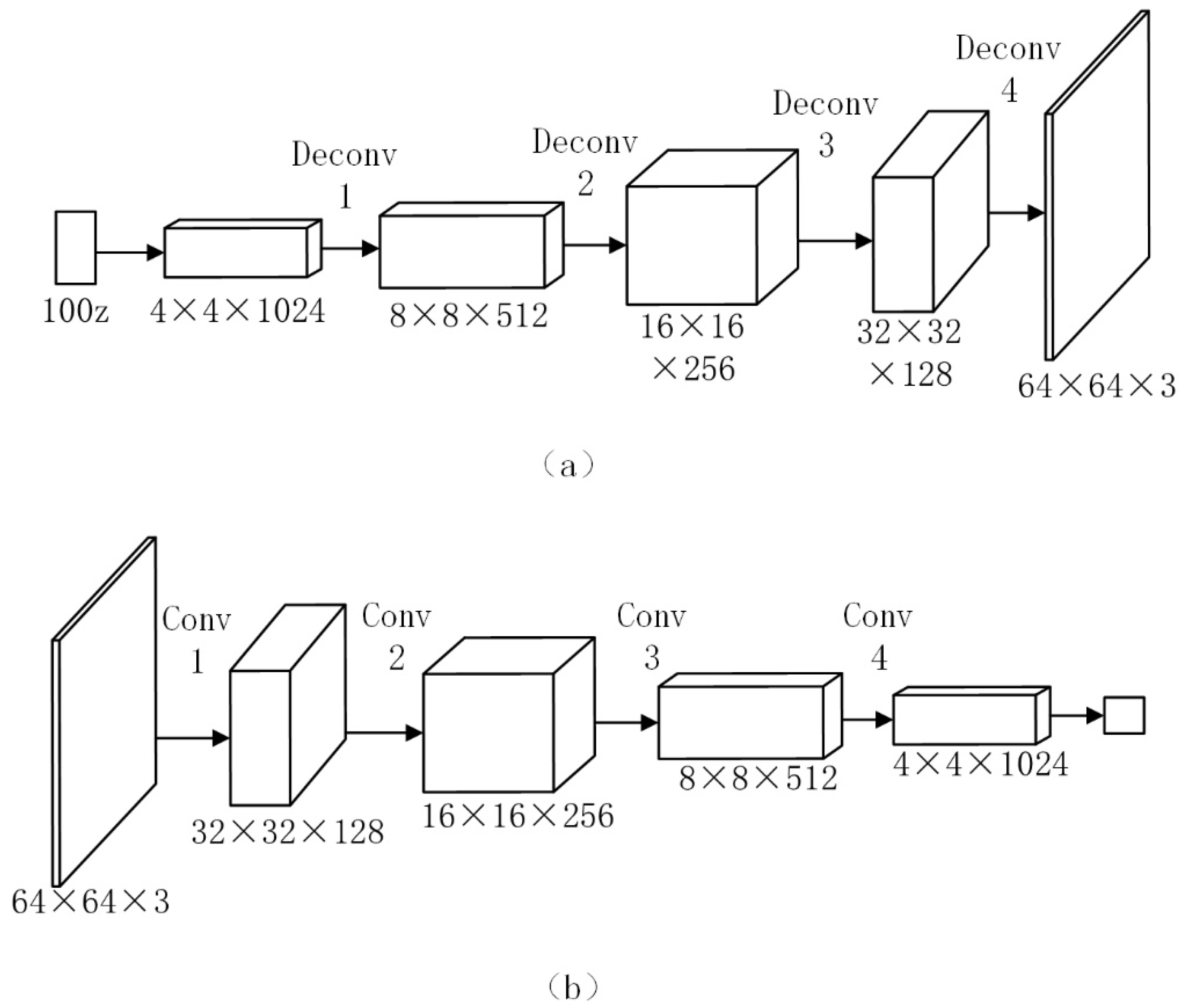
2.4. DCGAN
2.5. Image Recognition Network
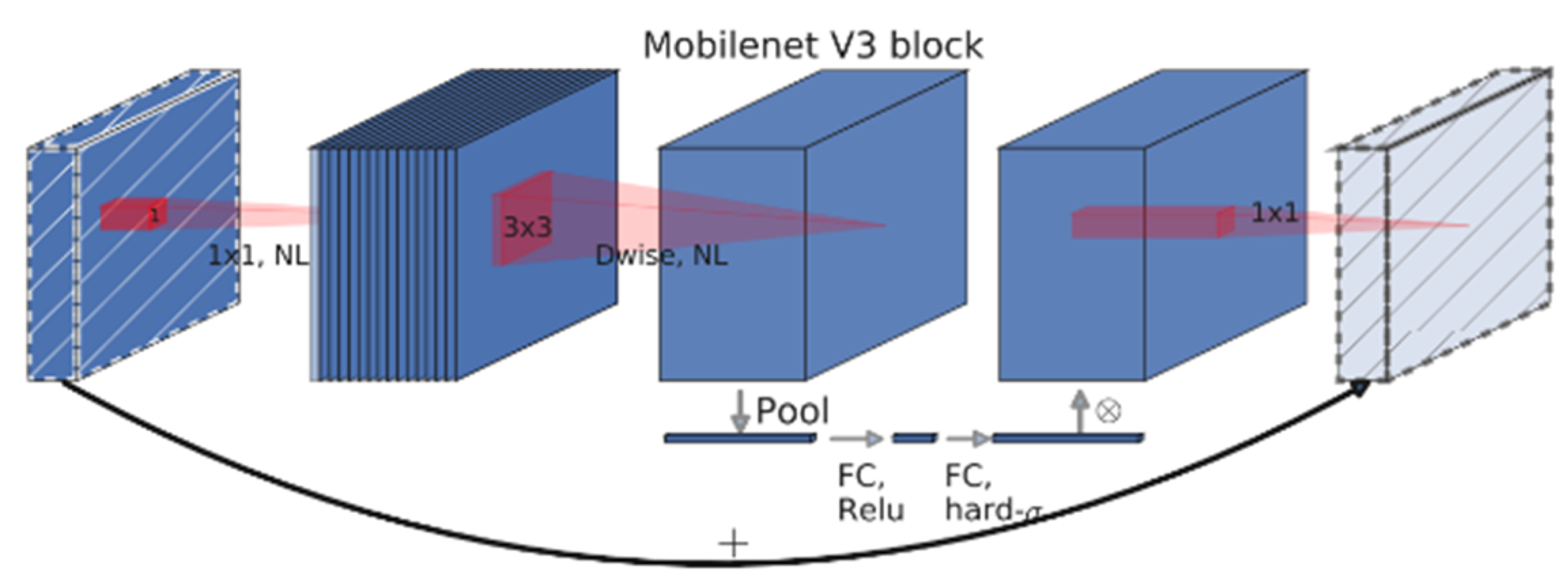
2.5.1. MobileNetV3-Large
2.5.2. AlexNet
2.5.3. ResNet
2.5.4. DenseNet
2.6. Evaluation Metric
3. Results
3.1. Image Segmentation Based on DeepLabV3+
3.2. Supervised Data Augmentation
3.3. Data Augmentation Based on DCGAN
3.4. Image Recognition Based on MobileNetV3-Large, Alexnet, Resnet, and Densenet
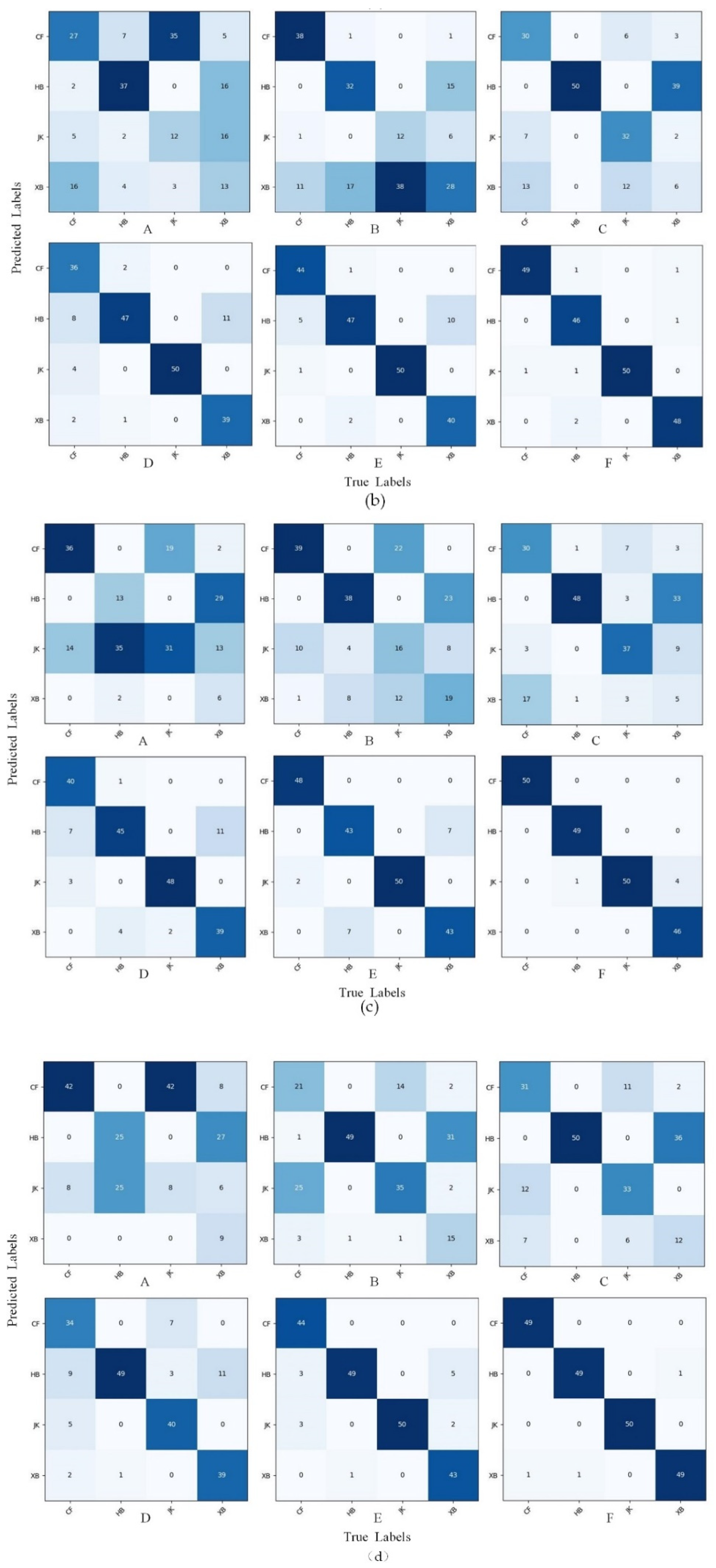
4. Data Analysis and Discussion
4.1. Data Analysis
4.2. Discussion
- (1)
- When using MobileNetV3-large to identify disease on sugarcane leaves, the accuracy of the validation set was only 53.50% when the training set had a complex background and the amount of data was small, and the accuracy of the validation set was increased from 53.50% to 87.50% by removing the complex background of the training set by DeepLabV3+. Then, the accuracy of the validation set was further increased from 87.50% to 99.00% by DCGAN data augmentation. These two methods can not only disregard the problem of image background but also save manpower and material resources in the process of sample collection.
- (2)
- The comparison shows that the recognition accuracy of the background-removed model trained by supervised data augmentation was only 91.50% and that the recognition accuracy of the network trained by the DCGAN augmented dataset could rise to 99.00%, indicating that the data augmentation using DCGAN was more effective than the supervised data augmentation. Supervised data augmentation does not generate a new sample, and it is difficult for the recognition model to learn more information because of the high similarity between the augmented sample and the original sample. Moreover, adding noise obscures the spots and affects the learning ability of the model, but data augmentation by DCGAN can generate a new spot color, spot shape, and spot distribution, which made the trained model have a better recognition ability. A modern, advanced form of augmentation is synthetic data augmentation, which overcomes the limitations of classical data augmentation.
- (3)
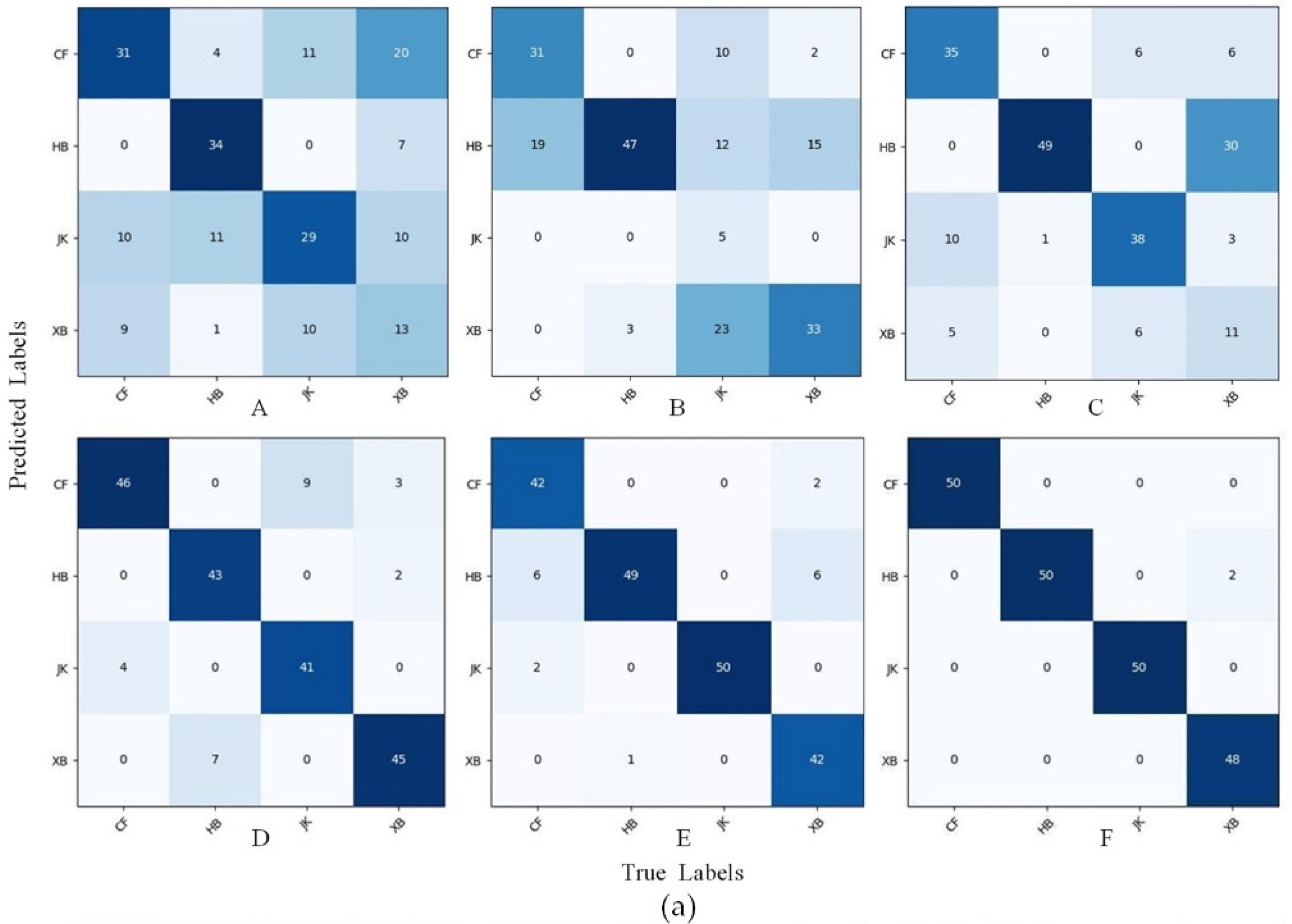
- In the recognition of sugarcane leaf spots, the morphological characteristics of the spots will affect the recognition ability of the model. In this study, we discovered through the confusion matrix that the misidentification problem caused by only a small area (leaf veins) of color difference between cane leaves with red rot and healthy cane leaves could be solved by data augmentation, but the spot color and spot distribution between the ring spots and rust were similar, especially when the rust infestation was serious, as well as when the rust spot areas were large and very similar to ring spots and when removing the background and data augmentation could reduce, but not completely resolve, the misidentification.
- (4)
- This analysis has a variety of limitations. Firstly, the variety of supervised data enhancements can continue to grow, and this paper proves that using these eight supervised data augmentations is inferior to using only a DCGAN, but not that using more and more supervised data augmentations is still inferior to using only a DCGAN. Secondly, we used a small dataset to train the DCGAN, and the quality of the synthetic samples produced in this research could be improved by integrating more labeled data, which would improve the learning process of the GAN. Thirdly, this experiment was performed in stages, with background removal, then data augmentation, and finally inputs to the recognition network, and we did not integrate these three parts into a single procedure. Lastly, in the future, we intend to improve not only the segmentation accuracy by enhancing the image segmentation model but also the quality of the synthetic crop images by training a D2PGGAN [34].
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Karamchandani, B.M.; Chakraborty, S.; Dalvi, S.G.; Satpute, S.K. Chitosan and its derivatives: Promising biomaterial in averting fungal diseases of sugarcane and other crops. J. Basic Microbiol. 2022, 62, 533–554. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.R.; Yang, L.T. Sugarcane Agriculture and Sugar Industry in China. Sugar Tech. 2015, 17, 1–8. [Google Scholar] [CrossRef]
- Zhang, K.; Xu, X.; Guo, X.; Ding, S.; Gu, T.; Qin, L.; He, Z. Sugarcane Streak Mosaic Virus P1 Attenuates Plant Antiviral Immunity and Enhances Potato Virus X Infection in Nicotiana benthamiana. Cells 2022, 11, 2870. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.; Tan, C.; Li, S.Q.; He, J.R.; Wang, H.Y. A Data Augmentation Method Based on Generative Adversarial Networks for Grape Leaf Disease Identification. IEEE Access 2020, 8, 102188–102198. [Google Scholar] [CrossRef]
- Khasawneh, N.; Faouri, E.; Fraiwan, M. Automatic Detection of Tomato Diseases Using Deep Transfer Learning. Appl. Sci. 2022, 12, 8467. [Google Scholar] [CrossRef]
- Kosamkar, P.K.; Kulkarni, V.Y.; Mantri, K.; Rudrawar, S.; Salmpuria, S.; Gadekar, N. Leaf Disease Detection and Recommendation of Pesticides using Convolution Neural Network. In Proceedings of the 4th International Conference on Computing Communication Control and Automation (ICCUBEA), Pune, India, 16–18 August 2018. [Google Scholar]
- Kaur, P.; Harnal, S.; Tiwari, R.; Upadhyay, S.; Bhatia, S.; Mashat, A.; Alabdali, A.M. Recognition of Leaf Disease Using Hybrid Convolutional Neural Network by Applying Feature Reduction. Sensors 2022, 22, 575. [Google Scholar] [CrossRef]
- Liang, W.J.; Zhang, H.; Zhang, G.F.; Cao, H.X. Rice Blast Disease Recognition Using a Deep Convolutional Neural Network. Sci. Rep. 2019, 9, 1–10. [Google Scholar] [CrossRef]
- Dhaka, V.S.; Meena, S.V.; Rani, G.; Sinwar, D.; Ijaz, M.F.; Wozniak, M. A Survey of Deep Convolutional Neural Networks Applied for Prediction of Plant Leaf Diseases. Sensors 2021, 21, 4749. [Google Scholar] [CrossRef]
- Jiang, P.; Chen, Y.H.; Liu, B.; He, D.J.; Liang, C.Q. Real-Time Detection of Apple Leaf Diseases Using Deep Learning Approach Based on Improved Convolutional Neural Networks. IEEE Access 2019, 7, 59069–59080. [Google Scholar] [CrossRef]
- Militante, S.V.; Gerardo, B.D.; Medina, R.P. Sugarcane Disease Recognition using Deep Learning. In Proceedings of the IEEE Eurasia Conference on IOT, Communication and Engineering (IEEE ECICE), Natl Formosa Univ, Yunlin, Taiwan, 3–6 October 2022; pp. 575–578. [Google Scholar]
- Yan, Q.; Yang, B.H.; Wang, W.Y.; Wang, B.; Chen, P.; Zhang, J. Apple Leaf Diseases Recognition Based on An Improved Convolutional Neural Network. Sensors 2020, 20, 3535. [Google Scholar] [CrossRef]
- Loti, N.N.A.; Noor, M.R.M.; Chang, S.W. Integrated analysis of machine learning and deep learning in chili pest and disease identification. J. Sci. Food Agric. 2021, 101, 3582–3594. [Google Scholar] [CrossRef] [PubMed]
- Brahimi, M.; Boukhalfa, K.; Moussaoui, A. Deep Learning for Tomato Diseases: Classification and Symptoms Visualization. Appl. Artif. Intell. 2017, 31, 299–315. [Google Scholar] [CrossRef]
- Adem, K.; Ozguven, M.M.; Altas, Z. A sugar beet leaf disease classification method based on image processing and deep learning. Multimed. Tools Appl. 2020, 18, 1–18. [Google Scholar] [CrossRef]
- Kianat, J.; Khan, M.A.; Sharif, M.; Akram, T.; Rehman, A.; Saba, T. A joint framework of feature reduction and robust feature selection for cucumber leaf diseases recognition. Optik 2021, 240, 166566. [Google Scholar] [CrossRef]
- Wang, C.S.; Du, P.F.; Wu, H.R.; Li, J.X.; Zhao, C.J.; Zhu, H.J. A cucumber leaf disease severity classification method based on the fusion of DeepLabV3+and U-Net. Comput. Electron. Agric. 2021, 189, 106373. [Google Scholar] [CrossRef]
- Bai, X.B.; Li, X.X.; Fu, Z.T.; Lv, X.J.; Zhang, L.X. A fuzzy clustering segmentation method based on neighborhood grayscale information for defining cucumber leaf spot disease images. Comput. Electron. Agric. 2017, 136, 157–165. [Google Scholar] [CrossRef]
- Lin, K.; Gong, L.; Huang, Y.X.; Liu, C.L.; Pan, J. Deep Learning-Based Segmentation and Quantification of Cucumber Powdery Mildew Using Convolutional Neural Network. Front. Plant Sci. 2019, 10, 155. [Google Scholar] [CrossRef]
- Wang, Z.; Zhang, S.; Zhao, B. Crop Diseases Leaf Segmentation Method Based on Cascade Convolutional Neural Network. Comput. Eng. Appl. 2020, 56, 242–250. [Google Scholar]
- Abbas, A.; Jain, S.; Gour, M.; Vankudothu, S. Tomato plant disease detection using transfer learning with C-GAN synthetic images. Comput. Electron. Agric. 2021, 187, 9. [Google Scholar] [CrossRef]
- Douarre, C.; Crispim-Junior, C.F.; Gelibert, A.; Tougne, L.; Rousseau, D. Novel data augmentation strategies to boost supervised segmentation of plant disease. Comput. Electron. Agric. 2019, 165, 9. [Google Scholar] [CrossRef]
- Zhang, M.; Liu, S.H.; Yang, F.Y.; Liu, J. Classification of Canker on Small Datasets Using Improved Deep Convolutional Generative Adversarial Networks. IEEE Access 2019, 7, 49680–49690. [Google Scholar] [CrossRef]
- Zhang, J.Y.; Rao, Y.; Man, C.; Jiang, Z.H.; Li, S.W. Identification of cucumber leaf diseases using deep learning and small sample size for agricultural Internet of Things. Int. J. Distrib. Sens. Netw. 2021, 17, 13. [Google Scholar] [CrossRef]
- Chen, L.C.E.; Zhu, Y.K.; Papandreou, G.; Schroff, F.; Adam, H. Encoder-Decoder with Atrous Separable Convolution for Semantic Image Segmentation. In Proceedings of the 15th European Conference on Computer Vision (ECCV), Munich, Germany, 8–14 September 2018; pp. 833–851. [Google Scholar]
- Chen, L.C.; Papandreou, G.; Kokkinos, I.; Murphy, K.; Yuille, A.L. DeepLab: Semantic Image Segmentation with Deep Convolutional Nets, Atrous Convolution, and Fully Connected CRFs. IEEE Trans. Pattern Anal. Mach. Intell. 2018, 40, 834–848. [Google Scholar] [CrossRef]
- Goodfellow, I.J.; Pouget-Abadie, J.; Mirza, M.; Xu, B.; Warde-Farley, D.; Ozair, S.; Courville, A.; Bengio, Y. Generative Adversarial Nets. In Proceedings of the 28th Conference on Neural Information Processing Systems (NIPS), Montreal, QC, Canada, 8–13 December 2015; pp. 2672–2680. [Google Scholar]
- Sandler, M.; Howard, A.; Zhu, M.L.; Zhmoginov, A.; Chen, L.C. MobileNetV2: Inverted Residuals and Linear Bottlenecks. In Proceedings of the 31st IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), Salt Lake City, UT, USA, 18–23 June 2022; pp. 4510–4520. [Google Scholar]
- Chollet, F. Xception: Deep Learning with Depthwise Separable Convolutions. In Proceedings of the 30th IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), Honolulu, HI, USA, 21–26 July 2017; pp. 1800–1807. [Google Scholar]
- Howard, A.; Sandler, M.; Chu, G.; Chen, L.C.; Chen, B.; Tan, M.X.; Wang, W.J.; Zhu, Y.K.; Pang, R.M.; Vasudevan, V.; et al. Searching for MobileNetV3. In Proceedings of the IEEE/CVF International Conference on Computer Vision (ICCV), Seoul, Republic of Korea, 27 October–2 November 2019; pp. 1314–1324. [Google Scholar]
- Litjens, G.; Toth, R.; van de Ven, W.; Hoeks, C.; Kerkstra, S.; van Ginneken, B.; Vincent, G.; Guillard, G.; Birbeck, N.; Zhang, J.D.; et al. Evaluation of prostate segmentation algorithms for MRI: The PROMISE12 challenge. Med. Image Anal. 2014, 18, 359–373. [Google Scholar] [CrossRef]
- Khan, Z.; Yahya, N.; Alsaih, K.; Meriaudeau, F. Zonal Segmentation of Prostate T2W-MRI using Atrous Convolutional Neural Network. In Proceedings of the 17th IEEE Student Conference on Research and Development (SCOReD), Seri Iskandar, Malaysia, 15–17 October 2019; pp. 95–99. [Google Scholar]
- Crum, W.R.; Camara, O.; Hill, D.L.G. Generalized overlap measures for evaluation and validation in medical image analysis. IEEE Trans. Med. Imaging 2006, 25, 1451–1461. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.Y.; Li, M.Y.; Yu, J. D2pggan: Two Discriminators Used in Progressive Growing of Gans. In Proceedings of the 44th IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP), Brighton, UK, 12–17 May 2049; pp. 3177–3181. [Google Scholar]

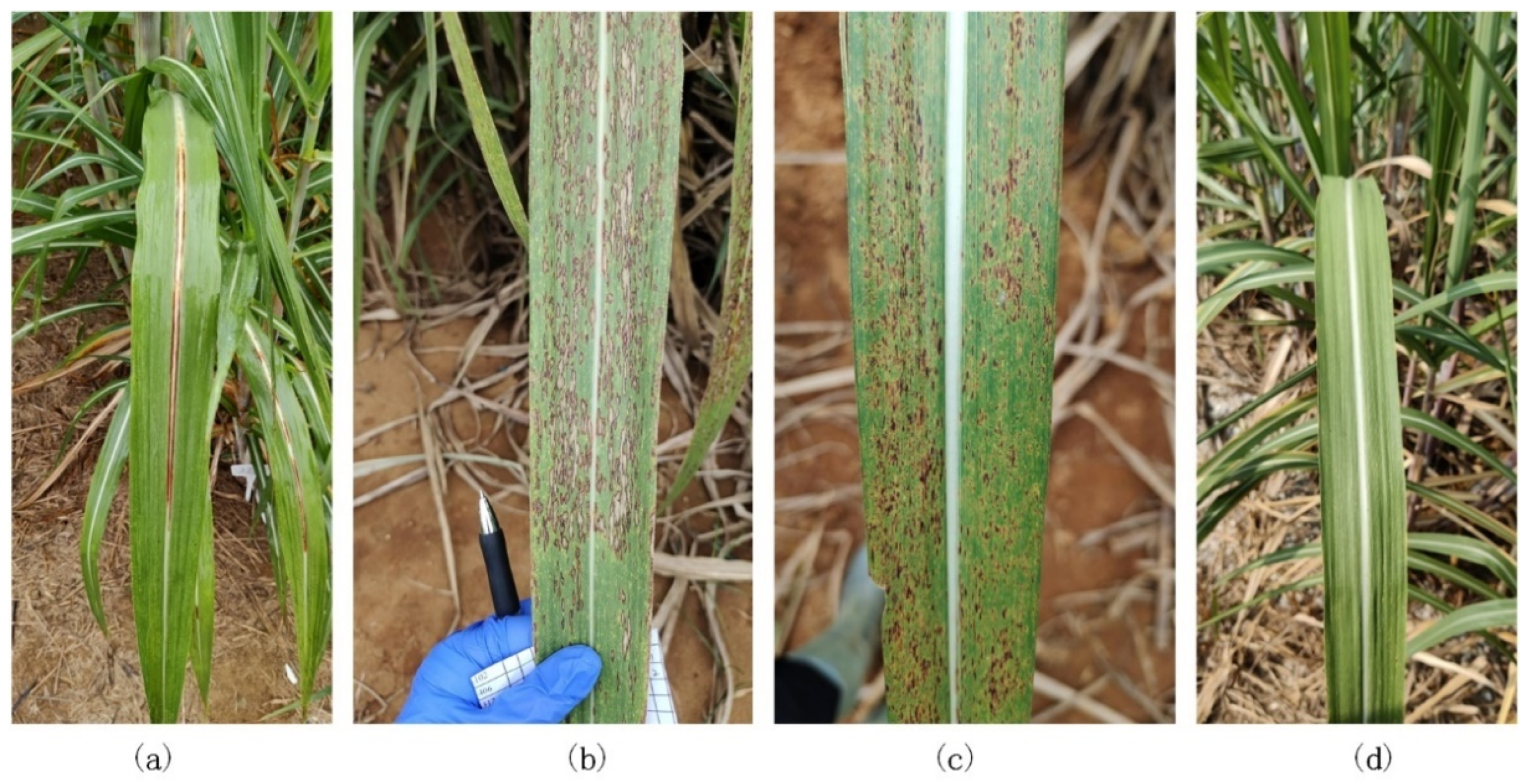


| Datasets | Classes | Number | Uses |
|---|---|---|---|
| Part 1 | Sugarcane leaves (without considering the disease type) | 790 | Train DeepLabV3+. |
| Part 2 | Leaver of red rot | 30 | Train for identification of classification models. |
| Leaver of ring spot | 30 | ||
| Healthy leaves | 30 | ||
| Leaver of rust | 30 |
| Stage | Hyperparameter | Value |
|---|---|---|
| Freezing | Epoch | 50 |
| Batch size | 4 | |
| Learning rate | 5 × 10−4 | |
| Unfreezing | Epoch | 50 |
| Batch size | 4 | |
| Learning rate | 5 × 10−5 |
| Sugarcane (Ture) | Background (Ture) | |
|---|---|---|
| Sugarcane (Predicted) | 315,282,153 | 2,369,606 |
| Background (Predicted) | 3,089,848 | 747,224,601 |
| Evaluation Metric | Value |
|---|---|
| IoU | 98.30% |
| DSC | 99.14% |
| PA | 99.50% |
| Precision | 99.25% |
| VOE | 0.86 |
| Original | 90° Rotation | 180° Rotation | 270° Rotation | Sp Noise | Gaussian Noise | Random Noise | Brightness Increase | Brightness Decrease |
|---|---|---|---|---|---|---|---|---|
 |  |  |  |  |  |  |  |  |
| Background | Class | Original | Generated |
|---|---|---|---|
| Removed by DeepLabV3+ | Red rot |    |    |
| Ring spot |    |    | |
| Rust |    |    | |
| Healthy |    |    | |
| Complex background | Red rot |    |    |
| Ring spot |    |    | |
| Rust |    |    | |
| Healthy |    |    |
| Sample | Number | Description | ||
|---|---|---|---|---|
| Train | Val | Test | ||
| A | 160 | 80 | 200 | Original images |
| B | 1440 | 720 | Supervised data augmentation for original images | |
| C | 2560 | 1280 | DCGAN data augmentation for back original images | |
| D | 160 | 80 | Background-removed images | |
| E | 1440 | 720 | Supervised data augmentation for background-removed images | |
| F | 2560 | 1280 | DCGAN data augmentation for group images | |
| Model | Group | Accuracy | Training Time/s |
|---|---|---|---|
| MobileNetV3-Large | A | 53.5% | 96.8 |
| B | 58.0% | 792.5 | |
| C | 66.5% | 1423.3 | |
| D | 87.5% | 94.2 | |
| E | 91.5% | 793.7 | |
| F | 99.0% | 1476.1 | |
| Alexnet | A | 44.5% | 87.3 |
| B | 55.0% | 457.6 | |
| C | 59.0% | 1405.2 | |
| D | 86.0% | 86.4 | |
| E | 90.5% | 444.9 | |
| F | 96.5% | 1394.3 | |
| Resnet | A | 43.0% | 192.3 |
| B | 56.0% | 1632.2 | |
| C | 60.0% | 2831.5 | |
| D | 86.0% | 197.3 | |
| E | 92.0% | 1613.5 | |
| F | 97.5% | 2976.4 | |
| Densenet | A | 42.0% | 175.1 |
| B | 60.0% | 1622.4 | |
| C | 63.0% | 2521.3 | |
| D | 81.0% | 176.3 | |
| E | 93.0% | 1659.4 | |
| F | 98.5% | 2423.6 |
| Group | Class | Precision | Recall | F1-Score | Specificity | Accuracy |
|---|---|---|---|---|---|---|
| A | Red rot | 0.47 | 0.62 | 0.53 | 0.68 | 53.50% |
| Ring spot | 0.83 | 0.68 | 0.75 | 0.91 | ||
| Healthy | 0.48 | 0.58 | 0.53 | 0.72 | ||
| Rust | 0.39 | 0.26 | 0.31 | 0.82 | ||
| B | Red rot | 0.72 | 0.62 | 0.67 | 0.88 | 58.00% |
| Ring spot | 0.51 | 0.94 | 0.66 | 0.60 | ||
| Healthy | 1.00 | 0.10 | 0.18 | 1.00 | ||
| Rust | 0.56 | 0.66 | 0.61 | 0.76 | ||
| C | Red rot | 0.74 | 0.70 | 0.72 | 0.89 | 66.50% |
| Ring spot | 0.62 | 0.98 | 0.76 | 0.74 | ||
| Healthy | 0.73 | 0.76 | 0.74 | 0.87 | ||
| Rust | 0.50 | 0.22 | 0.31 | 0.92 | ||
| D | Red rot | 0.79 | 0.92 | 0.85 | 0.91 | 87.50% |
| Ring spot | 0.95 | 0.86 | 0.90 | 0.99 | ||
| Healthy | 0.91 | 0.82 | 0.86 | 0.97 | ||
| Rust | 0.87 | 0.90 | 0.88 | 0.95 | ||
| E | Red rot | 0.95 | 0.84 | 0.89 | 0.99 | 91.50% |
| Ring spot | 0.80 | 0.98 | 0.88 | 0.92 | ||
| Healthy | 0.96 | 1.00 | 0.98 | 0.99 | ||
| Rust | 0.98 | 0.84 | 0.90 | 0.99 | ||
| F | Red rot | 1.00 | 1.00 | 1.00 | 1.00 | 99.00% |
| Ring spot | 0.96 | 1.00 | 0.98 | 0.99 | ||
| Healthy | 1.00 | 1.00 | 1.00 | 1.00 | ||
| Rust | 1.00 | 0.96 | 0.98 | 1.00 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Huang, Y.; Li, R.; Wei, X.; Wang, Z.; Ge, T.; Qiao, X. Evaluating Data Augmentation Effects on the Recognition of Sugarcane Leaf Spot. Agriculture 2022, 12, 1997. https://doi.org/10.3390/agriculture12121997
Huang Y, Li R, Wei X, Wang Z, Ge T, Qiao X. Evaluating Data Augmentation Effects on the Recognition of Sugarcane Leaf Spot. Agriculture. 2022; 12(12):1997. https://doi.org/10.3390/agriculture12121997
Chicago/Turabian StyleHuang, Yiqi, Ruqi Li, Xiaotong Wei, Zhen Wang, Tianbei Ge, and Xi Qiao. 2022. "Evaluating Data Augmentation Effects on the Recognition of Sugarcane Leaf Spot" Agriculture 12, no. 12: 1997. https://doi.org/10.3390/agriculture12121997
APA StyleHuang, Y., Li, R., Wei, X., Wang, Z., Ge, T., & Qiao, X. (2022). Evaluating Data Augmentation Effects on the Recognition of Sugarcane Leaf Spot. Agriculture, 12(12), 1997. https://doi.org/10.3390/agriculture12121997








