Comprehensive Evaluation of Morpho-Physiological and Ionic Traits in Wheat (Triticum aestivum L.) Genotypes under Salinity Stress
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials
2.2. Experimental Design
2.3. Plant Morphological Data
2.4. Chlorophyll Content and Photochemical Efficiency of PSII
2.5. Membrane Stability Index
2.6. Na+, K+, Ca2+ Content in Shoot and Root
2.7. Statistical Analysis
3. Results
3.1. Effects of Salt Stress on Morpho-Physiological and Ionic Traits
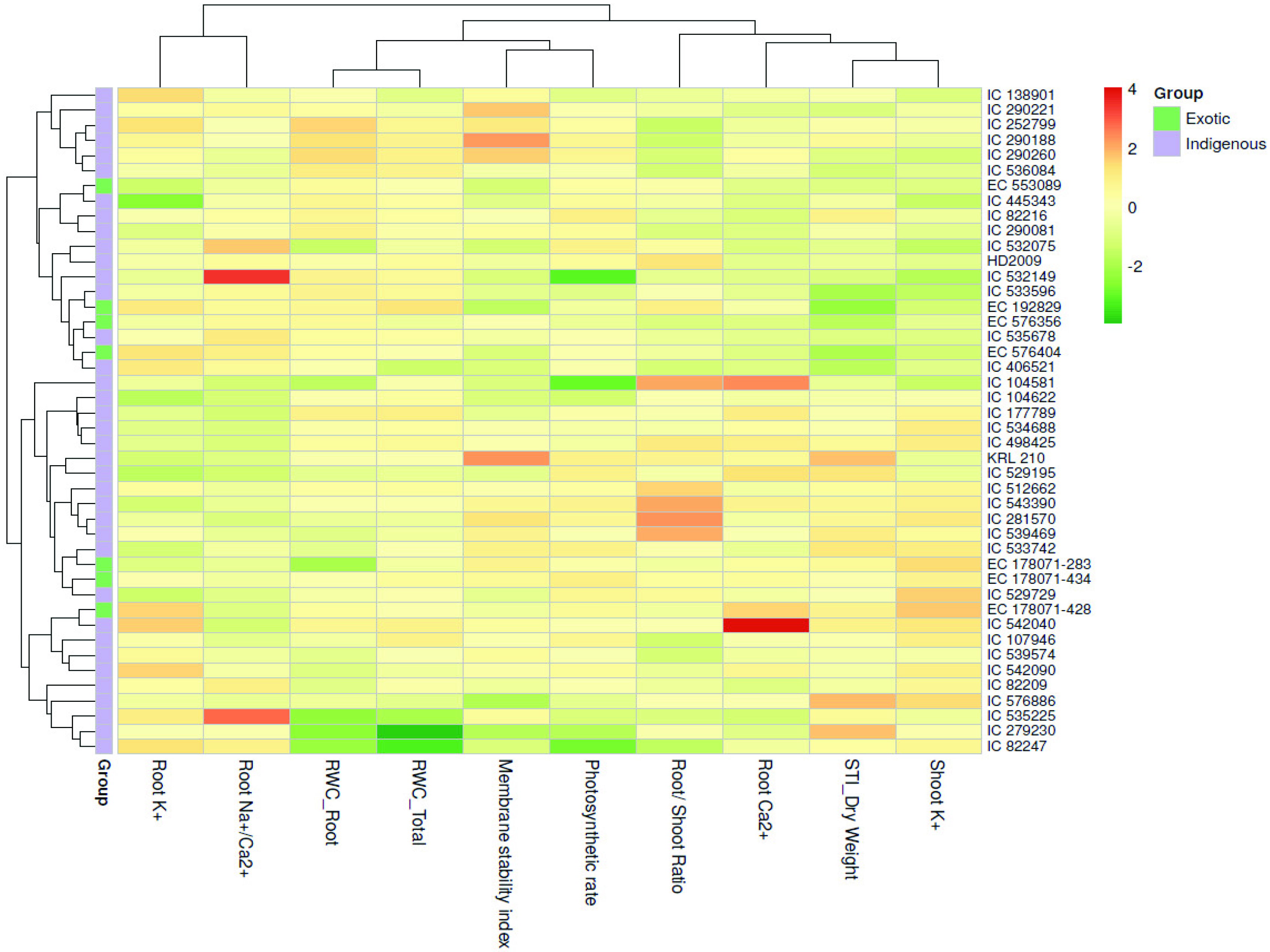
3.2. Categorization of Wheat Genotypes Based on Salt Tolerance Index (STI)
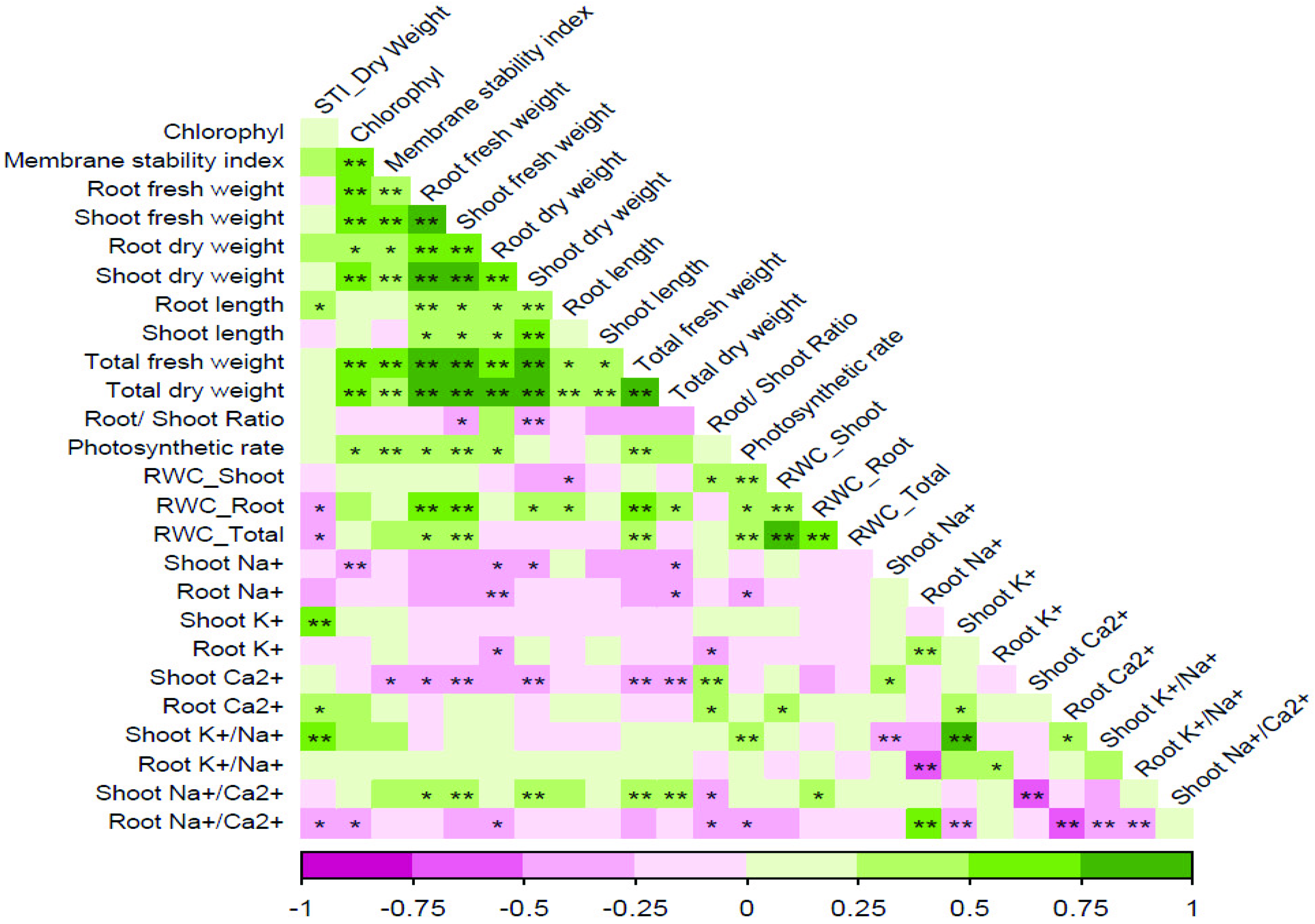
3.3. Salt Tolerance Index and Correlation between Traits under Salt Stress
3.4. Principal Component Analysis (PCA)
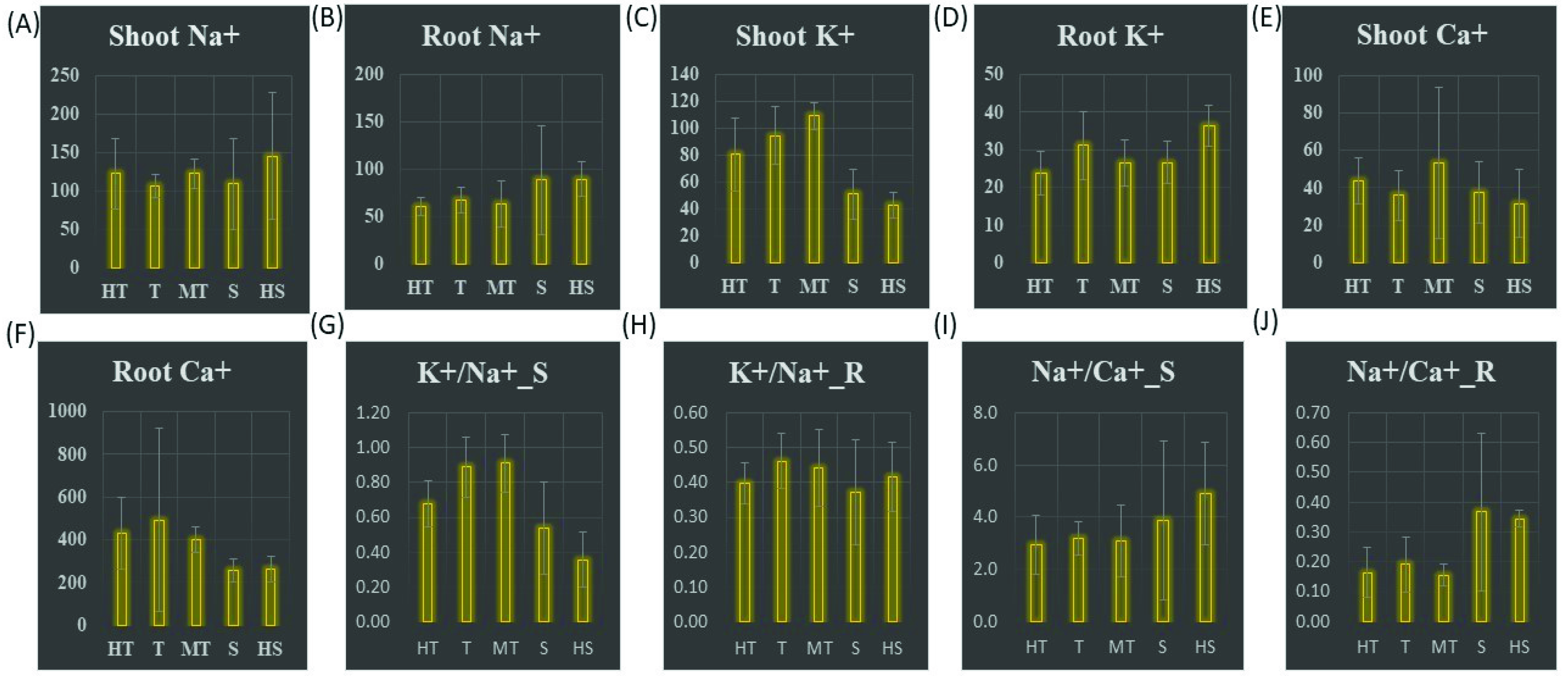
3.5. Ionic Concentrations among the Wheat Genotypes
3.6. Contribution of Traits in Salt Tolerance
4. Discussion
4.1. Phenotypic Trait Variation
4.2. Na+ and Ca2+ Interactions
4.3. Shoot K+/Na+ Response to Salt Stress
4.4. Important Traits for Salt Tolerance Evaluation
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| STI | Salt tolerance index based on TDW |
| CHL | Chlorophyll (SPAD-values) |
| MSI | Membrane stability index |
| RFW | Root fresh weight (g) |
| SFW | Shoot fresh weight (g) |
| RDW | Root dry weight (g) |
| SDW | Shoot dry weight (g) |
| TFW | Total fresh weight (g) |
| TDW | Total dry weight (g) |
| R/S | Root shoot ratio based on TDW |
| RWC_S | Relative water content of shoot (%) |
| RWC_R | Relative water content of root (%) |
| RWC_T | Total relative water content (%) |
| RL | Root length (cm) |
| SL | Shoot length (cm) |
| PS | Photochemical efficiency of PSII (μmols m−2 s−1) |
| Shoot Na+ | Na+ content of shoot (mg g−1) |
| Root Na+ | Na+ content of root (mg g−1) |
| Shoot K+ | K+ content of shoot (mg g−1) |
| Root K+ | K+ content of root (mg g−1) |
| Shoot Ca2+ | Ca2+ content of shoot (mg g−1) |
| Root Ca2+ | Ca2+ content of root (mg g−1) |
| K+/Na+_S | K+/Na+ ratio of shoot |
| K+/Na+_R | K+/Na+ ratio of root |
| Na+/Ca2+_S | Na+/Ca2+ ratio of shoot |
References
- Raman, H.; Stodart, B.J.; Cavanagh, C.; Mackay, M.; Morell, M.; Milgate, A.; Martin, P. Molecular diversity and genetic structure of modern and traditional landrace cultivars of wheat (Triticum aestivum L.). Crop. Pasture Sci. 2010, 61, 222–229. [Google Scholar] [CrossRef]
- Shewry, P.R.; Lafiandra, D.; Bedo, Z. Improving the nutritional quality and health benefits of wheat. Qual. Assur Saf. Crop. Foods 2012, 4, 136. [Google Scholar] [CrossRef]
- Ramadas, S.; Kumar, T.K.; Singh, G.P. Wheat Production in India: Trends and Prospects. In Recent Advances in Grain Crops Research; Shah, F., Khan, Z., Iqbal, A., Turan, M., Olgun, M., Eds.; IntechOpen: Rijeka, Croatia, 2019. [Google Scholar]
- FAO. The Special Challenge for Sub-Saharan Africa. In How to Feed the World 2050; FAO: Rome, Italy, 2009. [Google Scholar]
- Zeeshan, M.; Lu, M.; Sehar, S.; Holford, P.; Wu, F. Comparison of biochemical, anatomical, morphological, and physiological responses to salinity stress in wheat and barley genotypes deferring in salinity tolerance. Agronomy 2020, 10, 127. [Google Scholar] [CrossRef]
- Qadir, M.; Quillérou, E.; Nangia, V.; Murtaza, G.; Singh, M.; Thomas, R.J.; Drechsel, P.; Noble, A.D. Economics of salt-induced land degradation and restoration. Nat. Resour. Forum 2014, 38, 282–295. [Google Scholar] [CrossRef]
- Kumar, A.; Kumar, A.; Mann, A.; Devi, G.; Sharma, H.; Singh, R.; Sanwal, S.K. Phytoamelioration of the salt-affected soils through halophytes. In Ecophysiology, Abiotic Stress Responses and Utilization of Halophytes; Springer: Singapore, 2019; pp. 313–326. [Google Scholar]
- Munns, R.; Tester, M. Mechanisms of salinity tolerance. Annu. Rev. Plant Biol. 2008, 59, 651–681. [Google Scholar] [CrossRef]
- Munns, R.; Gilliham, M. Salinity tolerance of crops—What is the cost? New Phytol. 2015, 208, 668–673. [Google Scholar] [CrossRef] [PubMed]
- Hauser, F.; Horie, T. A conserved primary salt tolerance mechanism mediated by HKT transporters: A mechanism for sodium exclusion and maintenance of high K+/Na+ ratio in leaves during salinity stress. Plant Cell Environ. 2010, 33, 552–565. [Google Scholar] [CrossRef] [PubMed]
- Roy, S.J.; Negrao, S.; Tester, M. Salt resistant crop plants. Curr. Opin. Biotechnol. 2014, 26, 115–124. [Google Scholar] [CrossRef] [PubMed]
- Pires, I.S.; Negrao, S.; Oliveira, M.M.; Purugganan, M.D. Comprehensive phenotypic analysis of rice (Oryza sativa) response to salinity stress. Physiol. Plant 2015, 155, 43–54. [Google Scholar] [CrossRef]
- Singh, A.K.; Singh, R.; Kumar, R.; Chaurasia, S.; Yadav, S.; Kumar, S.; Wankhede, D.P. Genomics technologies for improving salt tolerance in wheat. In Engineering Practices for Management of Soil Salinity Agricultural, Physiological, and Adaptive Approaches; Gupta, S.K., Goyal, M., Singh, A., Eds.; Apple Academic Press: Waretown, NJ, USA, 2018. [Google Scholar]
- Singh, A.K.; Chaurasia, S.; Kumar, S.; Singh, R.; Kumari, J.; Yadav, M.C.; Singh, N.; Gaba, S.; Jacob, S.R. Identification, analysis and development of salt responsive candidate gene based SSR markers in wheat. BMC Plant Biol. 2018, 18, 249. [Google Scholar] [CrossRef] [PubMed]
- El-Hendawy, S.E.; Hu, Y.C.; Yakout, G.M.; Awad, A.M.; Hafiz, S.E.; Schmidhalter, U. Evaluating salt tolerance of wheat genotypes using multiple parameters. Eur. J. Agron. 2005, 22, 243–253. [Google Scholar] [CrossRef]
- Deng, Y.Q.; Bao, J.; Yuan, F.; Liang, X.; Feng, Z.T.; Wang, B.S. Exogenous hydrogen sulfide alleviates salt stress in wheat seedlings by decreasing Na+, content. Plant Growth Regul. 2016, 79, 391–399. [Google Scholar] [CrossRef]
- Neves-Piestun Beatriz, G.; Bernstein, N. Salinity-induced inhibition of leaf elongation in maize is not mediated by changes in cell wall acidification capacity. Plant Physiol. 2001, 125, 1419–1428. [Google Scholar] [CrossRef] [PubMed]
- Ashraf, M.; Harris, P.J.C. Potential biochemical indicators of salinity tolerance in plants. Plant Sci. 2004, 166, 3–16. [Google Scholar] [CrossRef]
- Jaleel, C.A.; Gopi, R.; Manivannan, P.; Panneerselvam, R. Antioxidative potentials as a protective mechanism in Catharanthus roseus (L.) plants under salinity stress. Turk. J. Bot. 2007, 31, 245–251. [Google Scholar]
- Qiu, L. Evaluation of salinity tolerance and analysis of allelic function of HvHKT1 and HvHKT2 in Tibetan wild barley. Theor. Appl. Genet. 2011, 122, 695–703. [Google Scholar] [CrossRef]
- Meneguzzo, S.; Navari-Izzo, F.; Izzo, R. NaCl effects on water relations and accumulation of minerals in shoots, roots and cell sap of wheat seedling. J. Plant Physiol. 2000, 156, 711–716. [Google Scholar] [CrossRef]
- Ma, L.Q.; Zhou, E.F.; Huo, N.X.; Zhou, R.H.; Wang, G.Y.; Jia, J.Z. Genetic analysis of salt tolerance in a recombinant inbred population of wheat (Triticum aestivum L.). Euphytica 2007, 153, 109–117. [Google Scholar] [CrossRef]
- James, R.A.; Blake, C.; Byrt, C.S.; Munns, R. Major genes for Na+ exclusion, Nax1 and Nax2 (wheat HKT1;4 and HKT1;5), decrease Na+ accumulation in bread wheat leaves under saline and waterlogged conditions. J. Exp. Bot. 2011, 62, 2939–2947. [Google Scholar] [CrossRef]
- Munns, R.; James, R.A.; Xu, B.; Athman, A.; Conn, S.J.; Jordans, C.; Byrt, C.S.; Hare, R.A.; Tyerman, S.D.; Tester, M.; et al. Wheat grain yield on saline soils is improved by an ancestral Na+ transporter gene. Nat. Biotechnol. 2012, 30, 360–364. [Google Scholar] [CrossRef]
- Oyiga, B.C.; Sharma, R.C.; Shen, J.; Baum, M.; Ogbonnaya, F.C.; Leon, J.; Ballvora, A. Identification and characterization of salt tolerance of wheat germplasm using a multivariable screening approach. J. Agron. Crop. Sci. 2016, 202, 472–485. [Google Scholar] [CrossRef]
- Chaurasia, S.; Singh, A.K.; Songachan, L.S.; Sharma, A.D.; Bhardwaj, R.; Singh, K. Multi-locus genome-wide association studies reveal novel genomic regions associated with vegetative stage salt tolerance in bread wheat (Triticum aestivum L.). Genomics 2020, 112, 4608–4621. [Google Scholar] [CrossRef]
- Chaurasia, S.; Singh, A.K.; Kumar, A.; Songachan, L.S.; Yadav, M.C.; Kumar, S.; Kumari, J.; Bansal, R.; Sharma, P.C.; Kuldeep Singh, K. Genome-wide association mapping reveals key genomic regions for physiological and yield-related traits under salinity stress in wheat (Triticum aestivum L.). Genomics 2021, 113, 3198–3215. [Google Scholar] [CrossRef] [PubMed]
- Ghogdi, E.A.; Izadi-Darbandi, A.; Borzouei, A. Effects of salinity on some physiological traits in wheat (Triticum aestivum L.) cultivars. Indian J. Sci. Technol. 2012, 5, 1901–1906. [Google Scholar] [CrossRef]
- Tao, R.; Ding, J.; Li, C.; Zhu, X.; Guo, W.; Zhu, M. Evaluating and screening of agro-physiological indices for salinity stress tolerance in wheat at the seedling stage. Front. Plant Sci. 2021, 12, 646175. [Google Scholar] [CrossRef]
- Quan, X.; Liang, X.; Li, H.; Xie, C.; He, W.; Qin, Y. Identification and Characterization of Wheat Germplasm for Salt Tolerance. Plants 2021, 10, 268. [Google Scholar] [CrossRef] [PubMed]
- Uzair, M.; Ali, M.; Fiaz, S.; Attia, K.; Khan, N.; Al-Doss, A.A.; Khan, M.R.; Ali, Z. The Characterization of Wheat Genotypes for Salinity Tolerance Using Morpho-Physiological Indices under Hydroponic Conditions. Saudi J. Biol. Sci. 2022, 29, 103299. [Google Scholar] [CrossRef] [PubMed]
- Kaya, C.; Higgs, D. Calcium nitrate as a remedy for salt-stressed cucumber plants. J. Plant Nutr. 2002, 25, 861–871. [Google Scholar] [CrossRef]
- Cheong, Y.H.; Kim, K.N.; Pandey, G.K.; Gupta, R.; Grant, J.J.; Luan, S. CBL1, a calcium sensor that differentially regulates salt, drought, and cold responses in Arabidopsis. Plant Cell 2003, 15, 1833–1845. [Google Scholar] [CrossRef]
- White, P.J.; Broadley, M.R. Calcium in plants. Ann. Bot. 2003, 92, 487–511. [Google Scholar] [CrossRef] [PubMed]
- Shabala, S.; Demidchik, V.; Shabala, L.; Cuin, T.A.; Smith, S.J.; Miller, A.J.; Davies, J.M.; Newman, I.A. Extracellular Ca2+ ameliorates NaCl-induced K+ loss from Arabidopsis root and leaf cells by controlling plasma membrane K+ permeable channels. Plant Physiol. 2006, 141, 1653–1665. [Google Scholar] [CrossRef]
- Tuna, A.L.; Kaya, C.; Ashraf, M.; Altunlu, H.; Yokas, I.; Yagmur, B. The effects of calcium sulfate on growth, membrane stability and nutrient uptake of tomato plants grown under salt stress. Environ. Exp. Bot. 2007, 59, 173–178. [Google Scholar] [CrossRef]
- Zhao, M.G.; Tian, Q.Y.; Zhang, W.H. Nitric oxide synthase-dependent nitric oxide production is associated with salt tolerance in Arabidopsis. Plant Physiol. 2007, 144, 206–217. [Google Scholar] [CrossRef] [PubMed]
- Tattini, M.; Traversi, M.L. Responses to changes in Ca2+ supply in two Mediterranean evergreens, Phillyrea latifolia and Pistacia lentiscus, during salinity stress and subsequent relief. Ann. Bot. 2008, 102, 609–622. [Google Scholar] [CrossRef]
- Dubcovsky, J.; Santa Maria, G.; Epstein, E.; Luo, M.C.; Dvořák, J. Mapping of the K+/Na+ discrimination locus Knal in wheat. Theor. Appl. Genet. 1996, 92, 448–454. [Google Scholar] [CrossRef] [PubMed]
- Zhu, M.; Shabala, S.; Shabala, L.; Fan, Y.; Zhou, M.X. Evaluating predictive values of various physiological indices for salinity stress tolerance in wheat. J. Agron. Crop. Sci. 2016, 202, 115–124. [Google Scholar] [CrossRef]
- Feng, K.; Licao, C.; Shuzuo, L.; Jianxin, B.; Wang, M.; Song, W.; Xiaojun, N. Comprehensive evaluating of wild and cultivated emmer wheat (Triticum turgidum L.) genotypes response to salt stress. Plant Growth Regul. 2018, 84, 261–273. [Google Scholar] [CrossRef]
- Kulshreshtha, N.; Kumar, A.; Kumar, A.; Lata, C. Genetic Interventions to Improve Salt and Microelement Toxicity Tolerance in Wheat. In New Horizons in Wheat and Barley Research; Springer: Singapore, 2022. [Google Scholar]
- Goudarzi, M.; Pakniyat, H. Evaluation of wheat cultivars under salinity stress based on some agronomic and physiological traits. J. Agric. Soc. Sci. 2008, 1, 35–38. [Google Scholar]
- Sairam, R.K. Effect of moisture stress on physiological activities of two contrasting wheat genotypes. Indian J. Exp. Biol. 1994, 3, 584–593. [Google Scholar]
- Vogel, A.I. A Text-Book of Quantitative INORGANIC analysis: Theory and Practice; Longmans Green, and Co.: London, UK, 1951. [Google Scholar]
- Nyquist, W.E.; Baker, R.J. Estimation of heritability and prediction of selection response in plant populations. Crit. Rev. Plant Sci. 1991, 10, 235–322. [Google Scholar] [CrossRef]
- Chandna, R.; Azooz, M.; Ahmad, P. Recent advances of metabolomics to reveal plant response during salt stress. In Salt Stress in Plants; Springer: New York, NY, USA, 2014; pp. 1–14. [Google Scholar]
- Gomes-Filho, E.; Lima, C.R.F.M.; Costa, J.H.; da Silva, A.C.M.; Lima, M.D.G.S.; de Lacerda, C.F.; Prisco, J.T. Cowpea ribonuclease: Properties and effect of NaCl-salinity on its activation during seed germination and seedling establishment. Plant Cell Rep. 2008, 27, 147–157. [Google Scholar] [CrossRef]
- Shah, S.H.; Houborg, R.; McCabe, M.F. Response of chlorophyll, carotenoid and SPAD-502 measurement to salinity and nutrient stress in wheat (Triticum aestivum L.). Agronomy 2017, 7, 61. [Google Scholar] [CrossRef]
- Davenport, R.; James, R.A.; Zakrisson-Plogander, A.; Tester, M.; Munns, R. Control of sodium transport in durum wheat. Plant Physiol. 2005, 137, 807–818. [Google Scholar] [CrossRef] [PubMed]
- James, R.A.; Davenport, R.J.; Munns, R. Physiological characterization of two genes for Na+ exclusion in durum wheat, Nax1 and Nax2. Plant Physiol. 2006, 142, 1537–1547. [Google Scholar] [CrossRef] [PubMed]
- Genc, Y.; Oldach, K.; Verbyla, A.P.; Lott, G.; Hassan, M.; Tester, M.; Wallwork, H.; McDonald, G.K. Sodium exclusion QTL associated with improved seedling growth in bread wheat under salinity stress. Theor. Appl. Genet. 2010, 121, 877–894. [Google Scholar] [CrossRef]
- Long, N.V.; Dolstra, O.; Malosetti, M.; Kilian, B.; Graner, A.; Visser, R.G.; van der Linden, C.G. Association mapping of salt tolerance in barley (Hordeum vulgare L.). Theor. Appl. Genet. 2013, 126, 2335–2351. [Google Scholar] [CrossRef]
- Buschmann, P.H.; Vaidyanathan, R.; Gassmann, W.; Schroeder, J.I. Enhancement of Na+ uptake currents, time-dependent inward-rectifying K+ channel currents, and K+ channel transcripts by K+ starvation in wheat root cells. Plant Physiol. 2000, 122, 1387–1398. [Google Scholar] [CrossRef]
- Reid, R.J.; Smith, F.A. The limits of sodium/calcium interactions in plant growth. Funct. Plant Biol. 2000, 27, 709–715. [Google Scholar] [CrossRef]
- Cramer, G.R. Sodium-calcium interactions under salinity stress. In Salinity, Environment-Plants-Molecules; Lauchli, A., Luttge, U., Eds.; Kluwer: Dordrecht, The Netherlands, 2002; pp. 205–227. [Google Scholar]
- Nedjimi, B.; Daoud, Y. Ameliorative effect of CaCl2 on growth, membrane permeability and nutrient uptake in Atriplex halimus subsp. schweinfurthii grown at high (NaCl) salinity. Desalination 2009, 249, 163–166. [Google Scholar] [CrossRef]
- Jin, Z.M.; Wang, C.H.; Liu, Z.P.; Gong, W.J. Physiological and ecological characters studies on Aloe vera under soil salinity and seawater irrigation. Process Biochem. 2007, 42, 710–714. [Google Scholar] [CrossRef]
- Murillo-Amado, B.; Jones, H.G.; Kaya, C.; Aguilar, R.L.; Garcia-Hernandez, J.L.; Troyo-Di’eguez, E.; Avila-Serrano, N.Y.; Rueda-Puente, E. Effects of foliar application of calcium nitrate on growth and physiological attributes of cowpea (Vigna unguiculata L. Walp.) grown under salt stress. Environ. Exp. Bot. 2006, 58, 188–196. [Google Scholar] [CrossRef]
- Allen, G.J.; Wyn-Jones, R.G.; Leigh, R.A. Sodium transport in plasma membrane vesicles isolated from wheat genotypes with differing K+/Na+ discrimination traits. Plant Cell Environ. 1995, 18, 105–115. [Google Scholar] [CrossRef]
- Hadi, M.R.; Khoush, K.S.N.; Khavarinezhad, R.A.; Khayam, N.S. The effect of elements accumulation on salinity tolerance in seven genotype durum wheat (Triticum turgidum L.) collected from the of middle east. Iran J. Biotechnol. 2008, 21, 1–15. [Google Scholar]
- Tester, M.; Davenport, R. Na+ tolerance and Na+ transport in higher plants. Ann. Bot. 2003, 91, 503–527. [Google Scholar] [CrossRef] [PubMed]
- Anschutz, U.; Becker, D.S. Going beyond nutrition: Regulation of potassium homoeostasis as a common denominator of plant adaptive responses to environment. J. Plant Physiol. 2014, 171, 670–687. [Google Scholar] [CrossRef] [PubMed]
- Cherel, I.; Lefoulon, C.; Boeglin, M.; Sentenac, H. Molecular mechanisms involved in plant adaptation to low K+ availability. J. Exp. Bot. 2014, 65, 833–848. [Google Scholar] [CrossRef] [PubMed]
- Kanawapee, N.; Sanitchon, J.; Lontom, W.; Threerakulpisut, P. Evaluation of salt tolerance at the seedling stage in rice genotypes by growth performance, ion accumulation, proline and chlorophyll content. Plant Soil. 2012, 358, 235–249. [Google Scholar] [CrossRef]
- Munns, R. Comparative physiology of salt and water stress. Plant Cell Environ. 2002, 25, 239–250. [Google Scholar] [CrossRef] [PubMed]
- Cuin, T.A.; Betts, S.A.; Chalmandrier, R.; Shabala, S. A root’s ability to retain K+ correlates with salt tolerance in wheat. J. Exp. Bot. 2008, 59, 2697–2706. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.W. Evolution of physiological responses to salt stress in hexaploid wheat. Proc. Natl. Acad. Sci. USA 2014, 111, 11882–11887. [Google Scholar] [CrossRef]
- Genc, Y.; McDonald, G.K.; Tester, M. Reassessment of tissue Na(+) concentration as a criterion for salinity tolerance in bread wheat. Plant Cell Environ. 2007, 30, 1486–1498. [Google Scholar] [CrossRef]
- Munns, R.; James, R.A. Screening methods for salinity tolerance: A case study with tetraploid wheat. Plant Soil 2003, 253, 201–218. [Google Scholar] [CrossRef]
- Shen, Q.; Fu, L.; Qiu, L.; Xue, F.; Zhang, G.P.; Wu, D. Time-course of ionic responses and proteomic analysis of a Tibetan wild barley at early stage under salt stress. Plant Growth Regul. 2016, 81, 11–21. [Google Scholar] [CrossRef]
- Shabala, S.; Shabala, S.; Cuin, T.A.; Pang, J.; Percey, W.; Chen, Z.; Conn, S.; Eing, C.; Wegner, L.H. Xylem ionic relations and salinity tolerance in barley. Plant J. 2010, 61, 839–853. [Google Scholar] [CrossRef] [PubMed]
- Farissi, M.; Faghire, M.; Bargaz, A.; Bouizgaren, A.; Makoudi, B.; Sentenac, H.; Growth, G.C. Nutrients concentrations, and enzymes involved in plants nutrition of alfalfa populations under saline conditions. J. Agric. Sci. Technol. 2014, 16, 301–314. [Google Scholar]
- Talei, D.; Valdiani, A.; Yusop, M.K.; Abdullah, M.P. Estimation of salt tolerance in Andrographis paniculata accessions using multiple regression model. Euphytica 2013, 189, 147–160. [Google Scholar] [CrossRef]
- Çiçek, N.; Çakirlar, H. The effect of salinity on some physiological parameters in two maize cultivars. Bulg. J. Plant Physiol. 2002, 28, 66–74. [Google Scholar]
- Hossain, A.A.; Halim, M.A.; Hossain, F.; Maher Niger, M.A. Effect of NaCl salinity on some physiological characters of wheat (Triticum aestivum L.). Bangladesh J. Bot. 2006, 35, 9–15. [Google Scholar]
- Asfaw, K.G. Effects of salinity on seedling biomass production and relative water content of twenty sorghum (Sorghum biolor L. Moench) accessions. Asian J. Agric. Sci. 2011, 3, 242–249. [Google Scholar]
- Win, K.T.; Oo, A.Z.; Hirasawa, T.; Ookawa, T.; Yutaka, H. Genetic analysis of Myanmar vigna species in responses to salt stress at the seedling stage. Afr. J. Biotechnol. 2011, 10, 1615–1624. [Google Scholar]

| S. No | Traits | Genotypes (G) | Salinity Treatments (T) | Genotype × Treatment |
|---|---|---|---|---|
| df | 43 | 1 | 43 | |
| 1 | CHL | 41.028 ** | 649.89 ** | 10.90 ** |
| 2 | MSI | 685.03 ** | 45900.15 ** | 209.11 ** |
| 3 | RFW | 0.48 ** | 5.53 ** | 0.13 ** |
| 4 | SFW | 5.62 ** | 279.36 ** | 3.71 ** |
| 5 | TFW | 8.92 ** | 363.52 ** | 4.94 ** |
| 6 | RDW | 0.006 ** | 0.074 ** | 0.002 ** |
| 7 | SDW | 0.07 ** | 1.73 ** | 0.04 ** |
| 8 | TDW | 0.11 ** | 2.52 ** | 0.05 ** |
| 9 | R/S | 0.13 ** | 1.27 ** | 0.04 ** |
| 10 | RL | 69.90 ** | 1392.74 ** | 44.68 ** |
| 11 | SL | 119.00 ** | 8684.74 ** | 46.58 ** |
| 12 | PS | 5234.92 ** | 227274.68 ** | 3654.37 ** |
| 13 | RWC_S | 78.90 ** | 2484.55 ** | 27.71 ** |
| 14 | RWC_R | 314.79 ** | 459.89 ** | 65.31 ** |
| 15 | RWC_T | 86.78 ** | 2293.51 ** | 22.53 ** |
| 16 | Shoot Na+ | 2344.95 ** | 692762.80 ** | 2064.73 ** |
| 17 | Root Na+ | 1593.13 ** | 215913.41 ** | 751.39 ** |
| 18 | Shoot K+ | 5014.66 ** | 233.04 * | 1568.55 ** |
| 19 | Root K+ | 1354.47 ** | 25986.23 ** | 952.10 ** |
| 20 | Shoot Ca+ | 1865.33 ** | 4383.89 ** | 556.15 ** |
| 21 | Root Ca+ | 166824.19 ** | 36555.03 ** | 43997.20 ** |
| 22 | K+/Na+_S | 7.75 ** | 1642.41 ** | 6.86 ** |
| 23 | K+/Na+_R | 4.39 ** | 529.30 ** | 4.30 ** |
| 24 | Na+/Ca+_S | 5.17 ** | 634.66 ** | 4.23 ** |
| 25 | Na+/Ca+_R | 0.05 ** | 2.08 ** | 0.02 ** |
| Trait | Treatment | STI | TDW | SDW | RDW |
|---|---|---|---|---|---|
| Shoot Na+ | Salt | NS | −0.371 ** | −0.355 ** | −0.317 ** |
| Control | −0.64 ** | −0.622 ** | −0.528 ** | ||
| Root Na+ | Salt | −0.244 * | −0.317 ** | −0.233 * | −0.443 ** |
| Control | −0.224 * | NS | −0.445 ** | ||
| Shoot K+ | Salt | 0.60 ** | NS | −0.197 * | NS |
| Control | −0.536 ** | −0.562 ** | −0.271 * | ||
| Root K+ | Salt | NS | NS | NS | −0.30 ** |
| Control | 0.436 ** | 0.473 ** | NS | ||
| Shoot Ca2+ | Salt | NS | −0.442 ** | −0.473 ** | −0.26* |
| Control | −0.632 ** | −0.649 ** | −0.379 ** | ||
| Root Ca2+ | Salt | 0.31 ** | NS | −0.189 * | NS |
| Control | NS | NS | NS | ||
| Shoot K+/Na+ | Salt | 0.533 ** | NS | NS | NS |
| Control | NS | NS | NS | ||
| Root K+/Na+ | Salt | NS | NS | NS | NS |
| Control | 0.51 ** | 0.491 ** | 0.443 ** | ||
| Shoot Na+/Ca2+ | Salt | −0.178 * | 0.40 ** | 0.459 ** | NS |
| Control | 0.245 * | 0.29 * | NS | ||
| Root Na+/Ca2+ | Salt | −0.361 ** | NS | NS | −0.351 ** |
| Control | −0.226 * | −0.181 * | −0.344 ** |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chaurasia, S.; Kumar, A.; Singh, A.K. Comprehensive Evaluation of Morpho-Physiological and Ionic Traits in Wheat (Triticum aestivum L.) Genotypes under Salinity Stress. Agriculture 2022, 12, 1765. https://doi.org/10.3390/agriculture12111765
Chaurasia S, Kumar A, Singh AK. Comprehensive Evaluation of Morpho-Physiological and Ionic Traits in Wheat (Triticum aestivum L.) Genotypes under Salinity Stress. Agriculture. 2022; 12(11):1765. https://doi.org/10.3390/agriculture12111765
Chicago/Turabian StyleChaurasia, Shiksha, Arvind Kumar, and Amit Kumar Singh. 2022. "Comprehensive Evaluation of Morpho-Physiological and Ionic Traits in Wheat (Triticum aestivum L.) Genotypes under Salinity Stress" Agriculture 12, no. 11: 1765. https://doi.org/10.3390/agriculture12111765
APA StyleChaurasia, S., Kumar, A., & Singh, A. K. (2022). Comprehensive Evaluation of Morpho-Physiological and Ionic Traits in Wheat (Triticum aestivum L.) Genotypes under Salinity Stress. Agriculture, 12(11), 1765. https://doi.org/10.3390/agriculture12111765







