1. Introduction
In the development of mankind, every country around the world needs sufficient agricultural products for sustainability. Insect attacks and plant diseases reduce food supplementation all over the world by 40% annually [
1]. Generally, the disease is spotted on the stems or leaves of the plants [
2]. Observing and finding the disease, pests, and traits by using visual patterns is a tedious process [
3]. Bacteria causes a plant disease that leads to considerable economic loss. Fungi, bacteria, and phytoplasmas are part of plant pathogens that cause leaf disease [
4]. Traditionally way of analyzing a plant disease severity is by manifesting the proportion of plant tissue symptoms to identify the disease [
5]. For instance, Xanthomonas creates bacterial diseases which cause noticeable damage to plants. Phytopathogenic fungi are a source of making nearly 30% of all crop diseases. Approximately 80 percent of agricultural production is generated by small-scale industries. Diseases in agriculture cause a lot of yield loss which is more than 50 percent [
6]. As per the analysis conducted in 2018, pathogens are responsible for diseased crop production resulting in a heavy economic loss in the global market [
7]. The major concern of identifying the leaf disease with naked human eyes will cost a lot of time in terms of large-scale agricultural fields. Pests and pathogens cause a global loss of around
$220 billion. To be more precise from the analysis of 2021, without crop production in the field of rice, wheat, maize, potato, and soybean, the total estimated loss is 44.5 metric tons which cost 122.3 billion dollars, and with crop production total estimated loss of 34.2 metric tons which cost of 93.3 billion dollars which proves crop production will reduce the yield lose [
8]. Crop production per unit area (yield) is increasing due to the incorporation advance technologies in farming and crop production [
9]. Various researchers are working on finding new systems that utilize deep learning techniques to provide better insights for farmers in diagnosing crop disease. The deep learning technology’s outputs are used to provide a metric for an approximative estimation of crop production [
10].
2. Related Work
A series of recent studies have indicated that the research topic of plant disease prediction discusses the major issues of plant pathology and provides solutions for the issues. Pantazi et al. have achieved an accuracy of 92% with the help of the Local Binary Patterns algorithm and image segmentation on identifying powdery mildew disease on the vine leaves, which initially gave an ideology for this research [
11]. The above study focuses on detecting the differences in health conditions analysis on the variety of crops. Simonyan et al. used a deep convolutional neural network (DCNN) approach that achieves the disease detection of plant leaves and recommends pesticide doses to cure disease in the plants [
12]. Interestingly, Ahmad et al. have explored several pre-trained models, such as VGG16, VGG19, ResNet, and InceptionV3, which are fine-tuned networks to identify different tomato leaf diseases [
13]. Goncharov et al. implemented the Deep Siamese network, which combines two similar convolution layers weight to handle the issue of minimal databases of plant leaves utilizing peculiar leaves of the grape in various groups. However, their research focuses on solving a convolutional neural network imbalance issue by using two combined CNN [
14]. Elaraby et al. proposed two different stages of deep CNN models to predict citrus-diseased leaves. The first stage focuses on the potential target of the diseased area, which is followed by classifying a similar area to the corresponding citrus diseased leaves, which resembles our channel and spatial attention model works. However, the model achieved up to 94.3% accuracy [
15].
By measuring variables, one can describe and define plant disease severity. These variables signify the percentage of infected plants per unit area to the total number of plants, the percentage of diseased leaves to the total number of leaves on the same plant, and the percentage of diseased spot area to the total leaf area on the same leaf [
16]. Guan Wang et al. and a group of botanists annotated mild, medium, severe, and healthy as ground truth and divided classes and then trained using a deep convolutional neural network to identify the disease severity [
17]. The following studies were conducted on detecting tomato leaf disease with an overall accuracy of 98.49%. Naresh et al. proposed an effective and high-performing deep learning network by using a Convolutional Neural Network to extract colors, texture, and edge to target the infected area in the image to segment from the original image. This tomato dataset consists of 3000 images affected by nine different diseases [
18]. Pethybridge et al. used color pictures to compute the proportion of disease severity and identify lesion regions from healthy tissues [
19]. Miaomiao Ji et al. suggest an efficient automatic detection and severity analysis technique based on deep learning to handles grape black measles and fuzzy logic [
20]. Mehmet et al. proposed a Faster R-CNN architecture for detecting the sugar beet plant disease severity by imaging-based expect systems. The overall accuracy for the method is 95.84% [
21]. Umesh Kumar et al. present an enhanced CNN model (ECNN) for detecting cassava leaf disease with an accuracy of 99.3%. The dataset consists of five different cassava leaf diseases with 6256 images. ECNN model is a gamma correction feature method that involves three Convolutional layers and four fully integrated layers with Global Average Election Pooling Layer to minimize overfitting [
22].
Interestingly, Chenghai Yin et al. proposed a novel deep learning network, DISE-Net, which uses dilated inception module for feature extraction [
23]. Ren et al. perform a pre-trained ImageNet to identify the region-based detector by extracting the features in the inputs, helping us to design our attention model to focus on extracting the correct feature in the given inputs [
24]. Vinayakumar Ravi et al. focus on classified cassava leaf disease using a transfer learning-based CNN model. The author states that the CNN-based pre-trained model failed to identify the tiny infected part in their overall cassava leaf [
25].
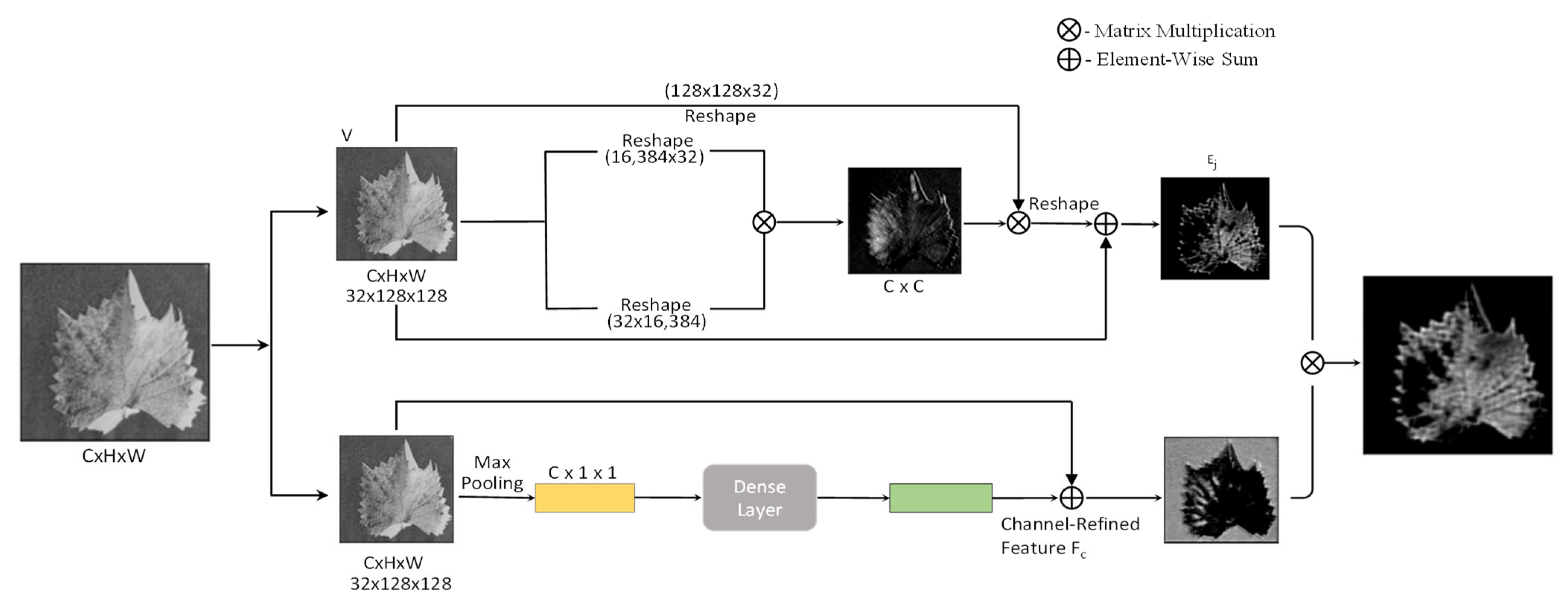
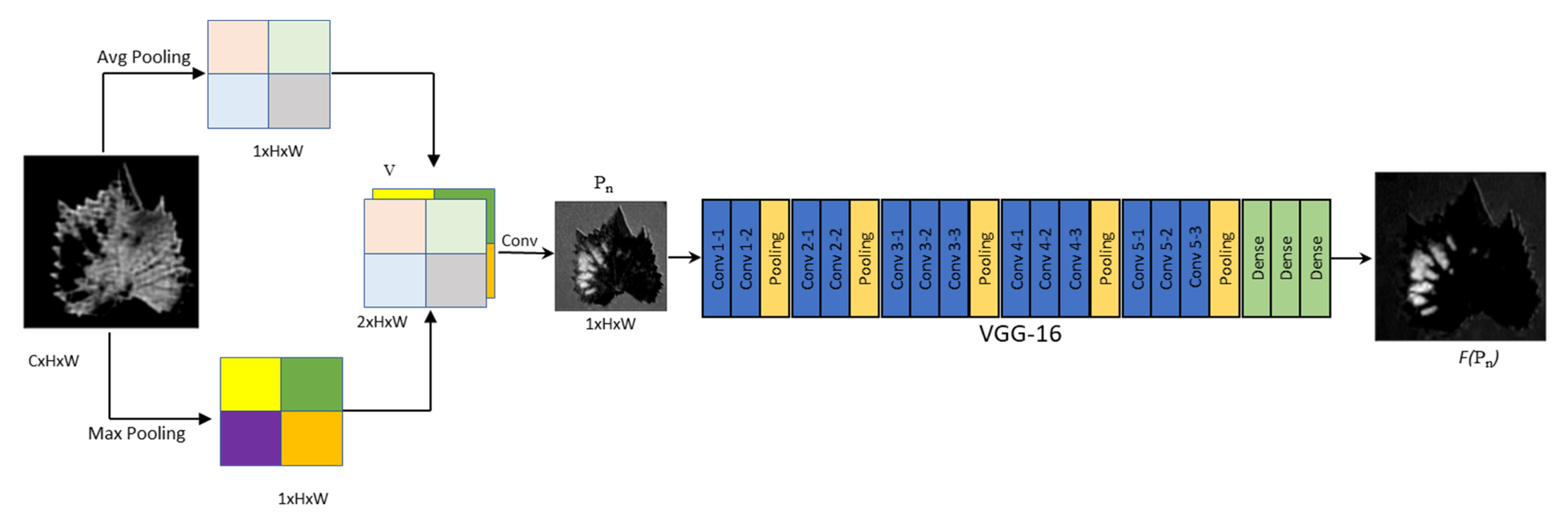
In recent years, researchers have been focusing on creating the attention model to solve imbalance network issues. The attention-based model has become more effective in recent years for various deep learning tasks, including natural language processing, image processing, and speech recognition. Woo et al. have introduced two separate mechanisms, such as channel and spatial attention mechanisms, in their study. Our neoteric method handles the plant disease prediction problem with two-phased architectures, such as channel-based attention mechanism and spatial attention mechanism. This work was inspired by the CBAM mechanism. CBAM involves channel and spatial attention mechanisms along with the CNN networks to find “what” and “where” in the region of a given leaf image for predicting the diseased leaves [
26]. The integration of attention strategies and deep learning makes the task of identifying and segmenting plant disease areas more attractive and detailed. CBAM (Convolutional Block Attention Module), SE-Net (Squeeze-and-Excitation Networks) VSG-Net (Visual-Spatial-Graph Network) are a few major common attention modules [
26,
27,
28].
On the other hand, Zhe Tang et al. used the lightweight CNN applied with a Channel-wise attention mechanism which holds the Shuffle Net as the Backbone. The squeeze-and-excitation is used to assist with the Channel-wise attention mechanism to improve the ShuffleNet architecture [
27]. Another study proposed a dual attention mechanism used to perform feature evaluation to find grape leaf disease.Their study has been limited to less than 4500 images which are very fewer data, and this is the limitation barrier of the study [
29]. Karthik et al. have applied two different significant architectures to detect the defected tomato leaves: 1. Initial architecture was designed by residual learning to classify the feature, 2. The attention mechanism is applied based on the classification to predict the leaf disease of early blight, late blight, and leaf mold. The accuracy of their study is 98%. The results show that some of the pre-trained neural networks with attention mechanisms are suitable for leaves diseases detection [
30]. Zhiwen et al. used an embedded network with CBAM in the densely connected convolutional network (DenseNet) to find the disease in the Wheat leaf image by utilizing C-DenseNet and CBAM as a Backbone network [
31]. The recent study employs pre-trained networks with their architectures to improve accuracy. Similarly, we have adopted pre-trained VGG16 with our spatial attention module. Yun Zhao et al. focus on predicting three different plant diseases, such as corn, potato, and tomato, with the help of inception and residual structure which is based on CNN embedded with CBAM [
32]. On the other hand, there are other technologies to identify plants and plant diseases using the mobile application. The flora incognita app helps to perceive biodiversity by determining plant species with the aid of a mobile application environment [
33]. There is a noticeable study that made an interactive mobile application called leaf doctor to measure the intensity of the disease [
19]. Another research study developed a new iPad application for assisting plant disease intensity and helping in treatment decisions [
34]. Abbas et al. established a conditional Generative Adversarial Network to generate an animated image of a tomato plant from the real image, and the DenseNet121 model uses both images to train the model. Moreover, their study uses transfer learning to classify tomato diseases. However, the accuracy achieved up to 98.65% [
35]. This work was intendedly created to support the real-time application.
The main significance of this work is described below:
- (i)
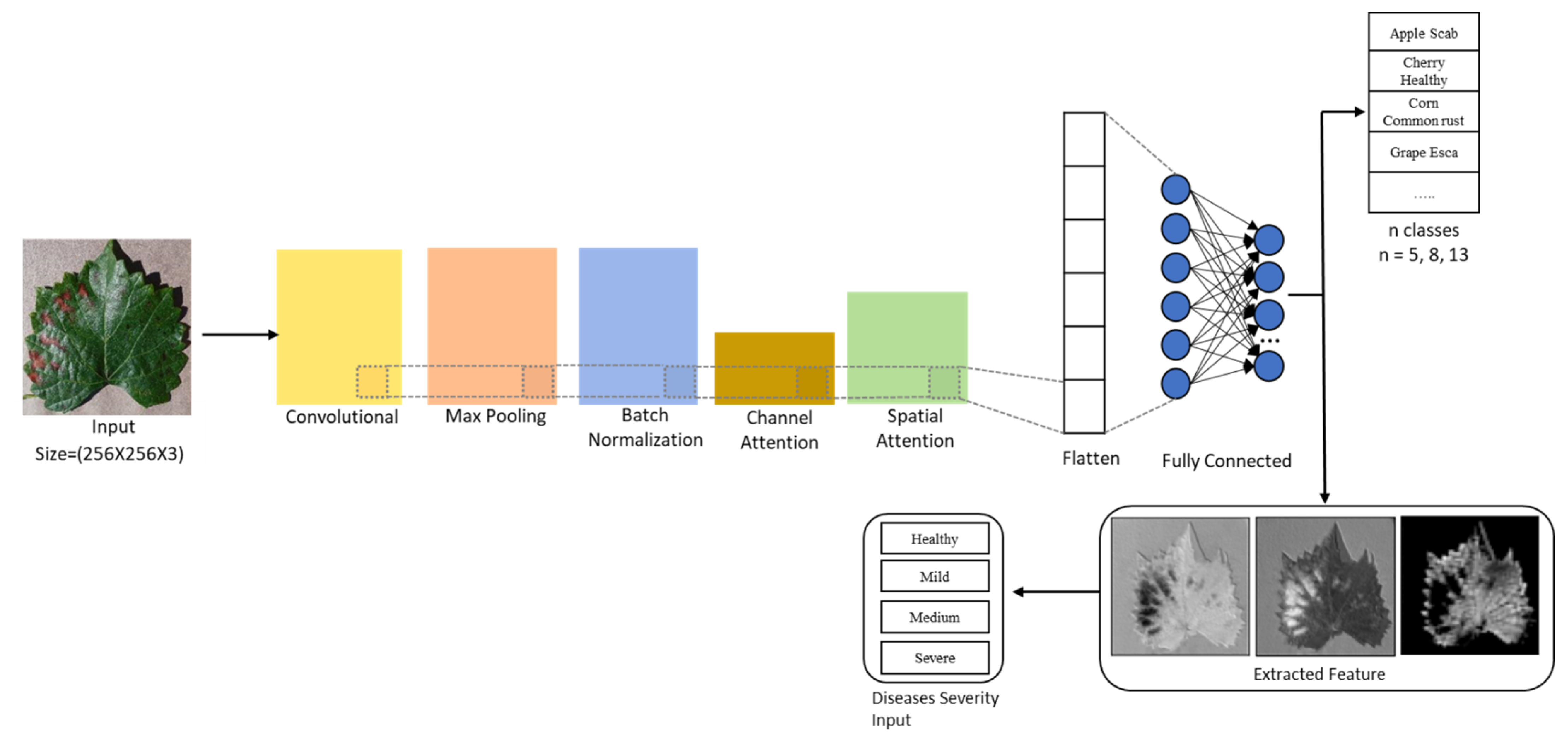
In this study, we have developed a Channel–Spatial segmentation network integrated with CBAM [
24] attention architecture to identify various healthy and unhealthy leaves. To understand the interconnection between the channels, we have implemented a channel attention module that enables the segmentation ability of the network.
- (ii)
This segmentation strategy of channel attention helps to separate the diseased portion of the leaf and also adds additional feature extraction strategies to spatial attention.
- (iii)
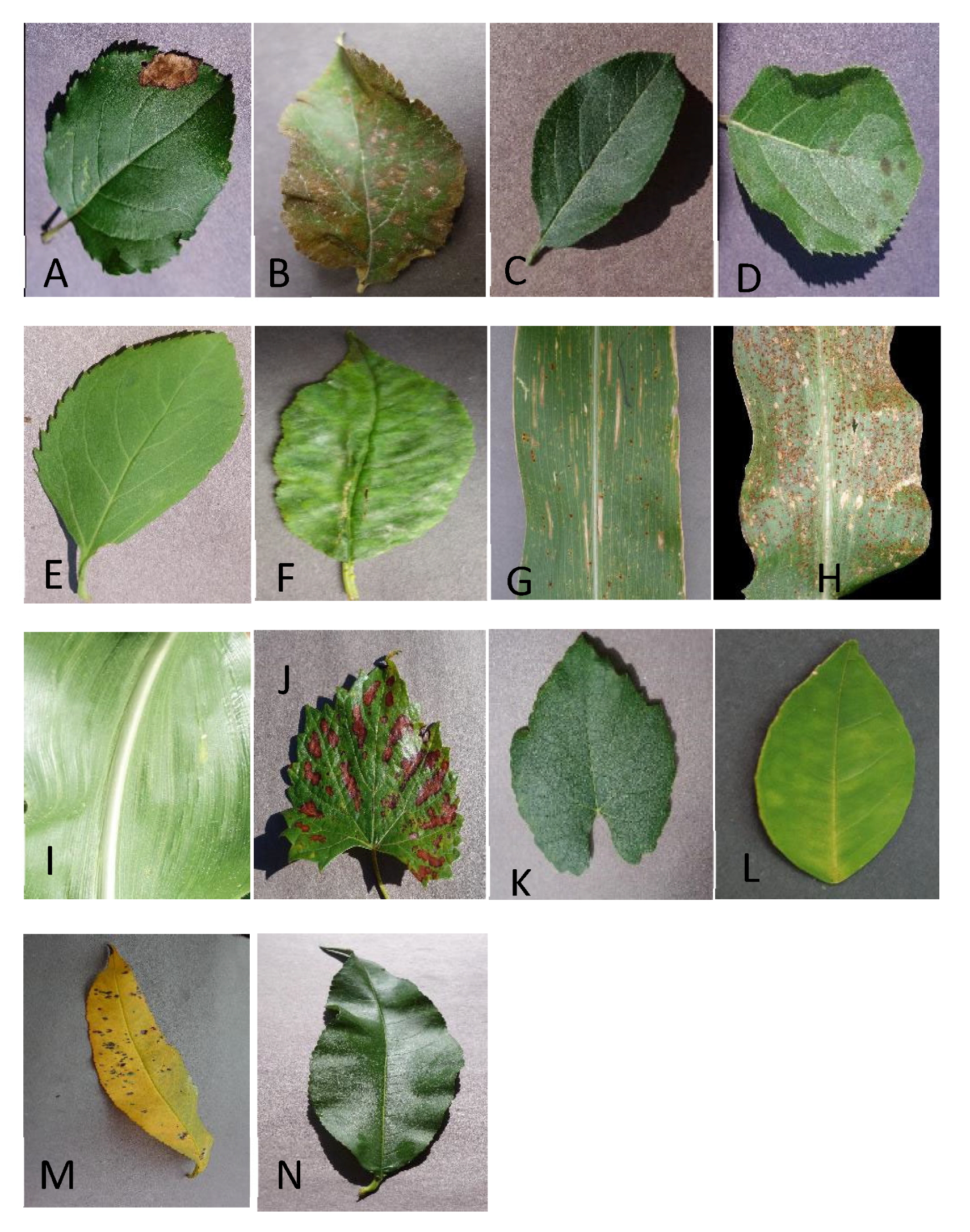
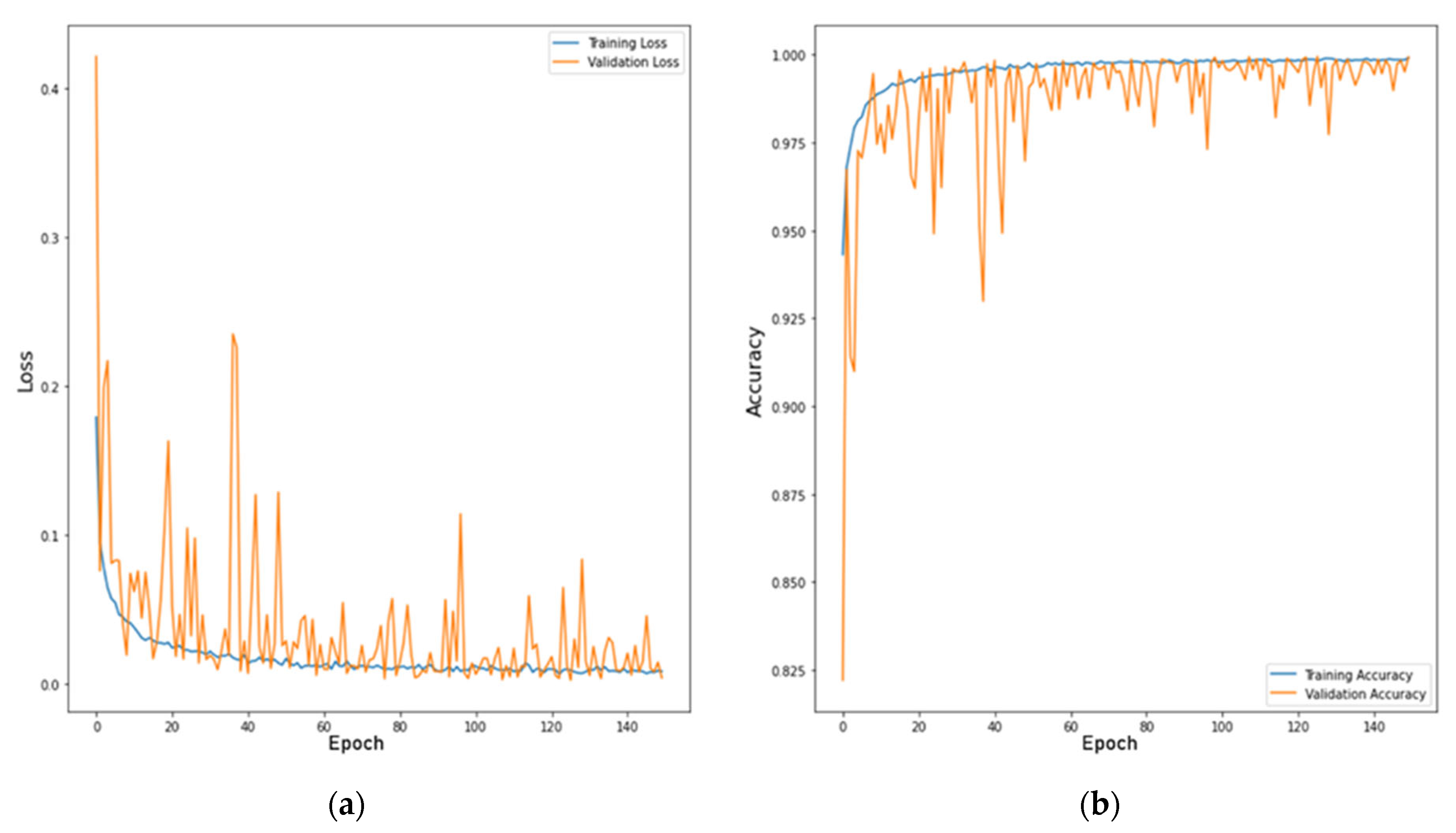
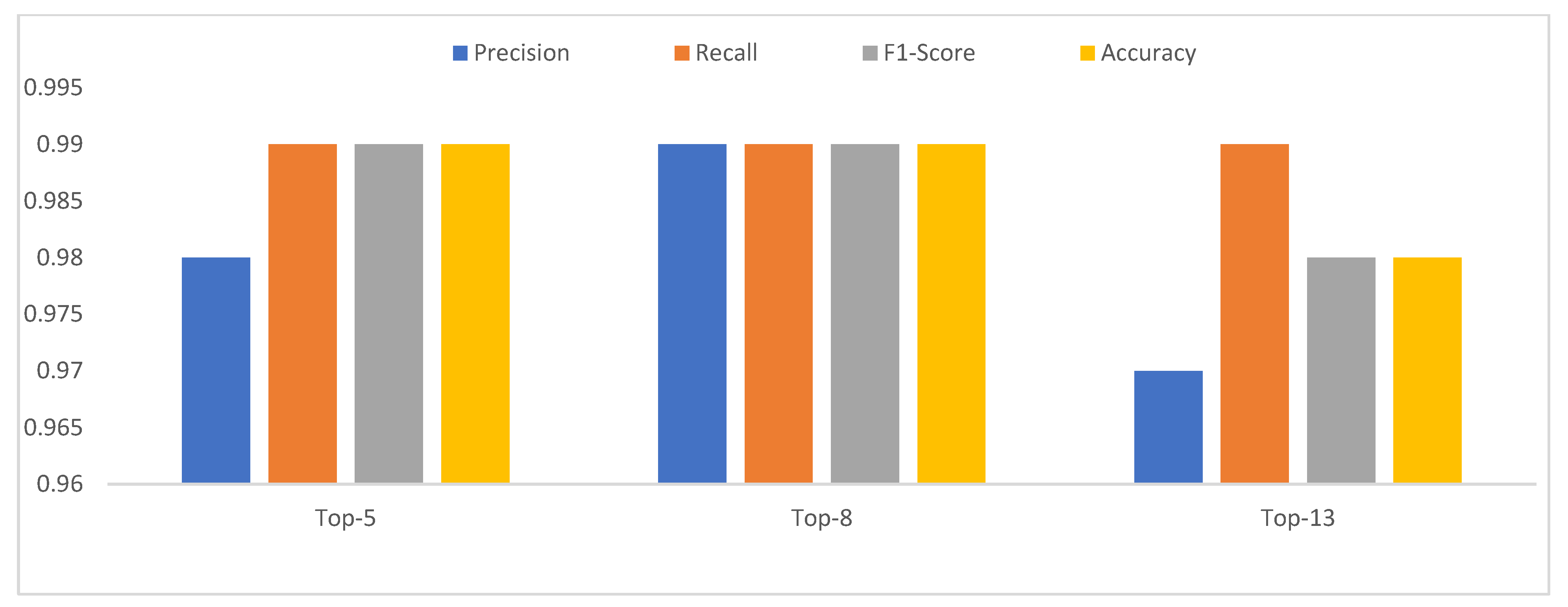
In this investigation, we also used a technique to validate the model by the total of 14 leave classes into the Top-13 leaf group, Top-8 leaf group, and Top-5 leaf group, which was discussed in the dataset with an overall accuracy of 99.76% [
35].
- (iv)
Our model identifies leaf disease with the added advantage of finding out negligible defects. Along with leaf disease prediction, we measure the severity by mild, medium, severe and healthy.