Distribution, Genetic Diversity and Population Structure of Aegilops tauschii Coss. in Major Wheat-Growing Regions in China
Abstract
1. Introduction
2. Materials and Methods
2.1. Aegilops tauschii Coss. Field Survey in China
2.2. Collection of Aegilops tauschii Coss. Seeds
2.3. DNA Extraction
2.4. SSR Primer Pairs Screening
2.5. SSR Detection
2.6. Data Analyses
3. Results
3.1. The Distribution and Occurrence of Aegilops tauschii Coss. in China
3.2. Polymorphism of the SSR Markers
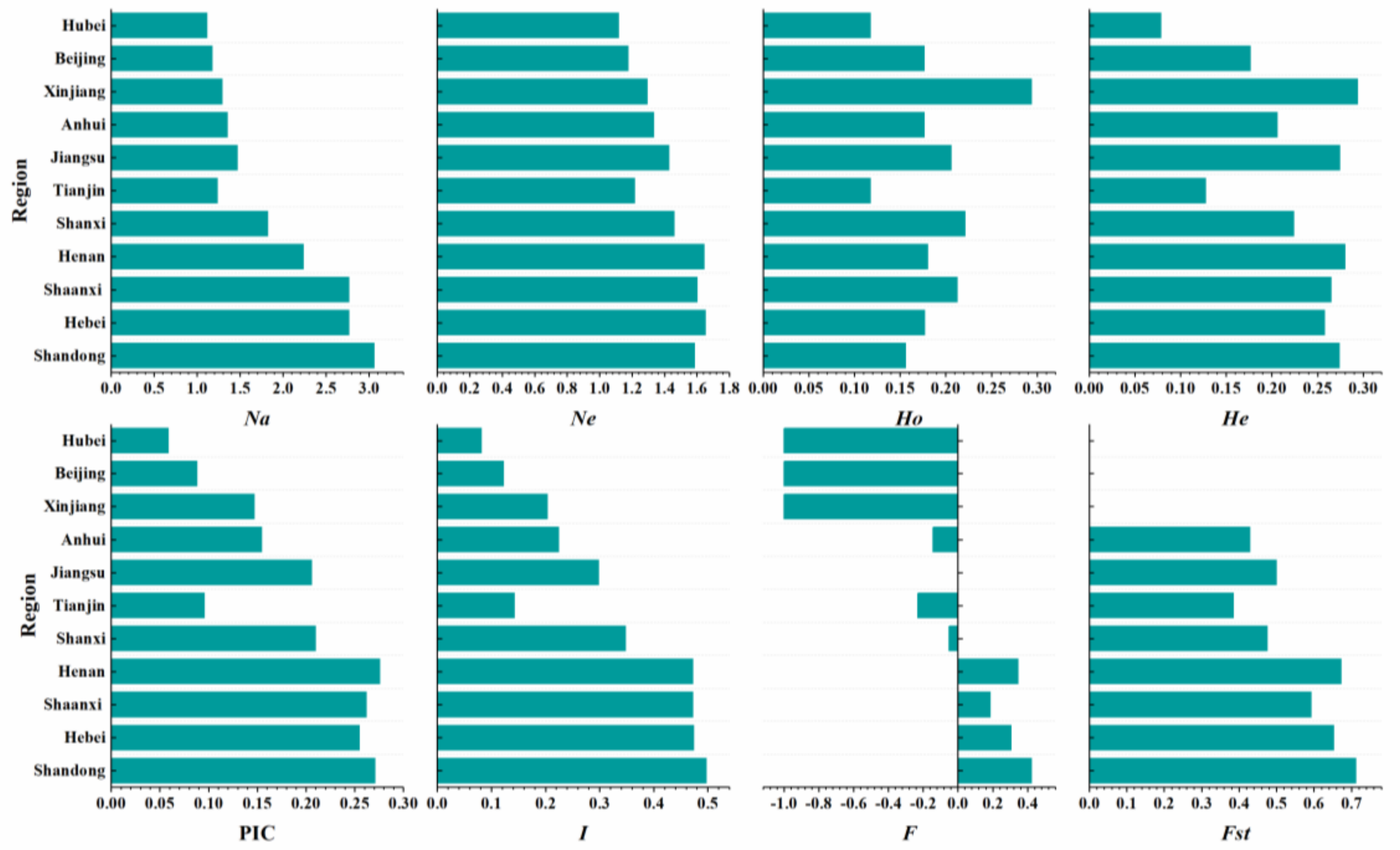
3.3. Comparative Genetic Diversity Analysis of Aegilops tauschii Coss. from Different Provinces
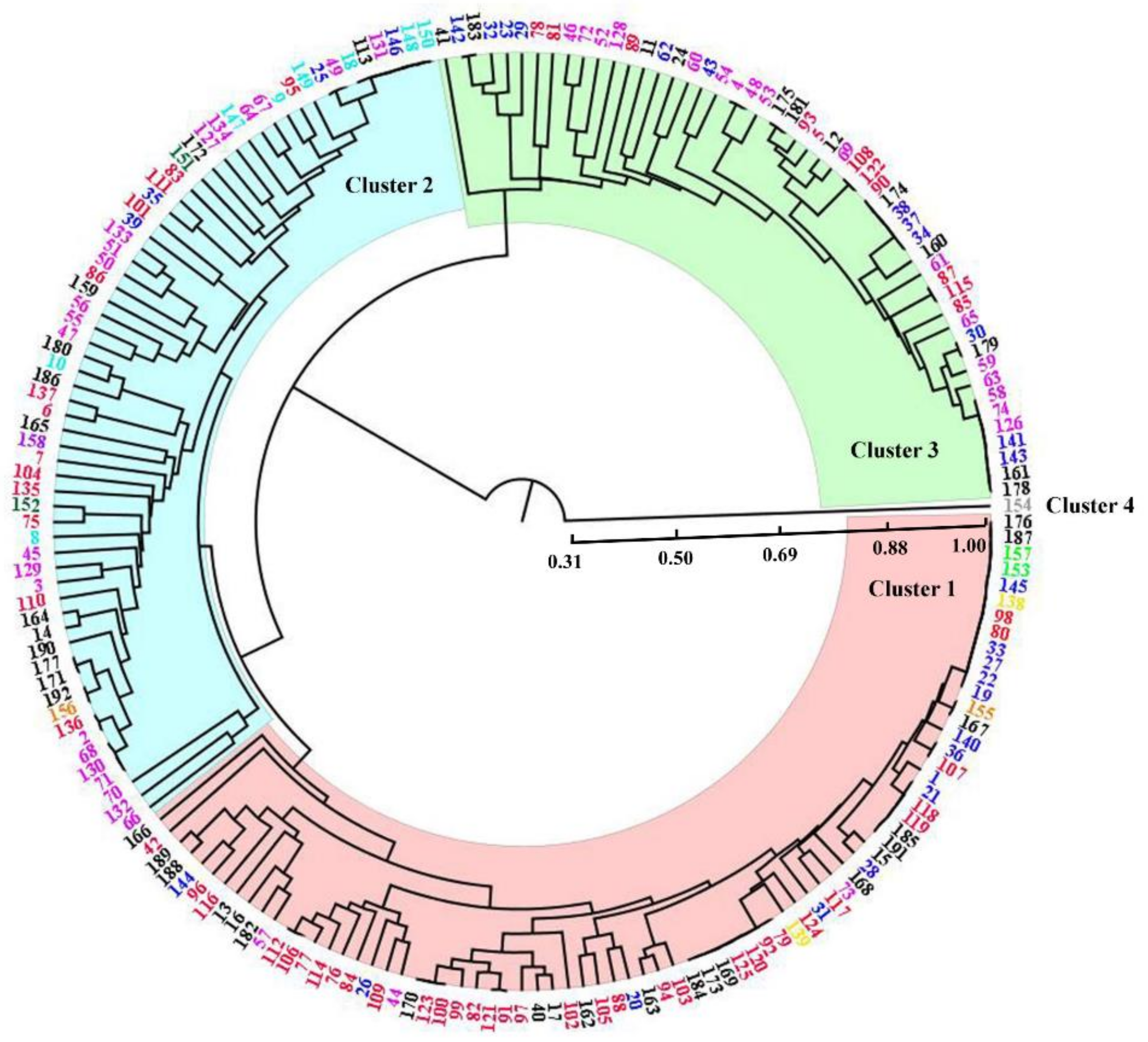
3.4. Cluster Analysis
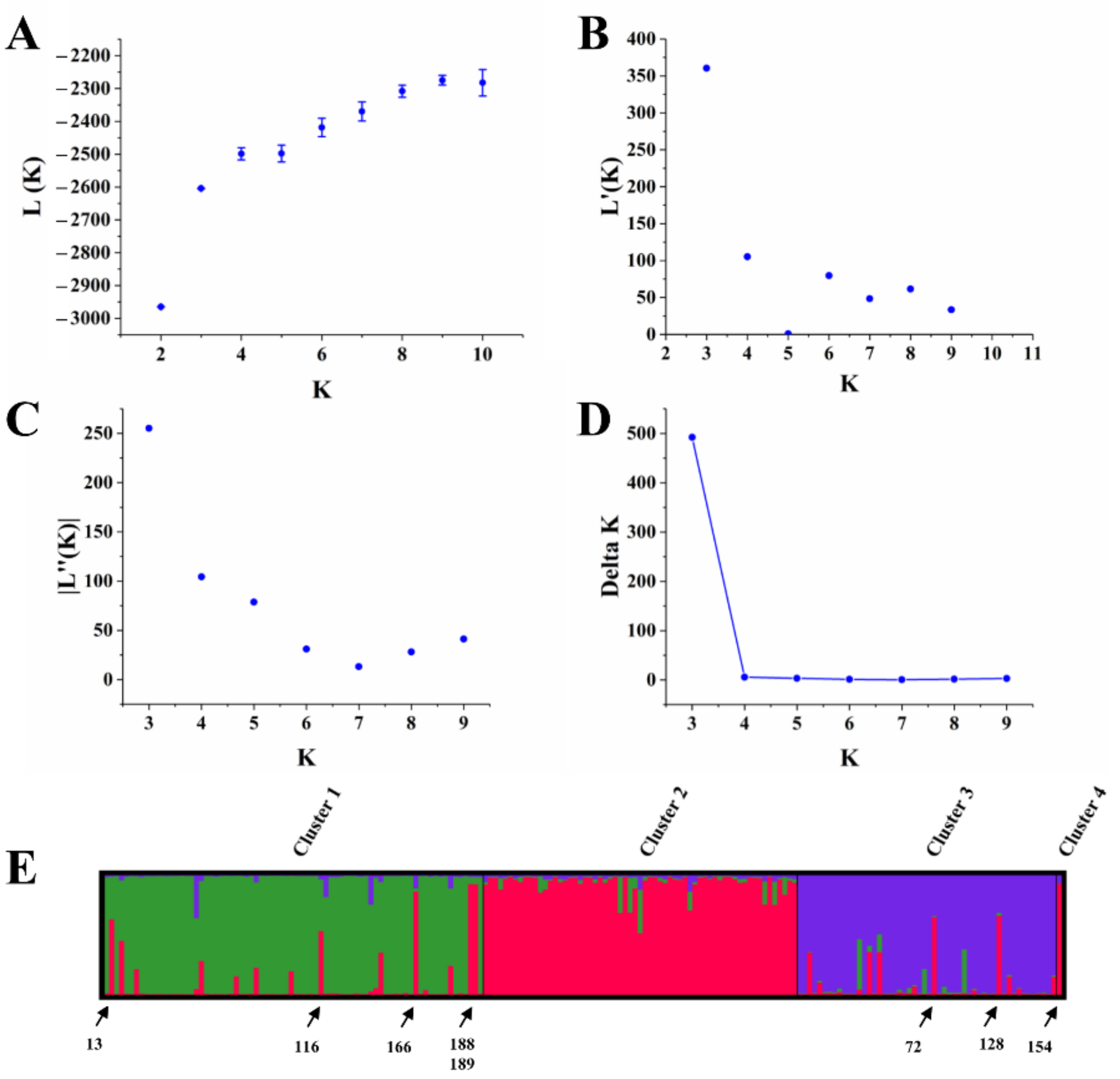
3.5. Population Structure Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zhang, C.X.; Li, X.J.; Huang, H.J.; Wei, S.H. Alert and prevention of the spreading of Aegilops tauschii, a worst weed in wheat field. Acta Phytophylacica Sin. 2007, 34, 103–106. [Google Scholar]
- Sohail, Q.; Shehzad, T.; Kilian, A.; Eltayeb, A.E.; Tanaka, H.; Tssujimoto, H. Development of diversity array technology (DArT) markers for assessment of population structure and diversity in Aegilops tauschii. Breed. Sci. 2012, 62, 38–45. [Google Scholar] [CrossRef]
- Yu, H.Y.; Li, X.J. Distribution of Aegilops tauschii Coss. China Res. Prog. J. Weed Sci. 2018, 36, 1–7. [Google Scholar]
- Fang, F.; Zhang, C.X.; Huang, H.J.; Li, M.; Gao, X.; Li, Y.; Wei, S.H. The occurrence of Tausch’s goatgrass (Aegilops tauschii Coss.) in wheat fields and its effect on wheat yield. Acta Ecol. Sin. 2014, 34, 3917–3923. [Google Scholar]
- Yang, J.; Yu, H.Y.; Li, X.J.; Dong, J.G. Genetic diversity and population structure of Commelina communis in China based on simple sequence repeat markers. J. Integr. Agr. 2018, 17, 2292–2301. [Google Scholar] [CrossRef]
- Zhao, S.Y.; Sun, S.G.; Dai, C.; Gituru, R.W.; Chen, J.M.; Wang, Q.F. Genetic variation and structure in native and incasive Solidago canadensis populations. Weed Res. 2015, 55, 163–172. [Google Scholar] [CrossRef]
- Matsuoka, Y.; Takumi, S.; Kawahara, T. Flowering time diversification and dispersal in central Eurasian wild wheat Aegilops tauschii Coss.: Genealogical and ecoligical framework. PLoS ONE 2008, 3, e3138. [Google Scholar] [CrossRef]
- Yan, J.; Yang, J.L.; Cui, N.R.; Zhong, J.P.; Dong, Y.C.; Sun, Y.Z.; Zhong, G.Y. The Aegilops tauschii Cosson from Yi-Li, Xinjiang, China. Acta Agron. Sin. 1984, 10, 1–8. [Google Scholar]
- Li, Y.G.; Sun, Y.Z.; Zhang, D.L.; Li, S.P. Genetic diversity of Aegilops tauschii accessions native to China revealed by ISSR markers. J. Triticeae Crop. 2017, 37, 30–39. [Google Scholar]
- Wei, H.; Li, J.; Peng, Z.; Lu, B.; Zhao, Z.; Yang, W. Relationships of Aegilops tauschii revealed by DNA fingerprints: The evidence for agriculture exchange between China and the West. Prog. Nat. Sci. 2008, 18, 1525–1531. [Google Scholar] [CrossRef]
- Staub, J.E.; Meglic, V. Molecular genetic markers and their legal relevance for cultivar discrimination: A case study in cucumber. HortTechnology 1993, 3, 291–300. [Google Scholar] [CrossRef]
- Liu, X.B.; Feng, B.; Li, J.; Yan, C.; Yang, Z.L. Genetic diversity and breeding history of Winter Mushroom (Flammulina velutipes) in China uncovered by genomic SSR markers. Gene 2016, 591, 227–235. [Google Scholar] [CrossRef] [PubMed]
- Zane, L.; Bargelloni, L.; Patarnello, T. Strategies for microsatellite isolation: A review. Mol. Ecol. 2002, 11, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Selkoe, K.A.; Toonen, R.J. Microsatellites for ecologists: A practical guide to using and evaluating microsatellite markers. Ecol. Lett. 2006, 9, 615–629. [Google Scholar] [CrossRef]
- Luo, M.; Gu, Y.Q.; Puiu, D.; Wang, H.; Twardziok, S.O.; Deal, K.R.; Huo, N.; Zhu, T.; Wang, L.; Wang, Y.; et al. Genome sequence of the progenitor of the wheat D genome Aegilops tauschii. Nature 2017, 551, 498–502. [Google Scholar] [CrossRef] [PubMed]
- Pandian, S.; Satish, L.; Rameshkumar, R.; Muthuramalingam, P.; Rency, A.S.; Rathinapriya, P.; Ramesh, M. Analysis of population structure and genetic diversity in an exotic germplasm collection of Eleusine coracana (L.) Gaertn. using genic-SSR markers. Gene 2018, 653, 80–90. [Google Scholar] [CrossRef] [PubMed]
- Noormohammadi, Z.; Trujillo, I.; Belaj, A.; Ataei, S.; Hosseini-Mazinan, M. Genetic structure of Iranian olive cultivars and their relationship with Mediterranean’s cultivars revealed by SSR markers. Sci. Hortic. 2014, 178, 175–183. [Google Scholar] [CrossRef]
- Pestsova, E.; Ganal, M.W.; Röder, M.S. Isolation and mapping of microsatellite markers specific for the D genome of bread wheat. Genome 2000, 43, 689–697. [Google Scholar] [CrossRef]
- Röder, M.S.; Korzun, V.; Wendehake, K.; Plaschke, J.; Tixier, M.; Leroy, P.; Ganal, M.W. A microsatellite map of wheat. Genetics 1998, 149, 2007–2023. [Google Scholar]
- Nei, M. Analysis of gene diversity in subdivided population. Proc. Natl. Acad. Sci. USA 1973, 70, 3321–3323. [Google Scholar] [CrossRef]
- Pritchard, J.K.; Stephens, M.; Donnelly, P. Inference of population structure using multilocus genotype data. Genetics 2000, 155, 945–959. [Google Scholar]
- Yu, H.; Yang, J.; Cui, H.; Li, Z.; Jia, F.; Chen, J.; Li, X. Effects of plant density on tillering in the weed grass Aegilops tauschii Coss. and its phytohormonal regulation. Plant Physiol. Bioch. 2020, 157, 70–78. [Google Scholar] [CrossRef]
- Tang, X.T.; Tao, H.H.; Du, Y.Z. Microsatellite-based analysis of the genetic structure and diversity of Aleurocanthus spiniferus (Hemiptera: Aleyrodidae) from tea plants in China. Gene 2015, 560, 107–113. [Google Scholar] [CrossRef]
- Botstein, D.; White, R.L.; Skolnick, M.; Davis, R.W. Construction of a genetic linkage map in man using restriction fragment length polymorphisms. Am. J. Hum. Genet. 1980, 32, 314–331. [Google Scholar]
- Lelley, T.; Stachel, M.; Grausgruber, H.; Vollmann, J. Analysis of relationships between Aegilops tauschii and the D genome of wheat utilizing microsatellites. Genome 2000, 43, 661–668. [Google Scholar] [CrossRef] [PubMed]
- Naghavi, M.R.; Nardi, M.; Pirseyedi, S.M.; Kazemi, M.; Potki, P.; Ghaffari, M.R. Comparison of genetic variation among accessions of Aegilops tauschii using AFLP and SSR markers. Genet. Resour. Crop Evol. 2007, 54, 237–240. [Google Scholar] [CrossRef]
- Saeidi, H.; Rahiminejad, M.R.; Vallian, S.; Heslop-Harrison, J.S. Biodiversity of diploid D-genome Aegilops tauschii Coss. in Iran measured using microsatellites. Genet. Resour. Crop Evol. 2006, 53, 1477–1484. [Google Scholar] [CrossRef]
- Hamrick, J.L.; Didt, M.J.W. Effects of life history traits on gentic diversity in plant species. Philos. Trans. R. Soc. Lond. Ser. B 1996, 351, 1291–1298. [Google Scholar]
- Wang, J.R.; Luo, M.C.; Chen, Z.X.; You, F.M.; Wei, Y.M.; Zheng, Y.L.; Dvorak, J. Aegilops tauschii single nucleotide polymorphisms shed light on the origins of wheat D-genome genetic diversity and pinpoint the geographic origin of hexaploid wheat. New Phytol. 2013, 198, 925–937. [Google Scholar] [CrossRef]
- Lan, X.J.; Liu, D.C.; Wei, Y.M.; Yan, Z.H.; Zheng, Y.L. Diversity of esterase isozyme in Aegilops tauschii Cosson. Guihaia 2001, 21, 77–80. [Google Scholar]
- Fang, F.; Gao, X.X.; Wei, S.H.; Li, Y.; Li, M.; Zhang, C.X. Occurrence and effects of Aegilops tauschii in China. Acta Prataculturae Sin. 2015, 24, 194–201. [Google Scholar]
- Breton, C.; Pinatel, C.; Médail, F.; Bonhomme, F.; Bervillé, A. Comparison between classical and Bayesian methods to investigate the history of olive cultivars using SSR-polymorphisms. Plant Sci. 2008, 175, 524–532. [Google Scholar] [CrossRef]
- Belaj, A.; Muñoz-diez, C.; Baldoni, L.; Porceddu, A.; Barranco, D.; Satovic, Z. Genetic diversity and population structure of wild olives from the North-western Mediterranean assessed by SSR markers. Ann. Bot. 2007, 100, 449–458. [Google Scholar] [CrossRef] [PubMed]
| Province | Year | Month | Occurrence Frequency (%) | Number of Populations |
|---|---|---|---|---|
| Shaanxi | 2017 | 5–6 | 56.96 | 43 |
| Henan | 2017 | 5–6 | 92.85 | 30 |
| Anhui | 2017 | 5 | 8.57 | 2 |
| Jiangsu | 2017 | 5 | 14.00 | 2 |
| Xinjiang | 2017 | 8 | 1.47 | 1 |
| Hubei | 2017 | 5 | 3.17 | 2 |
| Sichuan | 2017 | 4–5 | 0.91 | 0 |
| Shandong | 2018 | 6 | 87.07 | 58 |
| Hebei | 2018 | 6 | 75.24 | 43 |
| Shanxi | 2018 | 6 | 11.76 | 8 |
| Beijing | 2019 | 6 | 25.00 | 1 |
| Tianjin | 2019 | 6 | 12.50 | 2 |
| Total | —— | —— | —— | 192 |
| Primer Name | Primer Sequence (5′–3′) 1 | Tm (°C) | 5′ Modify | Allele Size (bp) |
|---|---|---|---|---|
| Xgdm153-5D | F-TATAGGCAAATTAATTAAGACG | 50.3 | hex | 208–277 |
| R-ATCTTTATGTGAGTACACTGC | ||||
| Xgwm314-3D | F-AGGAGCTCCTCTGTGCCAC | 61.1 | fam | 160–181 |
| R-TTCGGGACTCTCTTCCCTG | ||||
| Xgwm295-7D | F-GTGAAGCAGACCCACAACAC | 60 | fam | 232–254 |
| R-GACGGCTGCGACGTAGAG | ||||
| Xgwm102-2D | F-TCTCCCATCCAACGCCTC | 58 | hex | 138–147 |
| R-TGTTGGTGGCTTGACTATTG | ||||
| Xgdm111-1D | F-CACTCACCCCAAACCAAAGT | 55.3 | fam | 186–193 |
| R-GATGCAATCGGGTCGTTAGT | ||||
| Xgdm129-4D | F-GAGCAGGCAGCAGCTAGC | 60 | fam | 101–111 |
| R-TGCATCATCATCGGTCAAGT | ||||
| Xgwm165-4D | F-TGCAGTGGTCAGAGTTTTCC | 55.3 | hex | 189–193 |
| R-CTTTTCTTTCAGATTGCGCC | ||||
| Xgwm157-2D | F-GTCGTCGCGGTAAGCTTG | 56.7 | hex | 98–100 |
| R-GAGTGAACACACGAGGCTTG | ||||
| Xgwm383-3D | F-ACGCCAGTTGATCCGTAAAC | 60 | hex | 215–230 |
| R-GACATCAATAACCGTGGATGG | ||||
| Xgdm33-1D | F-GGCTCAATTCAACCGTTCTT | 60 | hex | 122–204 |
| R-TACGTTCTGGTGGCTGCTC | ||||
| Xgwm182-5D | F-TGATGTAGTGAGCCCATAGGC | 58 | fam | 148–171 |
| R-TTGCACACAGCCAAATAAGG | ||||
| Xgwm539-2D | F-CTGCTCTAAGATTCATGCAACC | 58.5 | fam | 98–106 |
| R-GAGGCTTGTGCCCTCTGTAG | ||||
| Xgwm469-6D | F-CAACTCAGTGCTCACACAACG | 57 | hex | 151–205 |
| R-CGATAACCACTCATCCACACC | ||||
| Xgwm55-6D | F-GCATCTGGTACACTAGCTGCC | 56.9 | hex | 124–136 |
| R-TCATGGATGCATCACATCCT | ||||
| Xgdm126-1D | F-TCCATCATATCCGTAGCACA | 54.1 | fam | 178–216 |
| R-CGTGGTTGATTTCAGGAGGT | ||||
| Xgwm205-5D | F-CGACCCGGTTCACTTCAG | 57.1 | fam | 126–138 |
| R-AGTCGCCGTTGTATAGTGCC | ||||
| Xgwm583-5D | F-TTCACACCCAACCAATAGCA | 57.5 | hex | 138–166 |
| R-TCTAGGCAGACACATGCCTG |
| Primer Name | Na | Ne | Ho | He | H | PIC | I | F | Fst | Nm |
|---|---|---|---|---|---|---|---|---|---|---|
| Xgdm153-5D | 4 | 1.021 | 0.005 | 0.021 | 0.021 | 0.021 | 0.069 | 0.748 | 0.874 | 0.036 |
| Xgwm314-3D | 7 | 3.534 | 0.609 | 0.719 | 0.717 | 0.717 | 1.385 | 0.151 | 0.575 | 0.185 |
| Xgwm295-7D | 7 | 1.851 | 0.010 | 0.461 | 0.460 | 0.460 | 0.852 | 0.977 | 0.989 | 0.003 |
| Xgwm102-2D | 3 | 1.320 | 0.005 | 0.244 | 0.243 | 0.243 | 0.460 | 0.979 | 0.989 | 0.003 |
| Xgdm111-1D | 3 | 1.042 | 0.000 | 0.041 | 0.041 | 0.041 | 0.113 | 1.000 | 1.000 | 0.000 |
| Xgdm129-4D | 3 | 1.021 | 0.000 | 0.021 | 0.021 | 0.021 | 0.065 | 1.000 | 1.000 | 0.000 |
| Xgwm165-4D | 2 | 1.587 | 0.000 | 0.371 | 0.370 | 0.360 | 0.557 | 1.000 | 1.000 | 0.000 |
| Xgwm157-2D | 2 | 1.011 | 0.000 | 0.010 | 0.010 | 0.010 | 0.033 | 1.000 | 1.000 | 0.000 |
| Xgwm383-3D | 5 | 2.656 | 0.912 | 0.625 | 0.624 | 0.623 | 1.102 | −0.462 | 0.269 | 0.679 |
| Xgdm33-1D | 11 | 2.641 | 0.010 | 0.623 | 0.621 | 0.621 | 1.239 | 0.983 | 0.992 | 0.002 |
| Xgwm182-5D | 5 | 1.328 | 0.000 | 0.248 | 0.247 | 0.257 | 0.487 | 1.000 | 1.000 | 0.000 |
| Xgwm539-2D | 2 | 1.011 | 0.000 | 0.010 | 0.010 | 0.010 | 0.032 | 1.000 | 1.000 | 0.000 |
| Xgwm469-6D | 10 | 2.786 | 1.000 | 0.643 | 0.641 | 0.644 | 1.214 | −0.560 | 0.220 | 0.886 |
| Xgwm55-6D | 6 | 2.596 | 0.505 | 0.616 | 0.615 | 0.616 | 1.118 | 0.178 | 0.589 | 0.174 |
| Xgdm126-1D | 5 | 1.094 | 0.010 | 0.086 | 0.086 | 0.086 | 0.231 | 0.879 | 0.939 | 0.016 |
| Xgwm205-5D | 2 | 1.011 | 0.000 | 0.010 | 0.010 | 0.010 | 0.033 | 1.000 | 1.000 | 0.000 |
| Xgwm583-5D | 3 | 1.105 | 0.005 | 0.095 | 0.095 | 0.095 | 0.219 | 0.945 | 0.973 | 0.007 |
| Mean | 4.706 | 1.683 | 0.181 | 0.285 | 0.284 | 0.284 | 0.542 | 0.695 | 0.848 | 0.117 |
| Total | 80 | —— | —— | —— | —— | —— | —— | —— | —— | —— |
| Source of Variation | Degree of Freedom (df) | Sum of Squares | Mean of Squares | Estimate Variance | Percentage of Variation (%) |
|---|---|---|---|---|---|
| Among groups | 2 | 130.464 | 65.232 | 0.439 | 7 |
| Among populations within groups | 189 | 1832.106 | 9.694 | 4.071 | 67 |
| Within populations | 192 | 298 | 1.552 | 1.552 | 26 |
| Total | 383 | 2260.57 | 6.062 | 6.062 | 100 |
| Group | 1 | 2 | 3 |
|---|---|---|---|
| 1 | 0 | 2.721 | 2.362 |
| 2 | 0.084 ** 1 | 0 | 8.264 |
| 3 | 0.096 ** | 0.029 ** | 0 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yu, H.; Yang, J.; Cui, H.; Abbas, A.; Wei, S.; Li, X. Distribution, Genetic Diversity and Population Structure of Aegilops tauschii Coss. in Major Wheat-Growing Regions in China. Agriculture 2021, 11, 311. https://doi.org/10.3390/agriculture11040311
Yu H, Yang J, Cui H, Abbas A, Wei S, Li X. Distribution, Genetic Diversity and Population Structure of Aegilops tauschii Coss. in Major Wheat-Growing Regions in China. Agriculture. 2021; 11(4):311. https://doi.org/10.3390/agriculture11040311
Chicago/Turabian StyleYu, Haiyan, Juan Yang, Hailan Cui, Adeel Abbas, Shouhui Wei, and Xiangju Li. 2021. "Distribution, Genetic Diversity and Population Structure of Aegilops tauschii Coss. in Major Wheat-Growing Regions in China" Agriculture 11, no. 4: 311. https://doi.org/10.3390/agriculture11040311
APA StyleYu, H., Yang, J., Cui, H., Abbas, A., Wei, S., & Li, X. (2021). Distribution, Genetic Diversity and Population Structure of Aegilops tauschii Coss. in Major Wheat-Growing Regions in China. Agriculture, 11(4), 311. https://doi.org/10.3390/agriculture11040311







