Modeling the Agricultural Soil Landscape of Germany—A Data Science Approach Involving Spatially Allocated Functional Soil Process Units
Abstract
1. Introduction
2. Materials and Methods
2.1. Data
2.1.1. Soil Profile Data—Consistency Check and Gap Filling
2.1.2. Data Cube of Covariates
| Soil Forming Factor | Abbreviation | Description | Data Source |
|---|---|---|---|
| Climate | PRESU | Average seasonal precipitation (summer) [raster, 1000 m] | [32] |
| PREWI | Average seasonal precipitation (winter) [raster, 1000 m] | ||
| TEMSU | Average seasonal temperature (summer) [raster, 1000 m] | [33] | |
| TEMWI | Average seasonal temperature (winter) [raster, 1000 m] | ||
| DINSU | Average seasonal drought index (summer) [raster, 1000 m] | [34] | |
| DINWI | Average seasonal drought index (winter) [raster, 1000 m] | ||
| Organisms/Soil | B0118, 0218, …B0818, B8A18, B1118, B1218 | Sentinel-2 spectral bands B1, B2, …B8, B8A, B11, and B12 composites of the 2nd yearly quartile of the year 2018 | |
| B0121, 0221, …B0821, B8A21, B1121, B1221 | Sentinel-2 spectral bands B1, B2, …B8, B8A, B11, and B12 composites of the 2nd yearly quartile of the year 2021 | ||
| EVI18, EVI21 | Enhanced vegetation index, calculated from Sentinel 2 band composites of 2nd quartile 2018 & 2021 (S2-Q2-18/21), EVI = G ∗ (B8A − B04)/(B8A + C1 ∗ B04 − C2 ∗ B02 + L), with G = 2.5, C1 = 6, C2 = 7.5 and L = 1 | ||
| MSI18, MSI21 | Moisture index: S2-Q2-18/21, MSI = B11/B08 | ||
| NDM18, NDM21 | Normalized difference moisture index: S2-Q2-18/21, NDMI = (B08 − B11)/(B08 + B11) | ||
| NDV18, NDV21 | Normalized difference vegetation index: S2-Q2-18/21, NDVI = (B08 − B04)/(B08 + B04) | ||
| NDW18, NDW21 | Normalized difference water index: S2-Q2-18/21, NDWI = (B03 − B08)/(B03 + B08) | ||
| PSR18, PSR21 | Plant senescence reflectance index: S2-Q2-18/21, PSRI = (B04 − B02)/B06 | ||
| DMP16 | Dry matter productivity, June 2016 [raster, 300 m] | [35] | |
| DMP18 | Dry matter productivity, June 2018 [raster, 300] | ||
| VPI16 | Vegetation Productivity Index, June 2016 [raster, 300 m] | [36] | |
| VPI18 | Vegetation Productivity Index, June 2018 [raster, 300 m] | ||
| Topography | GMK00 | Geomorphographic map of Germany [raster, 250 m resolution, map scale 1:1,000,000] | [37] |
| DEM00 | Digital elevation model [raster, 25 m resolution] | [38] | |
| SLO01, SLO05, SLO10 | Slope: calculated from DEM (cfD) with a search radius of 1, 5, 10 cells, using SAGA module Morphometric features | ||
| NOR01, NOR05, NOR10 | Northness: derived from aspect cfD with a search radius of 1, 5, 10 cells, using SAGA module Morphometric features | ||
| EAS01, EAS05, EAS10 | Eastness: derived from aspect cfD with a search radius of 1, 5, 10 cells, using SAGA module Morphometric features | ||
| TST01, TST05, TST10 | Terrain surface texture: cfD with a search radius of 1, 5, 10 cells, using SAGA module Terrain Surface Texture | ||
| TSR01, TSR05, TSR10 | Terrain surface ruggedness: cfD with a search radius of 1, 5, 10 cells, using SAGA module Terrain Ruggedness Index | ||
| CON01, CON05, CON10 | Convergence index: cfD with a search radius of 1, 5, 10 cells, using SAGA module Convergence Index (Search Radius) | ||
| SLH00 | Slope height: cfD using SAGA module Relative Heights and Slope Positions | ||
| VAD00 | Valley depth: cfD using SAGA module Relative Heights and Slope Positions | ||
| NOH00 | Normalized height: cfD using SAGA module Relative Heights and Slope Positions | ||
| WIN00 | Wind exposure: cfD using SAGA module Wind Effect | ||
| NOP00 | Negative openness: cfD using SAGA module Topographic Openness | ||
| POP00 | Positive openness: cfD using SAGA module Topographic Openness | ||
| VOF0S | Vertical overland flow distance to all river segments: cfD using SAGA module Terrain analysis/Channels | ||
| VOF0M | Vertical overland flow distance to major rivers: cfD using SAGA module Terrain analysis/Channels | ||
| HOF0S | Horizontal overland flow distance to all river segments: cfD using SAGA module Terrain analysis/Channels | ||
| HOFOM | Horizontal overland flow distance to major rivers: cfD using SAGA module Terrain analysis/Channels | ||
| SWI00 | SAGA wetness index: cfD using SAGA module SAGA Wetness Index | ||
| Parent material | LIT00 | Lithology, Hydrogeological map of Germany, HÜK [polygon shapefile, map scale 1:250,000] | [39] |
| STR00 | Stratigraphy, Hydrogeological map of Germany, HÜK [polygon shapefile, map scale 1:250,000] | ||
| BAG00 | Groups of soil parent material in Germany [polygon shapefile, map scale 1:5,000,000] | [40] | |
| Soil | BGL00 | Soil scapes in Germany [map scale 1:5,000,000] | [41] |
| DMP86 | Dry matter productivity, DMP18–DMP16 [raster, 300 m] | ||
| VPI86 | Vegetation Productivity Index, VPI18–VPI16 [raster, 300 m] | ||
| Geographic location | LAT00 | INSPIRE Latitude | [42] |
| LON00 | INSPIRE Longitude |
2.2. Differentiation of Functional SPUs
- The gap-filled, sliced (1 cm slices) profile data were used to calculate individual property distance matrices. First, the data were normalized to a range between 0 and 1, considering all slices in all profiles except for texture. For texture, the composites’ relation of sand, silt, and clay content were kept summing up to 1. Then, the mean of the slice-wise Euclidian profile distance was calculated for each variable and stored in separate distance matrices. Non-defined distances in case of differences in soil material causing missing data, e.g., missing texture data for organic horizons or slices assigned to bedrock, were assigned the maximum distance occurring between any two profile slices for the respective soil property. These property-wise distance matrices were then again normalized, resulting in a minimum distance of 0 and a maximum distance of 1. Hereafter, they will be referred to as normalized single-property distance matrices (nSPdist).
- The respective input parameter vectors of the involved optimization process to extract the SPUs are evaluated on behalf of a complex objective function. It seeks to identify those SPUs with the lowest possible internal variability and maximum inter-unit difference with regards to both their soil characteristics and landscape setting. The former is evaluated by using the Silhouette Index [47]. The latter requires the training of machine learning models to capture the soil-landscape relation and evaluate their predictive performance. A simple and fast learner is required to reduce the required computation time. The random forest (RF) algorithm [48] was chosen to suit this purpose. It is described in Section 2.3.1.
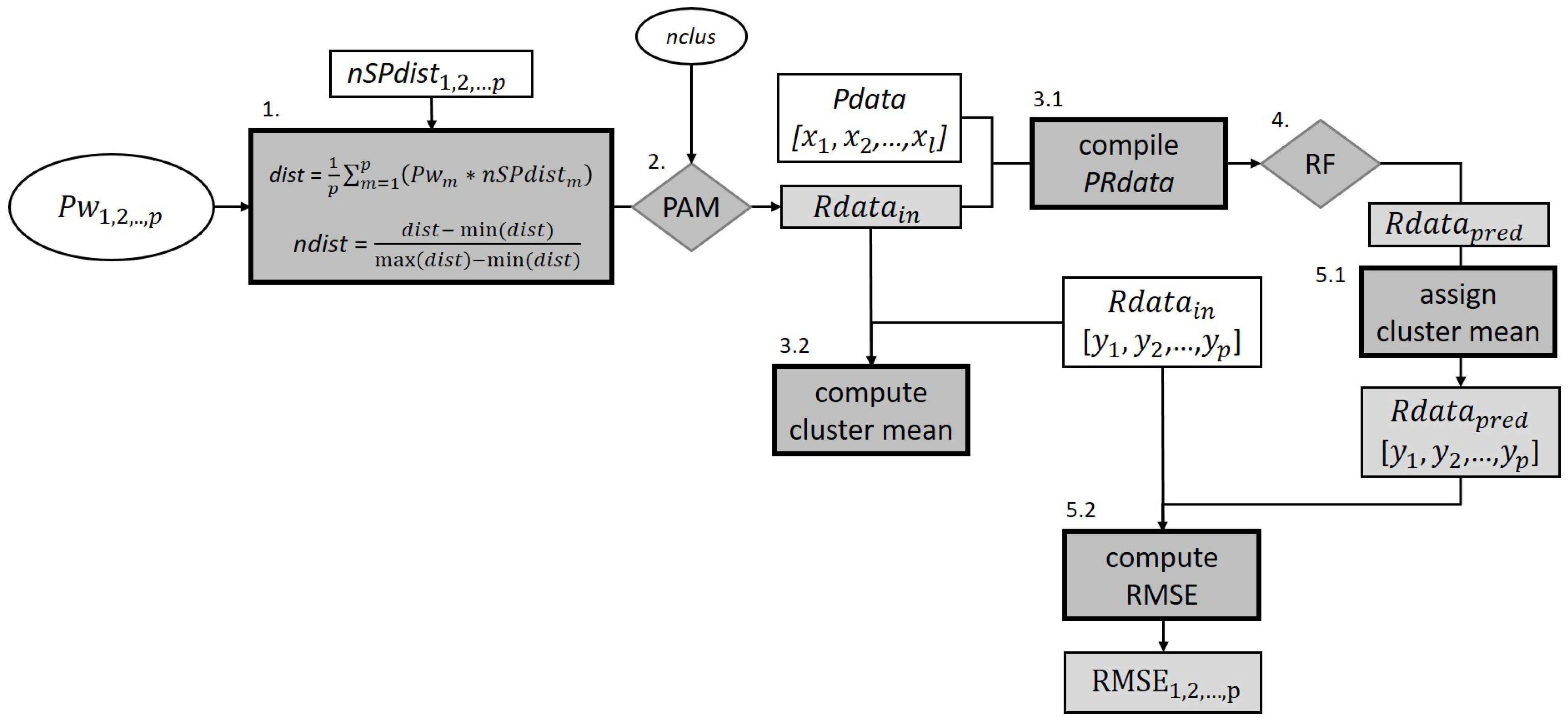
2.2.1. Approach 1 (PAMp)—SPU Extraction by P Weights Optimization
- 1.
- Pw1, Pw2, Pw3, …, Pwp in the range [0.1, 1] are assigned to each nSPdist, which are then combined by calculating the weighted average (dist). The values of the resulting distance matrix are normalized to the range [0, 1].
- 2.
- PAM clustering is conducted on the normalized distance matrix (ndist) with nclus = 8, 9, …, 100. The nclus minimum value was selected according to Ließ et al. [4]. For each input parameter vector including Pw1, Pw2, Pw3, …, Pwp and , the best cluster solution is selected on behalf of the Silhouette Index.
- 3.1.
- The resulting clustering solution Rdatain, which assigns each soil profile to one cluster, is then combined with the respective l covariates’ values x1, x2, …, xl of each profile (Pdata) to compile the predictor-response dataset (PRdata). The data were subdivided into 5 folds for a stratified 5-fold CV (Section 2.3.2). Categorical covariate values with zero data instances in any of the folds were removed.
- 3.2.
- Each profile’s property-wise mean along the depth profile, Rdatain [y1, y2, …, yp], was used to compute property-wise means per cluster.
- 4.
- An RF model was trained by 5-fold stratified CV using the PRdata [3.1] as input. The function ‘rfsrc’ of R package ‘randomForestSRC’ [49] was used with 1000 trees, a node size of five, and the default setting for the mtry parameter, while imputing no data values.
- 5.1.
- The previously computed property-wise cluster means [3.2] were assigned to each profile on behalf of the test set RF predictions (Rdatapred) generating Rdatapred [y1, y2, …, yp].
- 5.2.
- The property-wise RMSE was calculated using Rdatain [y1, y2, …, yp] and Rdatapred [y1, y2, …, yp]. The objective function value corresponds to the negative mean of the property-wise RMSE values. It is maximized in the optimization process.
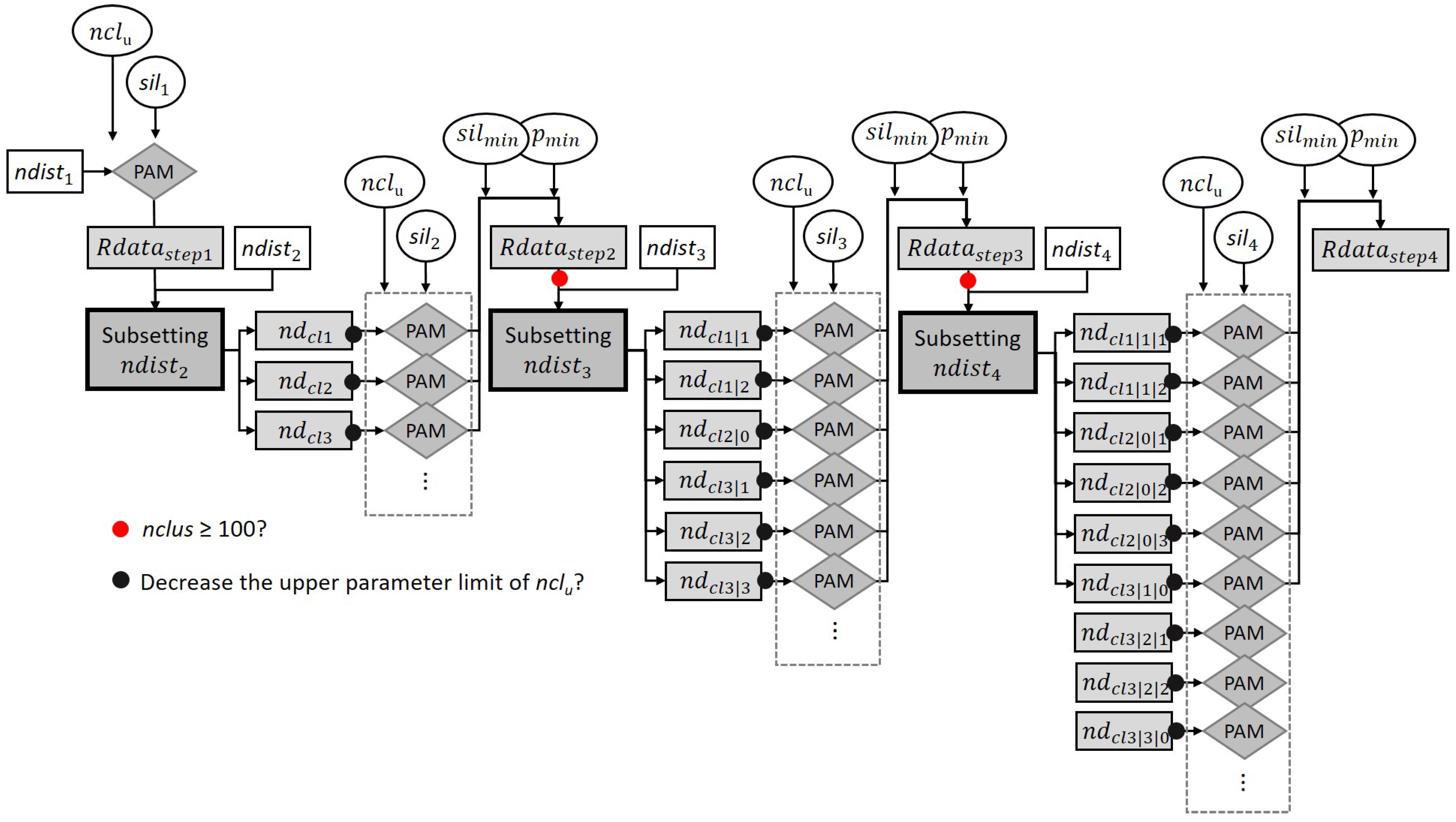
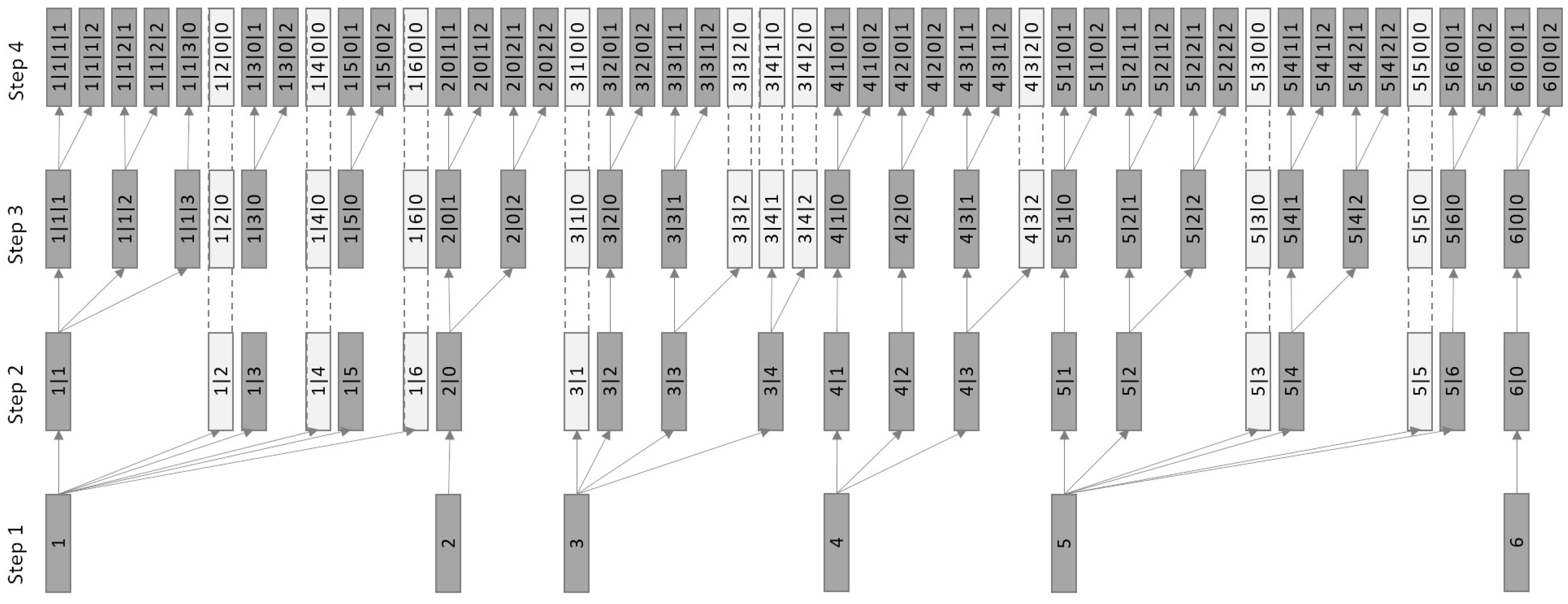
2.2.2. Approach 2 (PAMm)—SPU Extraction by Optimized Multistep Clustering
- nclu: The maximum number of clusters considered in each step.
- sil1,2,3,4: One Silhouette Index threshold value per step.
- silmin: The minimum Silhouette Index value tolerated to accept a lower-level clustering solution.
- pmin: The minimum number of profiles per cluster.
2.3. Modeling
- for gap filling,
- for SPU differentiation, and
- to train the pedometric model fathoming the soil–landscape relation to obtain nationwide and spatially continuous predictions (regionalization task).
2.3.1. Machine Learning Algorithms
2.3.2. Model Training, Tuning, and Evaluation
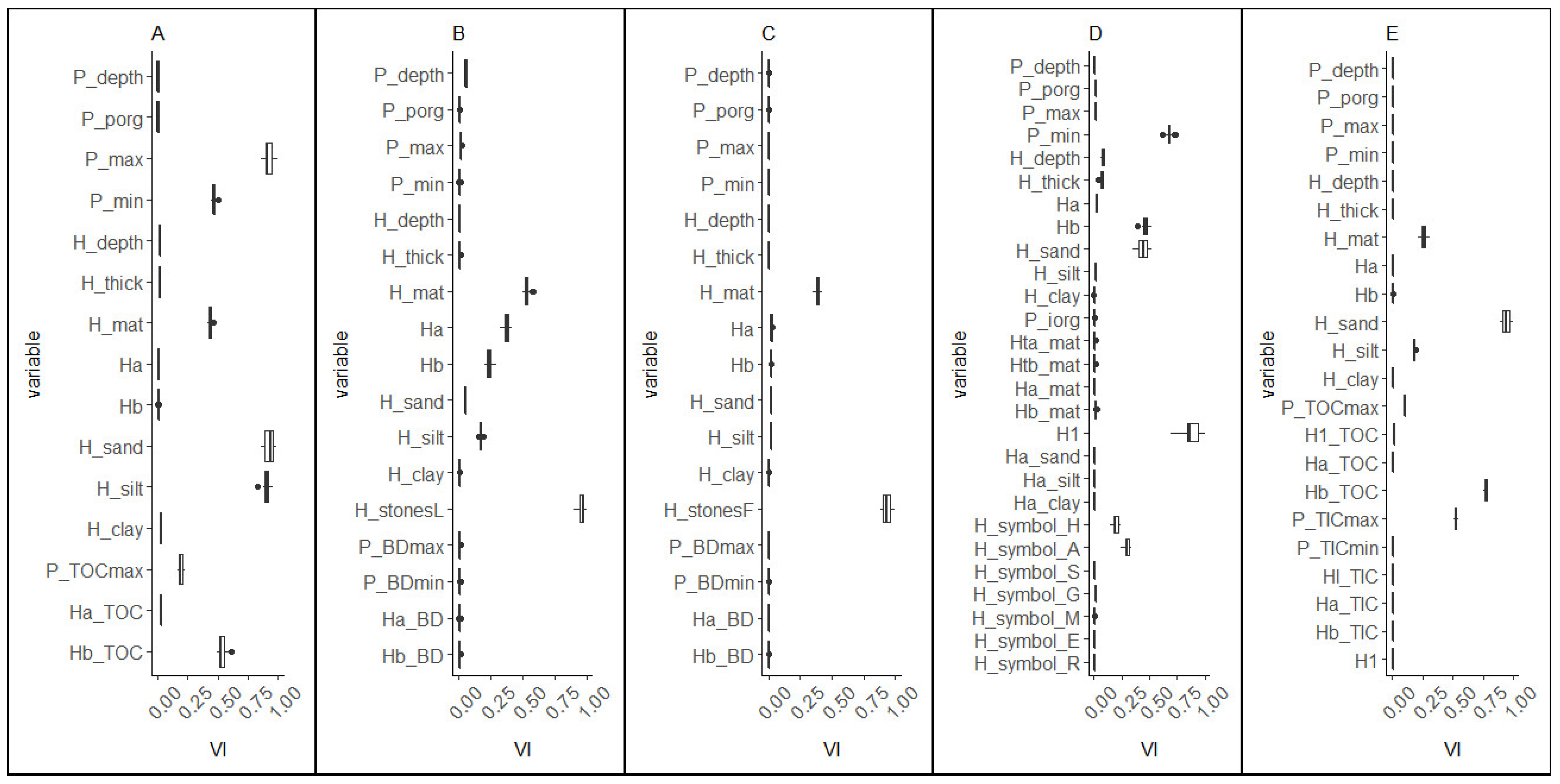
2.3.3. Variable Importance
2.4. Genetic Algorithm Optimization
3. Results and Discussion
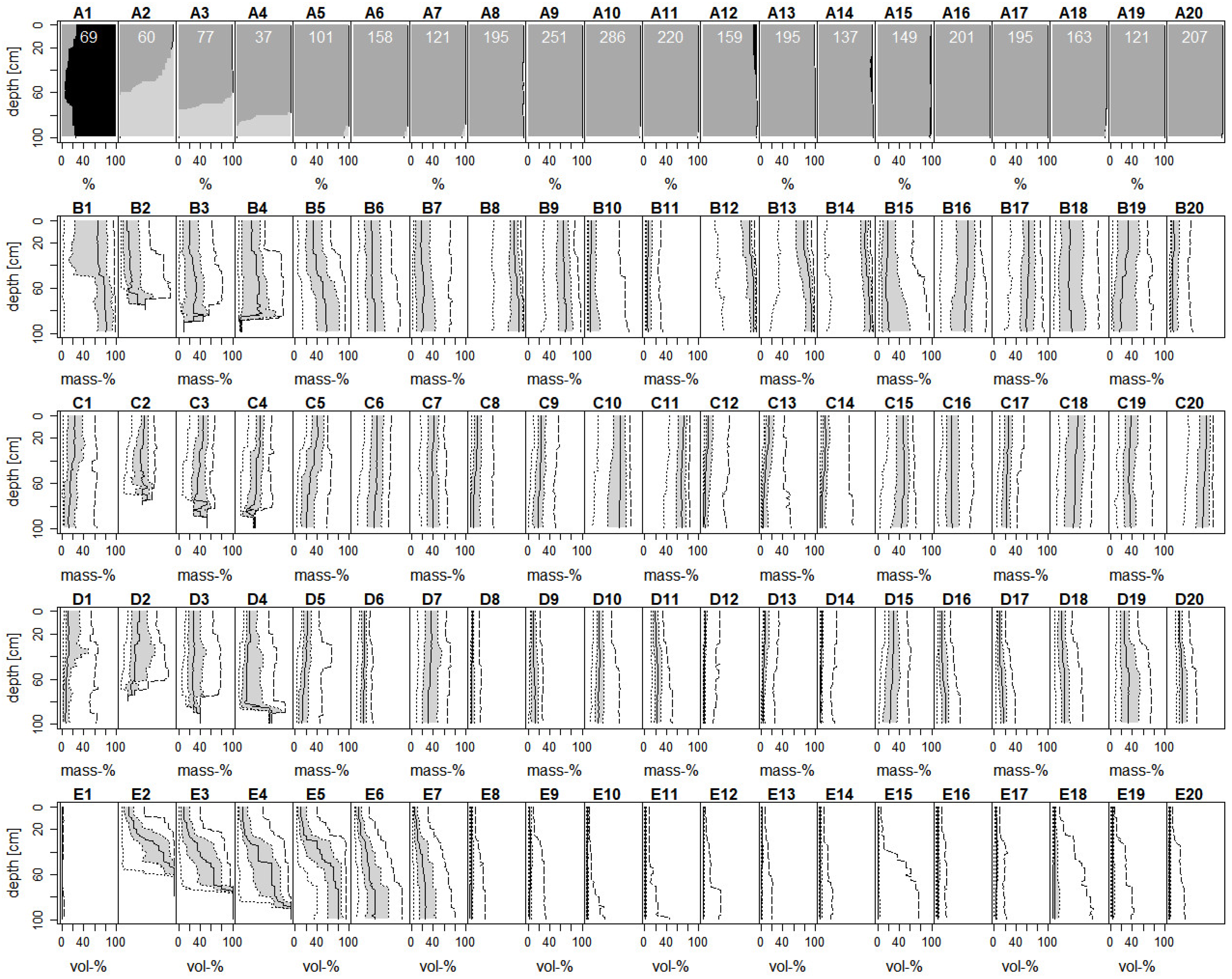
3.1. Gap Filling
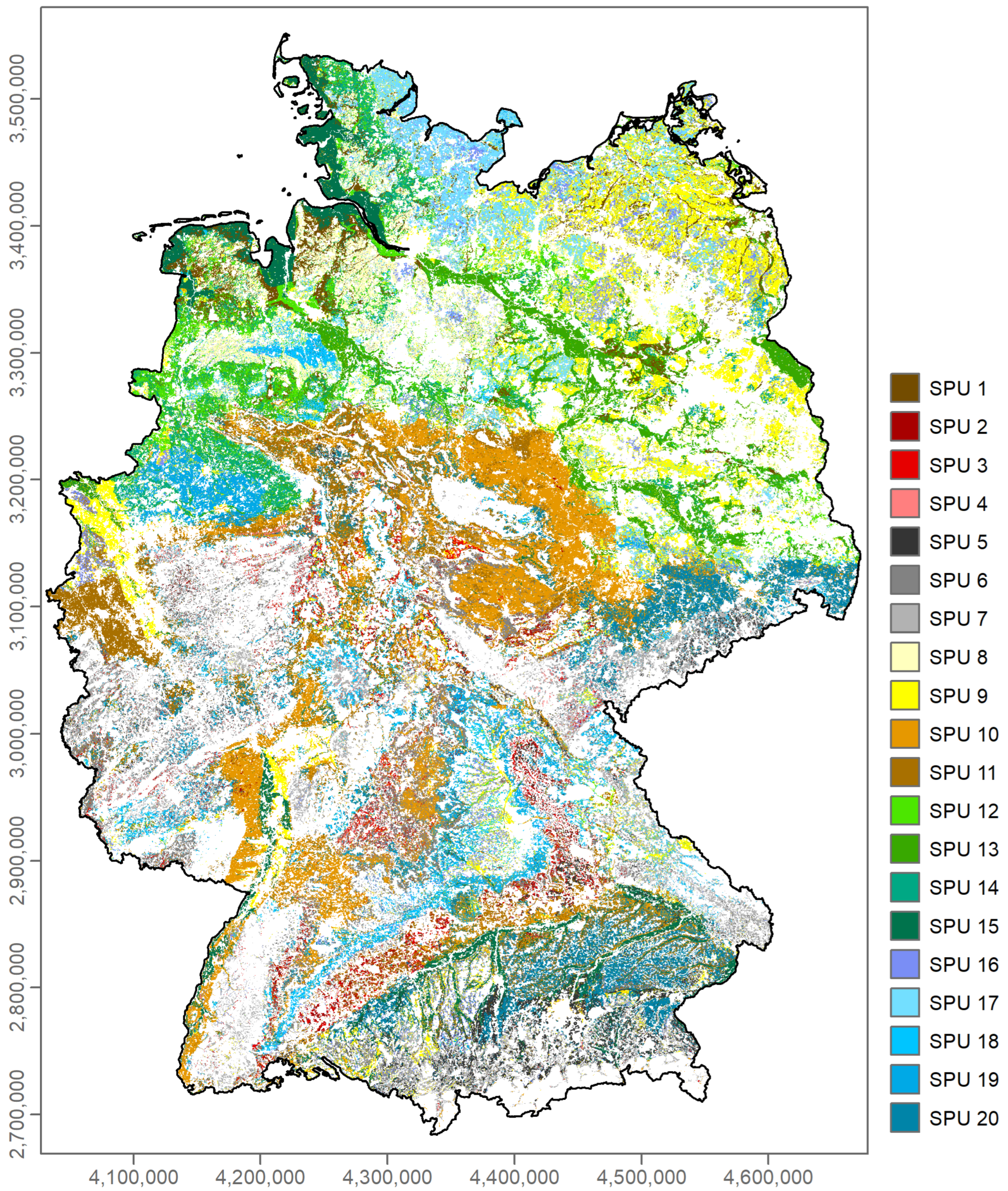
3.2. Differentiation of Functional SPUs
3.3. Pedometric Modeling to Capture the Soil–Landscape Relation
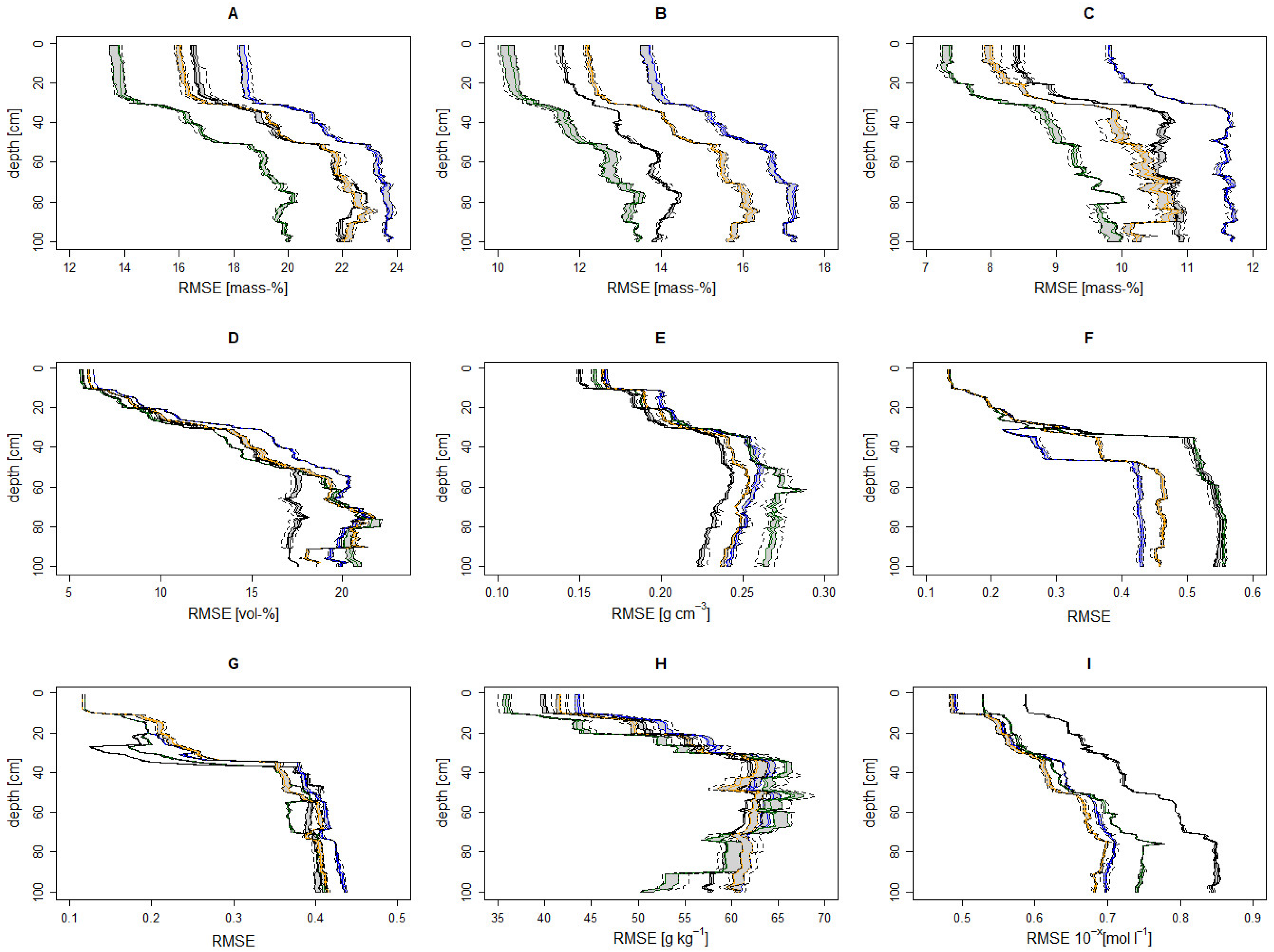
3.3.1. Model Performance
| Scale of the Data Product | Depth Interval [cm] | Sand Content [Mass-%] | Silt Content [Mass-%] | Clay Content [Mass-%] | Stone Content [Vol-%] | Bulk Density [g cm−3] | TOC [g kg−1] | pH 10−x mol L−1 |
|---|---|---|---|---|---|---|---|---|
| National | 0–30 | 15.0 [25] | 11.8 [25] | 8.2 [25] | - | - | 22 [64] | - |
| European | 0–20 | 17.6 [65] | 13.8 [65] | 9.8 [65] | 9 [65] | 0.26 [65] | 48.3 [66,68] | - |
| Global [67] | 0–5 | 19.3 | 16.5 | 11.4 | 7.1 | 0.30 | 43.6 | 1.2 |
| 5–15 | 19.4 | 16.4 | 11.0 | 7.8 | 0.30 | 46.2 | 1.2 | |
| 15–30 | 19.9 | 17.6 | 11.7 | 10.5 | 0.31 | 57.6 | 1.2 | |
| 30–60 | 22.9 | 18.7 | 13.8 | 17.5 | 0.35 | 62.4 | 1.3 | |
| 60–100 | 25.9 | 19.6 | 14.3 | 21.2 | 0.36 | 60.7 | 1.4 |
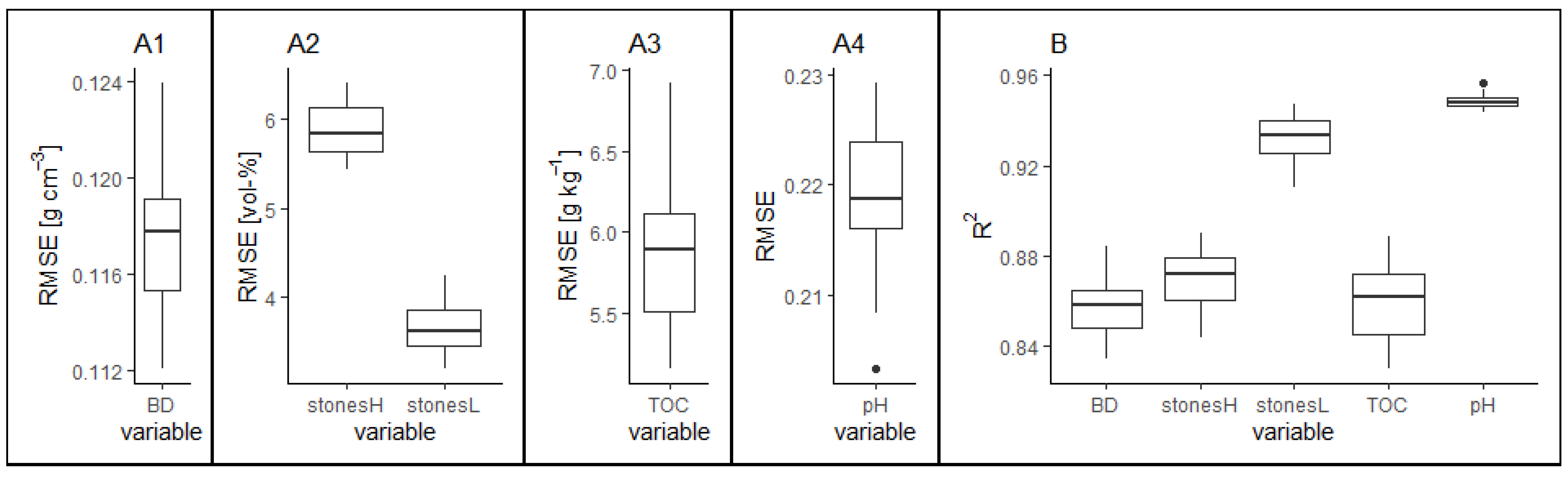
3.3.2. Variable Importance
3.3.3. Nationwide Prediction
4. Conclusions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Kapoor, D.; Bhardwaj, S.; Landi, M.; Sharma, A.; Ramakrishnan, M.; Sharma, A. The Impact of Drought in Plant Metabolism: How to Exploit Tolerance Mechanisms to Increase Crop Production. Appl. Sci. 2020, 10, 5692. [Google Scholar] [CrossRef]
- Magombeyi, M.S.; Taigbenu, A.E.; Barron, J. Effectiveness of Agricultural Water Management Technologies on Rainfed Cereals Crop Yield and Runoff in Semi-Arid Catchment: A Meta-Analysis. Int. J. Agric. Sustain. 2018, 16, 418–441. [Google Scholar] [CrossRef]
- Hatfield, J.L.; Dold, C. Water-Use Efficiency: Advances and Challenges in a Changing Climate. Front. Plant Sci. 2019, 10, 103. [Google Scholar] [CrossRef] [PubMed]
- Ließ, M.; Gebauer, A.; Don, A. Machine Learning With GA Optimization to Model the Agricultural Soil-Landscape of Germany: An Approach Involving Soil Functional Types With Their Multivariate Parameter Distributions Along the Depth Profile. Front. Environ. Sci. 2021, 9, 212. [Google Scholar] [CrossRef]
- Searle, R.; McBratney, A.; Grundy, M.; Kidd, D.; Malone, B.; Arrouays, D.; Stockman, U.; Zund, P.; Wilson, P.; Wilford, J.; et al. Digital Soil Mapping and Assessment for Australia and beyond: A Propitious Future. Geoderma Reg. 2021, 24, e00359. [Google Scholar] [CrossRef]
- Mueller, L.; Schindler, U.; Mirschel, W.; Graham Shepherd, T.; Ball, B.C.; Helming, K.; Rogasik, J.; Eulenstein, F.; Wiggering, H. Assessing the Productivity Function of Soils. A Review. Agron. Sustain. Dev. 2010, 30, 601–614. [Google Scholar] [CrossRef]
- Wallach, D.; Palosuo, T.; Thorburn, P.; Mielenz, H.; Buis, S.; Hochman, Z.; Gourdain, E.; Garcia, C.; Andrianasolo, F.; Dumont, B.; et al. Calibration of Crop Phenology Models: Going beyond Recommendations. bioRxiv 2022. [Google Scholar] [CrossRef]
- Boeing, F.; Rakovech, O.; Kumar, R.; Samaniego, L.; Schrön, M.; Hildebrandt, A.; Rebmann, C.; Thober, S.; Müller, S.; Zacharias, S.; et al. High-Resolution Drought Simulations and Comparison to Soil Moisture Observations in Germany. Hydrol. Earth Syst. Sci. 2022, 26, 5137–5161. [Google Scholar] [CrossRef]
- Bönecke, E.; Breitsameter, L.; Brüggemann, N.; Chen, T.W.; Feike, T.; Kage, H.; Kersebaum, K.C.; Piepho, H.P.; Stützel, H. Decoupling of Impact Factors Reveals the Response of German Winter Wheat Yields to Climatic Changes. Glob. Chang. Biol. 2020, 26, 3601–3626. [Google Scholar] [CrossRef]
- Webber, H.; Lischeid, G.; Sommer, M.; Finger, R.; Nendel, C.; Gaiser, T.; Ewert, F. No Perfect Storm for Crop Yield Failure in Germany. Environ. Res. Lett. 2020, 15, 104012. [Google Scholar] [CrossRef]
- Drastig, K.; Prochnow, A.; Libra, J.; Koch, H.; Rolinski, S. Irrigation Water Demand of Selected Agricultural Crops in Germany between 1902 and 2010. Sci. Total Environ. 2016, 569–570, 1299–1314. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Arrouays, D.; Angers, D.A.; Chenu, C.; Barré, P.; Martin, M.P.; Saby, N.P.A.; Walter, C. National Estimation of Soil Organic Carbon Storage Potential for Arable Soils: A Data-Driven Approach Coupled with Carbon-Landscape Zones. Sci. Total Environ. 2019, 666, 355–367. [Google Scholar] [CrossRef] [PubMed]
- Wiesmeier, M.; von Lützow, M.; Wollschlaeger, U.; Vogel, H.J.; Garcia-Franco, N.; Ließ, M.; Urbanski, L.; Hobley, E.; Lang, B.; Marin-Spiotta, E.; et al. Soil Organic Carbon Storage as a Key Function of Soils—A Review of Drivers and Indicators at Various Scales. Geoderma 2019, 333, 149–162. [Google Scholar] [CrossRef]
- Wang, C.; Amon, B.; Schulz, K.; Mehdi, B. Factors That Influence Nitrous Oxide Emissions from Agricultural Soils as Well as Their Representation in Simulation Models: A Review. Agronomy 2021, 11, 770. [Google Scholar] [CrossRef]
- Bouraoui, F.; Grizzetti, B. Modelling Mitigation Options to Reduce Diffuse Nitrogen Water Pollution from Agriculture. Sci. Total Environ. 2014, 468–469, 1267–1277. [Google Scholar] [CrossRef]
- Sundermann, G.; Wägner, N.; Cullmann, A.; von Hirschhausen, C.R.; Kemfert, C. Nitrate Pollution of Groundwater Long Exceeding Trigger Value: Fertilization Practices Require More Transparency and Oversight, DIW Weekly Report; DIW Weekly; Deutsches Institut für Wirtschaftsforschung (DIW): Berlin, Germany, 2020. [Google Scholar]
- BGR. Soil Map of Germany 1:250,000; Federal Institute for Geosciences and Natural Resources: Hanover, Germany, 2018.
- Ad-hoc-AG Boden. Bodenkundliche Kartieranleitung. KA5, 5th ed.; Bundesanstalt für Geowissenschaften und Rohstoffe in Zusammenarbeit mit den Staatlichen Geologischen Diensten: Stuttgart, Germany, 2005; ISBN 978-3-510-95920-4.
- Jenny, H. Factors of Soil Formation. A System of Quantitative Pedology; Dover Publications: New York, NY, USA, 1941. [Google Scholar]
- McBratney, A.B.; Mendonca Santos, M.L.; Minasny, B. On Digital Soil Mapping. Geoderma 2003, 117, 352. [Google Scholar] [CrossRef]
- Padarian, J.; Minasny, B.; McBratney, A.B. Machine Learning and Soil Sciences: A Review Aided by Machine Learning Tools. Soil 2020, 6, 35–52. [Google Scholar] [CrossRef]
- Arrouays, D.; Mulder, V.L.; Richer-de-Forges, A.C. Soil Mapping, Digital Soil Mapping and Soil Monitoring over Large Areas and the Dimensions of Soil Security—A Review. Soil Secur. 2021, 5, 100018. [Google Scholar] [CrossRef]
- Chen, S.; Arrouays, D.; Leatitia Mulder, V.; Poggio, L.; Minasny, B.; Roudier, P.; Libohova, Z.; Lagacherie, P.; Shi, Z.; Hannam, J.; et al. Digital Mapping of GlobalSoilMap Soil Properties at a Broad Scale: A Review. Geoderma 2022, 409, 115567. [Google Scholar] [CrossRef]
- Daniel, Ž.; Minařík, R.; Skála, J.; Beitlerová, H.; Juřicová, A.; Rojas, J.R.; Penížek, V.; Zádorová, T. High-Resolution Agriculture Soil Property Maps from Digital Soil Mapping Methods, Czech Republic. Catena 2022, 212, 106024. [Google Scholar] [CrossRef]
- Gebauer, A.; Sakhaee, A.; Don, A.; Poggio, M.; Ließ, M. Topsoil Texture Regionalization for Agricultural Soils in Germany—An Iterative Approach to Advance Model Interpretation. Front. Soil Sci. 2022, 1, 25. [Google Scholar] [CrossRef]
- Malone, B.; Searle, R. Updating the Australian Digital Soil Texture Mapping (Part 2): Spatial Modelling of Merged Field and Lab Measurements. Soil Res. 2021, 59, 419–434. [Google Scholar] [CrossRef]
- Reddy, N.N.; Chakraborty, P.; Roy, S.; Singh, K.; Minasny, B.; McBratney, A.B.; Biswas, A.; Das, B.S. Legacy Data-Based National-Scale Digital Mapping of Key Soil Properties in India. Geoderma 2021, 381, 114684. [Google Scholar] [CrossRef]
- Padarian, J.; Minasny, B.; McBratney, A.B. Using Deep Learning for Digital Soil Mapping. Soil 2019, 5, 79–89. [Google Scholar] [CrossRef]
- Ma, Y.; Minasny, B.; McBratney, A.; Poggio, L.; Fajardo, M. Predicting Soil Properties in 3D: Should Depth Be a Covariate? Geoderma 2021, 383, 114794. [Google Scholar] [CrossRef]
- Poeplau, C.; Don, A.; Flessa, H.; Heidkamp, A.; Jacobs, A.; Prietz, R. First German Agricultural Soil Inventory–Core Dataset; Open Agrar Repositorium: Göttingen, Germany, 2020. [Google Scholar] [CrossRef]
- Jacobs, A.; Flessa, H.; Don, A.; Heidkamp, A.; Prietz, R.; Gensior, A.; Poeplau, C.; Riggers, C.; Tiemeyer, B.; Vos, C.; et al. Landwirtschaftlich Genutzte Böden in Deutschland–Ergebnisse Der Bodenzustandserhebung, Thünen Report 64; Johann Heinrich von Thünen-Institut: Braunschweig, Germany, 2018; ISBN 9783865761927. [Google Scholar]
- DWD. Seasonal Grids of Sum of Precipitation over Germany, Version v1.0. Available online: https://opendata.dwd.de/climate_environment/CDC/grids_germany/seasonal/precipitation/ (accessed on 23 October 2022).
- DWD. Seasonal Grids of Monthly Averaged Daily Air Temperature (2m) over Germany, Version v1.0. Available online: https://opendata.dwd.de/climate_environment/CDC/grids_germany/seasonal/air_temperature_mean/ (accessed on 23 October 2022).
- DWD. Seasonal Grids of Sum of Drought Index (de Martonne) over Germany, Version v1.0. Available online: https://opendata.dwd.de/climate_environment/CDC/grids_germany/seasonal/drought_index/ (accessed on 23 October 2022).
- Swinnen, E.; Van Hoolst, R. Copernicus Global Land Operations ”Vegetation and Energy”. Issue I1.12, Version 1. Available online: https://land.copernicus.eu/global/sites/cgls.vito.be/files/products/CGLOPS1_ATBD_DMP300m-V1_I1.12.pdf (accessed on 23 October 2022).
- Swinnen, E.; Dierckx, W.; Toté, C. Gio Global Land Component–Lot I ”Operation of the Global Land Component”. Quality Assessment Report Proba-V NDVI, VCI and VPI. Issue 1.21. Available online: https://land.copernicus.eu/global/sites/cgls.vito.be/files/products/GIOGL1_QAR_NDVI-VCI-VPI_I1.21.pdf (accessed on 23 October 2022).
- BGR. Geomorphographic Map of Germany, GMK1000; Federal Institute for Geosciences and Natural Resources: Hanover, Germany, 2007.
- European Environment Agency (EEA). Copernicus Land Monitoring Service—EU-DEM, European Digital Elevation Model Version 1.1.; EEA: Copenhagen, Denmark, 2017.
- BGR; SDG. Hydrogeological Map of Germany 1:250,000 (HÜK250); Federal Institute for Geosciences and Natural Resources (BGR): Hanover, Germany; German State Geological Surveys (SGD): Hanover, Germany, 2019.
- BGR. Groups of Soil Parent Material in Germany 1:5,000,000. BAG5000, Version 3.0; Federal Institute for Geosciences and Natural Resources: Hanover, Germany, 2008.
- BGR. Soil Scapes in Germany 1:5,000,000. BGL5000; Federal Institute for Geosciences and Natural Resources: Hanover, Germany, 2008.
- INSPIRE Thematic Working Group. INSPIRE–Infrastructure for Spatial Information in Europe. D2.8.I.2 Data Specification on Geographical Grid Systems–Technical Guidelines; INSPIRE Thematic Working Group Coordinate Reference Systems & Geographical Grid Systems: Brussels, Belgium, 2014. [Google Scholar]
- Conrad, O.; Bechtel, B.; Bock, M.; Dietrich, H.; Fischer, E.; Gerlitz, L.; Wehberg, J.; Wichmann, V.; Böhner, J. System for Automated Geoscientific Analyses (SAGA) v. 2.1.4. Geosci. Model Dev. 2015, 8, 1991–2007. [Google Scholar] [CrossRef]
- Kaufman, L.; Rousseeuw, P.J. Finding Groups in Data: An Introduction to Cluster Analysis; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 1990; ISBN 9780471878766. [Google Scholar]
- Ahmad, A.; Khan, S.S. Survey of State-of-the-Art Mixed Data Clustering Algorithms. IEEE Access 2019, 7, 31883–31902. [Google Scholar] [CrossRef]
- Van Mechelen, I.; Boulesteix, A.-L.; Dangl, R.; Dean, N.; Guyon, I.; Hennig, C.; Leisch, F.; Steinley, D. Benchmarking in Cluster Analysis: A White Paper. arXiv 2018, arXiv:1809.10496. [Google Scholar]
- Rousseeuw, P.J. Silhouettes: A Graphical Aid to the Interpretation and Validation of Cluster Analysis. J. Comput. Appl. Math. 1987, 20, 53–65. [Google Scholar] [CrossRef]
- Breiman, L. Random Forests. J. Chem. Inf. Model. 2001, 53, 1689–1699. [Google Scholar] [CrossRef]
- Ishwaran, H.; Kogalur, U.B. Package ‘RandomForestSRC’. Fast Unified Random Forests for Survival, Regression, and Classification (RF-SRC). Version 3.1.1. 2022. Available online: https://www.randomforestsrc.org/ (accessed on 23 October 2022).
- Hothorn, T.; Hornik, K.; Strobl, C.; Zeileis, A. Package ‘Party’. A Laboratory for Recursive Partytioning. Version 1.3-11. 2022. Available online: http://party.r-forge.r-project.org/ (accessed on 23 October 2022).
- Cortes, C.; Vapnik, V. Support-Vector Networks. Mach. Leaming 1995, 20, 273–297. [Google Scholar] [CrossRef]
- Chang, C.-C.; Lin, C.-J. LIBSVM: A Library for Support Vector Machines. ACM Trans. Intell. Syst. Technol. 2011, 2, 1–39. [Google Scholar] [CrossRef]
- Meyer, D. Support Vector Machines—The Interface to Libsvm in Package E1071. FH Tech. Wien 2019, 16, 130. [Google Scholar]
- Hastie, T.; Tibshirani, R.; Friedman, J. The Elements of Statistical Learning–Data Mining, Inference, and Prediction, 2nd ed.; Springer Science+Business Media, LLC: New York, NY, USA, 2009; ISBN 978-0-387-84857-0. [Google Scholar]
- Affenzeller, M.; Winkler, S.; Wagner, S.; Beham, A. Genetic Algorithms and Genetic Programming; Taylor and Francis Group: Boca Raton, FL, USA, 2009; ISBN 978-1-58488-629-7. [Google Scholar]
- Batjes, N. A Taxotransfer Rule Based Approach for Filling Gaps in Measured Soil Data in Primary SOTER Databases (Version 1.1); World Soil Information: Wageningen, The Netherlands, 2003. [Google Scholar]
- Hugelius, G.; Bockheim, J.G.; Camill, P.; Elberling, B.; Grosse, G.; Harden, J.W.; Johnson, K.; Jorgenson, T.; Koven, C.D.; Kuhry, P.; et al. A New Data Set for Estimating Organic Carbon Storage to 3 m Depth in Soils of the Northern Circumpolar Permafrost Region. Earth Syst. Sci. Data 2013, 5, 393–402. [Google Scholar] [CrossRef]
- Almendra-Martín, L.; Martínez-Fernández, J.; Piles, M.; González-Zamora, Á. Comparison of Gap-Filling Techniques Applied to the CCI Soil Moisture Database in Southern Europe. Remote Sens. Environ. 2021, 258, 112377. [Google Scholar] [CrossRef]
- Wang, Q.; Wang, L.; Zhu, X.; Ge, Y.; Tong, X.; Atkinson, P.M. Remote Sensing Image Gap Filling Based on Spatial-Spectral Random Forests. Sci. Remote Sens. 2022, 5, 100048. [Google Scholar] [CrossRef]
- Taki, R.; Wagner-Riddle, C.; Parkin, G.; Gordon, R.; VanderZaag, A. Comparison of Two Gap-Filling Techniques for Nitrous Oxide Fluxes from Agricultural Soil. Can. J. Soil Sci. 2019, 99, 12–24. [Google Scholar] [CrossRef]
- Kim, Y.; Johnson, M.S.; Knox, S.H.; Black, T.A.; Dalmagro, H.J.; Kang, M.; Kim, J.; Baldocchi, D. Gap-Filling Approaches for Eddy Covariance Methane Fluxes: A Comparison of Three Machine Learning Algorithm Algorithms and Algorithm a Traditional Method with Principal Component Analysis. Glob. Chang. Biol. 2020, 26, 1499–1518. [Google Scholar] [CrossRef]
- Ghanbarian, B.; Pachepsky, Y. Machine Learning in Vadose Zone Hydrology: A Flashback. Vadose Zo. J. 2022, 21, e20212. [Google Scholar] [CrossRef]
- Lamichhane, S.; Kumar, L.; Wilson, B. Digital Soil Mapping Algorithms and Covariates for Soil Organic Carbon Mapping and Their Implications: A Review. Geoderma 2019, 352, 395–413. [Google Scholar] [CrossRef]
- Sakhaee, A.; Gebauer, A.; Ließ, M.; Don, A. Spatial Prediction of Organic Carbon in German Agricultural Topsoil Using Machine Learning Algorithms. Soil 2022, 8, 587–604. [Google Scholar] [CrossRef]
- Ballabio, C.; Panagos, P.; Monatanarella, L. Mapping Topsoil Physical Properties at European Scale Using the LUCAS Database. Geoderma 2016, 261, 110–123. [Google Scholar] [CrossRef]
- Aksoy, E.; Yigini, Y.; Montanarella, L. Combining Soil Databases for Topsoil Organic Carbon Mapping in Europe. PLoS ONE 2016, 11, 2022. [Google Scholar] [CrossRef] [PubMed]
- Poggio, L.; De Sousa, L.M.; Batjes, N.H.; Heuvelink, G.B.M.; Kempen, B.; Ribeiro, E.; Rossiter, D. SoilGrids 2.0: Producing Soil Information for the Globe with Quantified Spatial Uncertainty. Soil 2021, 7, 217–240. [Google Scholar] [CrossRef]
- Van Liedekerke, M.; Panagos, P. Predicted Distribution of SOC Content in Europe (Based on LUCAS, BioSoil and CZO) in the Context of the EU-Funded SoilTrEC Project. PLoS ONE 2016, 11, e0152098. [Google Scholar]
- BGR. General Geological Map of the Federal Republic of Germany 1:200,000; Federal Institute for Geosciences and Natural Resources: Hanover, Germany, 2007.
- Probst, P.; Wright, M.N.; Boulesteix, A.L. Hyperparameters and Tuning Strategies for Random Forest. Wiley Interdiscip. Rev. Data Min. Knowl. Discov. 2019, 9, e1301. [Google Scholar] [CrossRef]
- Strobl, C.; Boulesteix, A.-L.; Zeileis, A.; Hothorn, T. Bias in Random Forest Variable Importance Measures: Illustrations, Sources and a Solution. BMC Bioinformatics 2007, 8, 25. [Google Scholar] [CrossRef]
- BGR. Soil Map of Germany 1:1,000,000. BÜK1000; Federal Institute for Geosciences and Natural Resources: Hanover, Germany, 2013.


| Parameter | Lower Limit | Upper Limit |
|---|---|---|
| nclu | 3 | 10 |
| sil1, | 0.3 | 0.4 |
| sil2,3,4 | 0.4 | 0.8 |
| silmin | 0.25 | 0.4 |
| pmin | 5 | 10 |
| Parameter | P weights | nclus | ||||||
|---|---|---|---|---|---|---|---|---|
| Texture | Stone Content | Bulk Density | Symbol_S | Symbol_G | TOC | pH | ||
| value | 0.24 | 0.56 | 0.70 | 0.51 | 0.44 | 0.64 | 0.86 | 20 |
| Parameter | |||||||
|---|---|---|---|---|---|---|---|
| value | 6 | 0.31 | 0.74 | 0.62 | 0.53 | 0.34 | 13 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ließ, M. Modeling the Agricultural Soil Landscape of Germany—A Data Science Approach Involving Spatially Allocated Functional Soil Process Units. Agriculture 2022, 12, 1784. https://doi.org/10.3390/agriculture12111784
Ließ M. Modeling the Agricultural Soil Landscape of Germany—A Data Science Approach Involving Spatially Allocated Functional Soil Process Units. Agriculture. 2022; 12(11):1784. https://doi.org/10.3390/agriculture12111784
Chicago/Turabian StyleLieß, Mareike. 2022. "Modeling the Agricultural Soil Landscape of Germany—A Data Science Approach Involving Spatially Allocated Functional Soil Process Units" Agriculture 12, no. 11: 1784. https://doi.org/10.3390/agriculture12111784
APA StyleLieß, M. (2022). Modeling the Agricultural Soil Landscape of Germany—A Data Science Approach Involving Spatially Allocated Functional Soil Process Units. Agriculture, 12(11), 1784. https://doi.org/10.3390/agriculture12111784