Induced Polyploidy: A Tool for Forage Species Improvement
Abstract
1. Introduction
2. Production of Polyploid Plants
3. Impact of Polyploidy on Forage Yield and Contributing Traits
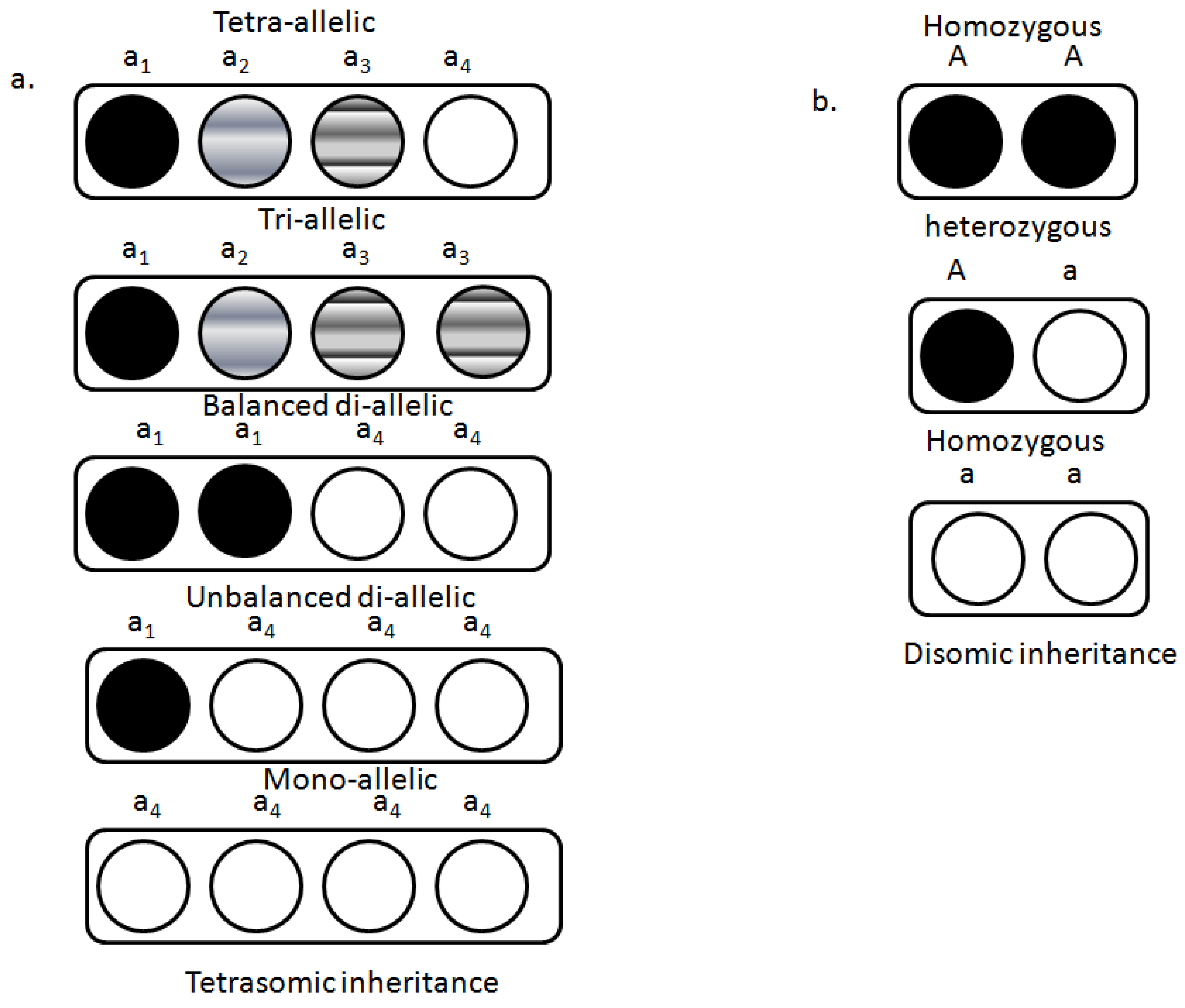
4. Genetics of Polyploidy
5. “OMICS” Analysis of Polyploidy
6. Restoration of Fertility
7. Stability of Induced Polyploidy
8. Stress Tolerance after Induced Polyploidy
9. Species: An Example of Commercial Success of Induced Polyploidy
10. Factors Affecting Commercialization of Induced-Polyploid Cultivars
11. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Chen, Z.J. Molecular mechanisms of polyploidy and hybrid vigor. Trends Plant Sci. 2010, 15, 57–71. [Google Scholar] [CrossRef]
- Shimizu-Inatsugi, R.; Terada, A.; Hirose, K.; Kudoh, H.; Sese, J.; Shimizu, K.K. Plant adaptive radiation mediated by polyploid plasticity in transcriptomes. Mol. Ecol. 2017, 26, 193–207. [Google Scholar] [CrossRef]
- Estep, M.C.; McKain, M.R.; Diaz, D.V.; Zhong, J.; Hodge, J.G.; Hodkinson, T.R.; Layton, D.J.; Malcomber, S.T.; Pasquet, R.; Kellogg, E.A. Allopolyploidy, diversification, and the Miocene grassland expansion. Proc. Natl. Acad. Sci. USA 2014, 111, 15149–15154. [Google Scholar] [CrossRef]
- Randhawa, M.S.; Singh, R.P.; Dreisigacker, S.; Bhavani, S.; Huerta-Espino, J.; Rouse, M.N.; Sandoval-Sanchez, M. Identification and validation of a common stem rust resistance locus in two bi-parental populations. Front. Plant Sci. 2018, 9, 1788. [Google Scholar] [CrossRef]
- Rao, S.; Tian, Y.; Xia, X.; Li, Y.; Chen, J. Chromosome doubling mediates superior drought tolerance in Lycium ruthenicum via abscisic acid signaling. Hort. Res. 2020, 7, 40. [Google Scholar] [CrossRef]
- Te Beest, M.; Le Roux, J.J.; Richardson, D.M.; Brysting, A.K.; Suda, J.; Kubešová, M.; Pyšek, P. The more the better? The role of polyploidy in facilitating plant invasions. Annal. Bot. 2012, 109, 19–45. [Google Scholar] [CrossRef] [PubMed]
- Venial, L.R.; Mendonça, M.A.C.; Amaral-Silva, P.M.; Canal, G.B.; Passos, A.B.R.J.; Ferreira, A.; Soares, T.C.B.; Clarindo, W.R. Autotetraploid Coffeacanephora and auto-alloctaploid Coffeaa rabicafrom in vitro chromosome set doubling: New germplasms for Coffea. Front. Plant Sci. 2020, 11, 154. [Google Scholar] [CrossRef] [PubMed]
- Dhooghe, E.; Van Laere, K.; Eeckhaut, T.; Leus, L.; Van Huylenbroeck, J. Mitotic chromosome doubling of plant tissues in vitro. Plant Cell Tissue Organ Cult. 2011, 104, 359–373. [Google Scholar] [CrossRef]
- Bretagnolle, F.A.; Thompson, J.D. Gametes with the somatic chromosome number: Mechanisms of their formation and role in the evolution of autopolyploid plants. New Phytol. 1995, 129, 1–22. [Google Scholar] [CrossRef]
- Sattler, M.C.; Carvalho, C.R.; Clarindo, W.R. The polyploidy and its key role in plant breeding. Planta 2016, 243, 281–296. [Google Scholar] [CrossRef] [PubMed]
- Ciprian-Salcedo, G.C.; Jimenez-Davalos, J.; Zolla, G. Flow-cytometry applications in plant breeding. Rev. Perua. Biol. 2020, 27, 79–84. [Google Scholar] [CrossRef]
- Rauf, S.; Sienkiewicz-Paderewska, D.; Malinowski, D.P.; Hussain, M.M.; Niazi, I.A.K.; Kausar, M. Forages: Ecology, breeding objectives and procedure. In Advances in Plant Breeding Strategies: Agronomic, Abiotic and Biotic Stress Traits; Al-Khayri, J.M., Jain, S.M., Johnson, D.V., Eds.; Springer: Cham, Switzerland, 2016; pp. 149–201. [Google Scholar]
- Stebbins, G.L. The origin of the complex of Bromus carinatus and its phylogeographic implications. Gray Herb. Harv. Univ. 1947, 165, 42–55. [Google Scholar]
- Stebbins, G.L. Chromosomes and evolution in the genus Bromus (Gramineae). Bot. Jahrb. Syst. Pflanzengesch. Pflanzengeogr. 1981, 102, 359–379. [Google Scholar]
- Rao, S.R.; Kumar, A.; Purohit, J.; Khedasana, R.; Bewal, S. Cytogenetical investigations in colchicine induced tetraploids of Cyamopsis tetragonoloba L. Czech J. Genet. Plant Breed. 2010, 45, 143–154. [Google Scholar]
- Niazi, I.A.K.; Rauf, S.; Teixeira da Silva, J.A.; Iqbal, Z.; Munir, H. Induced polyploidy in inter-subspecific maize hybrids to reduce heterosis breakdown and restore reproductive fertility. Grass Forage Sci. 2015, 70, 682–694. [Google Scholar] [CrossRef]
- Corneillie, S.; De Storme, N.; Van Acker, R.; Fangel, J.U.; De Bruyne, M.; De Rycke, R.; Boerjan, W. Polyploidy affects plant growth and alters cell wall composition. Plant Physiol. 2019, 179, 74–87. [Google Scholar] [CrossRef] [PubMed]
- Ardabili, G.S.; Zakaria, R.A.; Zare, N. In vitro induction of polyploidy in Sorghum bicolor L. Cytologia 2015, 80, 495–503. [Google Scholar] [CrossRef]
- Poosamart, W.; Siniri, N.; Simia, S. Effects of colchicine concentration level and soaking period to induce polyploid mutation on five forage sorghum (Sorghum bicolor L. Moench) cultivars. J. Mahanakorn Veter. Med. 2015, 10, 99–110. [Google Scholar]
- Simioni, C.; Valle, C.B. Meiotic analysis in induced tetraploids of Brachiari Adecumbens Stapf. Crop Breed. Appl. Biotechnol. 2011, 11, 43–49. [Google Scholar] [CrossRef][Green Version]
- De Souza-Kaneshima, A.M.; Simioni, C.; Felismino, M.F.; Mendes-Bonato, A.B.; Risso-Pascotto, C.; Pessim, C.; Pagliarini, M.S.; Do Valle, C.B. Meiotic behaviour in the first interspecific hybrids between Brachiaria brizantha and Brachiaria decumbens. Plant Breed. 2010, 129, 186–191. [Google Scholar] [CrossRef]
- Pagliarini, M.S.; Carneiro Vieira, M.L.; Borges do Valle, C. Meiotic Behavior in Intra-and Interspecific Sexual and Somatic Polyploid Hybrids of Some Tropical Species. In Meiosis Molecular Mechanisms and Cytogenetic Diversity. 2012. Available online: https://www.intechopen.com/books/meiosis-molecular-mechanisms-and-cytogenetic-diversity/meiotic-behavior-in-intra-and-interspecific-tetraploid-sexual-and-somatic-hybrids-of-some-tropical-s (accessed on 28 October 2019).
- Yu, C.Y.; Kim, H.S.; Rayburn, A.L.; Widholm, J.M.; Juvik, J.A. Chromosome doubling of the bioenergy crop, Miscanthus× giganteus. GCB Bioenergy 2009, 1, 404–412. [Google Scholar] [CrossRef]
- Faleiro, F.G.; Kannan, B.; Altpeter, F. Regeneration of fertile, hexaploid, interspecific hybrids of elephant grass and pearl millet following treatment of embryogenic calli with antimitotic agents. Plant Cell Tissue Organ Cult. 2016, 124, 57–67. [Google Scholar] [CrossRef]
- Dabkevičienė, G.; Statkevičiūtė, G.; Mikaliūnienė, J.; Norkevičienė, E.; Kemešytė, V. Production of Trifolium pratense L. and T. hybridum L. tetraploid populations and assessment of their agrobiological charac-teristics. Zemdir. Agric. 2016, 103, 377–384. [Google Scholar] [CrossRef][Green Version]
- Dabkevičienė, G.; Kemešytė, V.; Statkevičiūtė, G.; Lemežienė, N.; Brazauskas, G. Autopolyploids in fodder grass breeding: Induction and field performance. Span. J. Agric. Res. 2017, 15, 20. [Google Scholar] [CrossRef]
- Castillo, A.; Lopez Carro, B.; Dalla Rizza, M.; Reyno, R. Use of in vitro methods to induce autotetraploids in the native forage legume Trifolium polymorphum. In VII International Symposium on Production and Establishment of Micropropagated Plants; ISHS: Lavras, Minas Gerais, Brazil, 2017; pp. 73–80. [Google Scholar]
- Silva, A.J.; Carvalho, C.R.; Clarindo, W.R. Chromosome set doubling and ploidy stability in synthetic auto- and allotetraploid of Eucalyptus: From in vitro condition to the field. Plant Cell Tissue Organ Cult. 2019, 138, 387–394. [Google Scholar] [CrossRef]
- Dirihan, S.; Terho, P.; Helander, M.; Saikkonen, K. Efficient analysis of ploidy levels in plant evolutionary ecology. Caryologia 2013, 66, 251–256. [Google Scholar] [CrossRef]
- Yun, L.; Yun, J.; Li, J.; Zheng, L.; Zhao, W.; Qi, L. Callus polyploidy induction and identification of Russian wild ryegrass. Acta Pratac. Sin. 2010, 19, 126–131. [Google Scholar]
- Joshi, P.; Verma, R.C. High frequency production of colchicine induced autotetraploids in faba bean (Viciafaba L.). Cytologia 2004, 69, 141–147. [Google Scholar] [CrossRef]
- Pereira, R.C.; Ferreira, M.T.M.; Davide, L.C.; Pasqual, M.; Mittelmann, A.; Techio, V.H. Chromosome duplication in Lolium multiflorum Lam. Crop Breed. Appl. Biotechnol. 2014, 14, 251–255. [Google Scholar] [CrossRef][Green Version]
- Yoon, S.; Aucar, S.; Hernlem, B.J.; Edme, S.; Palmer, N.; Sarath, G.; Mitchell, R.; Blumwald, E.; Tobias, C.M. Generation of octaploid switchgrass by seedling treatment with mitotic inhibitors. BioEnergy Res. 2017, 10, 344–352. [Google Scholar] [CrossRef]
- Campos, J.M.S.; Davide, L.C.; Salgado, C.C.; Santos, F.C.; Costa, P.N.; Silva, P.S.; Pereira, A.V. In vitro induction of hexaploid plants from triploid hybrids of Pennisetum purpureum and Pennisetum glaucum. Plant Breed. 2009, 128, 101–104. [Google Scholar] [CrossRef]
- Ishigaki, G.; Gondo, T.; Suenaga, K.; Akashi, R. Induction of tetraploid ruzigrass (Brachiaria ruziziensis) plants by colchicine treatment of in vitro multiple-shoot clumps and seedlings. Grassl. Sci. 2009, 55, 164–170. [Google Scholar] [CrossRef]
- Timbó, A.L.D.O.; Souza, P.N.D.C.; Pereira, R.C.; Nunes, J.D.; Pinto, J.E.B.P.; SouzaSobrinho, F.D.; Davide, L.C. Obtaining tetraploid plants of ruzigrass (Brachiaria ruziziensis). R. Bras. Zootec. 2014, 43, 127–131. [Google Scholar] [CrossRef][Green Version]
- Głowacka, K.; Jeżowski, S.; Kaczmarek, Z. In vitro induction of polyploidy by colchicine treatment of shoots and preliminary characterisation of induced polyploids in two Miscanthus species. Ind. Crop. Prod. 2010, 32, 88–96. [Google Scholar] [CrossRef]
- Niazi, I.A.K. Evaluation of Zea mays × Zea mexicana for High Fodder Yield. Ph.D. Thesis, University of Sargodha, Sargodha, Pakistan, 2016. [Google Scholar]
- Abd El-Naby, Z.M.; Mohamed, N.A.; Radwan, K.H.; El-Khishin, D.A. Colchicine induction of polyploidy in Egyptian clover genotypes. J. Am. Sci. 2012, 8, 221–227. [Google Scholar]
- Wilkins, P.W.; Lovatt, J.A. Gains in dry matter yield and herbage quality from breeding perennial ryegrass. Ir. J. Agric. Food Res. 2011, 50, 23–30. [Google Scholar]
- Dhawan, O.P.; Lavania, U.C. Enhancing the productivity of secondary metabolites via induced polyploidy: A review. Euphytica 1996, 87, 81–89. [Google Scholar] [CrossRef]
- Tulay, E.; Unal, M. Production of colchicine induced tetraploids in Vicia villosa roth. Caryologia 2010, 63, 292–303. [Google Scholar] [CrossRef]
- Dabkevičienė, G.; Kemešytė, V.; Lemežienė, N.; Butkutė, B. Production of slender cocksfoot (Dactylis polygama H.) tetraploid populations and their assessment for agromorphological characteristics. Zemdir. Agric. 2013, 100, 303–310. [Google Scholar] [CrossRef][Green Version]
- Balocchi, O.A.; López, I.F. Herbage production, nutritive value and grazing preference of diploid and tetraploid perennial ryegrass cultivars (Lolium perenne L.). Chil. J. Agric. Res. 2009, 69, 331–339. [Google Scholar] [CrossRef]
- Price, S.C. Tetraploid corn and methods of its production. U.S. Patent No. 4,75,910, 10 November 1987. [Google Scholar]
- Nadel, D.; Nadel, M.; Nadel, B. High Yield Maize Derivatives. U.S. Patent Application 12/004,894, 25 June 2009. [Google Scholar]
- Smith, K.F.; McFarlane, N.M.; Croft, V.M.; Trigg, P.J.; Kearney, G.A. The effects of ploidy and seed mass on the emergence and early vigour of perennial ryegrass (Lolium perenne L.) cultivars. Aust. J. Exp. Agric. 2003, 43, 481–486. [Google Scholar] [CrossRef]
- Bartek, M.S.; Hodnett, G.L.; Burson, B.L.; Stelly, D.M.; Rooney, W.L. Pollen Tube Growth After Intergeneric Pollinations of iap-Homozygous Sorghum. Crop Sci. 2012, 52, 1553–1560. [Google Scholar] [CrossRef]
- Ardabili, G.S.; Zakaria, R.A.; Zare, N. Polyploidy induction and its effects on some morpho-physiologic characteristics in sorghum (Sorghum bicolor cv. KFS2). Iran. J. Crop Sci. 2014, 16, 151–164. [Google Scholar]
- Pathak, S.; Malaviya, D.R.; Roy, A.K.; Dwivedi, K.; Kaushal, P. Multifoliate Leaf Formation in Induced Tetraploids of Trifolium alexandrinum L. Cytologia 2015, 80, 59–66. [Google Scholar] [CrossRef][Green Version]
- Chaves, A.L.A.; Raquel, B.; Chiavegatto, M.L.; Gavilanes, F.R.G.B.; Vânia, H.T. Effect of polyploidy on the leaf epidermis structure of Cynodondactylon (L.) Pers. (Poaceae). Biologia 2018, 73, 1007–1013. [Google Scholar] [CrossRef]
- Zielinski, M.L.; Scheid, O.M. Meiosis in polyploid plants. In Polyploidy and Genome Evolution; Springer: Berlin/Heidelberg, Germany, 2012; pp. 33–55. [Google Scholar]
- Comai, L. The advantages and disadvantages of being polyploid. Nat. Rev. Genet. 2005, 6, 836–846. [Google Scholar] [CrossRef]
- Skiebe, K.; Seliger, P. Isoenzymes and their importance for breeding autopolyploids. Plant Breed. 1990, 105, 106–111. [Google Scholar] [CrossRef]
- Valizadeh, M.; Mohayeji, M.; Yasinzadeh, N.; Nasrullazadeh, S.; Moghaddam, M. Genetic diversity of synthetic alfalfa generations and cultivars using tetrasomic inherited allozyme markers. J. Agric. Sci. Tech. 2011, 13, 425–430. [Google Scholar]
- Corts, M.R.; Morales, M.C.; Martinez, C. Variation of PGM and IDH isozymes for identification of alfalfa varieties. Euphytica 2000, 112, 137–143. [Google Scholar] [CrossRef]
- Jalaly, H.M.; Valizadeh, M.; Ahmadi, M.; Nabizadeh, H.; Moharramnejad, S.; Moghaddam, M. Discrimination of alfalfa half-sib families by allozyme banding pattern and its relationship with forage yield attributes. J. Biodiv. Environ. Sci. 1990, 6, 344–350. [Google Scholar]
- Fjellstrom, R.G.; Steiner, J.J.; Beuselinck, P.R. Tetrasomic linkage mapping of RFLP, PCR, and isozyme loci in Lotus corniculatus L. Crop Sci. 2003, 43, 1006–1020. [Google Scholar] [CrossRef]
- Diwan, N.; Bouton, J.H.; Kochert, G.; Cregan, P.B. Mapping of simple sequence repeat (SSR) DNA markers in diploid and tetraploid alfalfa. Theoret. Appl. Genet. 2000, 101, 165–172. [Google Scholar] [CrossRef]
- Julier, B.; Flajoulot, S.; Barre, P.; Cardinet, G.; Santoni, S.; Huguet, T.; Huyghe, C. Construction of two genetic linkage maps in cultivated tetraploid alfalfa (Medicago sativa) using microsatellite and AFLP markers. BMC Plant Biol. 2003, 3, 9. [Google Scholar] [CrossRef][Green Version]
- Stein, J.; Quarin, C.L.; Martínez, E.J.; Pessino, S.C.; Ortiz, J.P.A. Tetraploid races of Paspalum notatum show polysomic inheritance and preferential chromosome pairing around the apospory-controlling locus. Theor. Appl. Genet. 2004, 109, 186–191. [Google Scholar] [CrossRef] [PubMed]
- Begheyn, R.F.; Lübberstedt, T.; Studer, B. Haploid and doubled haploid techniques in perennial ryegrass (Lolium perenne L.) to advance research and breeding. Agronomy 2016, 6, 60. [Google Scholar] [CrossRef]
- Capomaccio, S.; Veronesi, F.; Rosellini, D. Polyploidization and gene expression in Medicago sativa. In Sustainable Use of Genetic Diversity in Forage and Turf Breeding; Springer: Dordrecht, The Netherlands, 2010; pp. 397–401. [Google Scholar]
- Rosellini, D.; Ferradini, N.; Allegrucci, S.; Capomaccio, S.; Zago, E.D.; Leonetti, P.; Veronesi, F. Sexual polyploidization in Medicago sativa L.: Impact on the phenotype, gene transcription, and genome methylation. G3-Genes Genom. Genet. 2016, 6, 925–938. [Google Scholar]
- Molin, W.T.; Meyers, S.P.; Baer, G.R.; Schrader, L.E. Ploidy effects in isogenic populations of alfalfa. II. Photosynthesis, chloroplast number, ribulose-1,5-bisphosphate carboxylase, chlorophyll, and DNA in protoplasts. Plant Physiol. 1982, 70, 1710–1714. [Google Scholar] [CrossRef]
- Mo, L.; Chen, J.; Lou, X.; Xu, Q.; Dong, R.; Tong, Z.; Huang, H.; Lin, E. Colchicine-induced polyploidy in Rhododendron fortune Lindl. Plants 2020, 9, 424. [Google Scholar] [CrossRef]
- Pei, Y.; Yao, N.; He, L.; Deng, D.; Li, W.; Zhang, W. Comparative study of the morphological, physiological and molecular characteristics between diploid and tetraploid radish (Raphunas sativus L.). Scient. Hort. 2019, 257, 108739. [Google Scholar] [CrossRef]
- Santos, F.C.; Guyot, R.; Do Valle, C.B.; Chiari, L.; Techio, V.H.; Heslop-Harrison, P.; Vanzela, A.L.L. Chromosomal distribution and evolution of abundant retrotransposons in plants: Gypsy elements in diploid and polyploid Brachiaria forage grasses. Chromosome Res. 2015, 23, 571–582. [Google Scholar] [CrossRef] [PubMed]
- Oberprieler, C.; Talianova, M.; Griesenbeck, J. Effects of polyploidy on the coordination of gene expression between organellar and nuclear genomes in Leucanthemum Mill. (Compositae, Anthemideae). Ecol. Evol. 2019, 9, 9100–9110. [Google Scholar] [CrossRef] [PubMed]
- Murashige, T.; Skoog, F. A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol. Plant. 1962, 15, 473–497. [Google Scholar] [CrossRef]
- Nair, R.M. Developing tetraploid perennial ryegrass (Lolium perenne L.) populations. N. Z. J. Agric. Res. 2004, 47, 45–49. [Google Scholar] [CrossRef]
- Mendes-Bonato, A.B.; Felismino, M.F.; Kaneshima, A.S.; Pessim, C.; Calisto, V.; Pagliarini, M.S.; do Valle, C.B. Abnormal meiosis in tetraploid genotypes of Brachiaria brizantha (Poaceae) induced by colchicine: Its implications for breeding. J. Appl. Genet. 2009, 50, 83–87. [Google Scholar] [CrossRef]
- Dos Reis, G.B.; Ishii, T.; Fuchs, J.; Houben, A.; Davide, L.C. Tissue-specific genome instability in synthetic interspecific hybrids of Pennisetum purpureum (Napier grass) and Pennisetum glaucum (pearl millet) is caused by micronucleation. Chromosome Res. 2016, 24, 285–297. [Google Scholar] [CrossRef]
- Simioni, C.; Wittmann, M.T.S.; Dall, M.; Guerra, D. Selection for increasing 2n gamete production in red clover. Crop Breed. Appl. Biotech. 2004, 4, 217–220. [Google Scholar] [CrossRef]
- Pereira, R.C.; Davide, L.C.; Techio, V.H.; Timbó, A.L.O. Chromosome doubling of grasses: An alternative to plant breeding. Ciência Rural 2012, 42, 1278–1285. [Google Scholar] [CrossRef]
- Baduel, P.; Bray, S.; Vallejo-Marin, M.; Kolář, F.; Yant, L. The “polyploid hop”: Shifting challenges and opportunities over the evolutionary lifespan of genome duplications. Front. Ecol. Evol. 2018, 6, 117. [Google Scholar] [CrossRef]
- Frank, A.B.; Berdahl, J.D. Gas exchange and water relations in diploid and tetraploid Russian wildrye. Crop Sci. 2001, 41, 87–92. [Google Scholar] [CrossRef]
- Ribotta, A.N.; Griffa, S.M.; Díaz, D.; Carloni, E.J.; Colomba, E.L.; Tommasino, E.A.; Grunberg, K. Selecting salt-tolerant clones and evaluating genetic variability to obtain parents of new diploid and tetraploid germplasm in rhodesgrass (Chloris gayana L.). S. Afr. J. Bot. 2013, 84, 88–93. [Google Scholar] [CrossRef][Green Version]
- Tozer, K.N.; Carswell, K.; Griffiths, W.M.; Crush, J.R.; Cameron, C.A.; Chapman, D.F.; Popay, A.; King, W. Growth responses of diploid and tetraploid perennial ryegrass (Lolium perenne) to soil-moisture deficit, defoliation and a root-feeding invertebrate. Crop Pasture Sci. 2017, 68, 632–642. [Google Scholar] [CrossRef]
- Sugiyama, S. Differentiation in competitive ability and cold tolerance between diploid and tetraploid cultivars in Lolium perenne. Euphytica 1998, 103, 55–59. [Google Scholar] [CrossRef]
- Chandra, A.; Dube, A. Assessment of ploidy level on stress tolerance of Cenchrus species based on leaf photosynthetic characteristics. Acta Physiol. Plant. 2009, 31, 1003–1013. [Google Scholar] [CrossRef]
- Sugiyama, S.I. Responses of shoot growth and survival to water stress gradient in diploid and tetraploid populations of Lolium multiflorum and L. perenne. Grassl. Sci. 2006, 52, 155–160. [Google Scholar] [CrossRef]
- Anwar, S. Genetic-phenotypic variability and correlation between morphology anatomy physiology characteristics and dry matter yield of polyploidized forage grasses under aluminum stressed condition. Anim. Product. 2007, 9, 23–29. [Google Scholar]
- Marzougui, N.; Boubaya, A.; Guasmi, F.; Elfalleh, W.; Touil, L.; Ferchichi, A.; Beji, M. Influence of artificial polyploidy on the salt stress tolerance of Trigonella foenum-graecum L. in Tunisia. Acta Bot. Gall. 2010, 157, 295–303. [Google Scholar] [CrossRef]
- Meng, H.B.; Jiang, S.S.; Hua, S.J.; Lin, X.Y.; Li, Y.L.; Guo, W.L.; Jiang, L.X. Comparison between a tetraploid turnip and its diploid progenitor (Brassica rapa L.): The adaptation to salinity stress. Agric. Sci. China 2011, 10, 363–375. [Google Scholar] [CrossRef]
- Franco, M.F.; Colabelli, M.N.; Petigrosso, L.R.; De Battista, J.P.; Echeverría, M.M. Evaluation of infection with endophytes in seeds of forage species with different levels of ploidy. N. Z. J. Agric. Res. 2015, 58, 181–189. [Google Scholar] [CrossRef]
- Kemesyte, V.; Statkeviciute, G.; Brazauskas, G. Perennial ryegrass yield performance under abiotic stress. Crop Sci. 2017, 57, 1935–1940. [Google Scholar] [CrossRef]
- Lee, M.A.; Howard-Andrews, V.; Chester, M. Resistance of multiple diploid and tetraploid perennial ryegrass (Lolium perenne L.) varieties to three projected drought scenarios for the UK in 2080. Agronomy 2019, 9, 159. [Google Scholar] [CrossRef]
- Chandra, A.; Dubey, A. Effect of ploidy levels on the activities of Δ1-pyrroline-5-carboxylate synthetase, superoxide dismutase and peroxidase in Cenchrus species grown under water stress. Plant Physiol. Biochem. 2010, 48, 27–34. [Google Scholar] [CrossRef]
- Arseniuk, E. Triticale abiotic stresses—An overview. Commun. Agric. Appl. Biol. Sci. 2015, 79, 82–100. [Google Scholar]
- Ryan, P.R.; Dong, D.; Teuber, F.; Wendler, N.; Mühling, K.H.; Liu, J.; Xu, M.; Salvador Moreno, N.; You, J.; Maurer, H.P.; et al. Assessing how the aluminum-resistance traits in wheat and rye transfer to hexaploid and octoploid triticale. Front. Plant Sci. 2018, 9, 1334. [Google Scholar] [CrossRef]
- Niazi, I.A.K.; Avais, R.; Rauf, S.; Teixeira da Silva, J.A.; Afzal, M. Simultaneous selection for stem borer resistance and forage related traits in maize (Zea mays ssp. mays L.) × teosinte (Zea mays ssp. mexicana L.) derived populations. Crop Prot. 2014, 57, 27–34. [Google Scholar]
- Dewey, D.R. Some applications and misapplications of induced polyploidy to plant breeding. In Polyploidy; Springer: Boston, MA, USA, 1980; pp. 445–470. [Google Scholar]
- Den Nijs, A.P.M.; Stephenson, A.G. Potential of unreduced pollen for breeding tetraploid perennial ryegrass. In Sexual Reproduction in Higher Plants; Springer: Berlin/Heidelberg, Germany, 1988; pp. 131–136. [Google Scholar]
- Rios, E.F.; Kenworthy, K.E.; Munoz, P.R. Association of phenotypic traits with ploidy and genome size in annual ryegrass. Crop Sci. 2015, 55, 2078–2090. [Google Scholar] [CrossRef]
- Sugiyama, S.I. Polyploidy and cellular mechanisms changing leaf size: Comparison of diploid and autotetraploid populations in two species of Lolium. Ann. Bot. 2005, 96, 931–938. [Google Scholar] [CrossRef]
- Wang, Y.; Bigelow, C.A.; Jiang, Y. Ploidy level and DNA content of perennial ryegrass germplasm as determined by flow cytometry. HortScience 2009, 44, 2049–2052. [Google Scholar] [CrossRef]
- Burns, G.A.; Gilliland, T.J.; Grogan, D.; O’Kiely, P. Comparison of the agronomic effects of maturity and ploidy in perennial ryegrass. In Grassland: An European Resource; Polskie Towarzystwo Łąkarskie: Lublin, Poland, 2012; pp. 349–351. [Google Scholar]
- Akinroluyo, O.K.; Jaškūnė, K.; Kemešytė, V.; Statkevičiūtė, G. Drought stress response of Westerwolths ryegrass (Lolium multiflorum ssp. multiflorum) cultivars differing in their ploidy level. Zemdir. Agric. 2020, 107, 221–230. [Google Scholar]
- O’Donovan, M.; Delaby, L.A. comparison of perennial ryegrass cultivars differing in heading date and grass ploidy with spring calving dairy cows grazed at two different stocking rates. Anim. Res. 2005, 54, 337–350. [Google Scholar] [CrossRef]
- Solomon, J.K.; Macoon, B.; Lang, D.J.; Vann, R.C.; Ward, S. Cattle grazing preference among tetraploid and diploid annual ryegrass cultivars. Crop Sci. 2014, 54, 430–438. [Google Scholar] [CrossRef]
- Hume, D.E.; Hickey, M.J.; Lyons, T.B.; Baird, D.B. Agronomic performance and water-soluble carbohydrate expression of selected ryegrasses at two locations in New Zealand. N. Z. J. Agric. Res. 2010, 53, 37–57. [Google Scholar] [CrossRef][Green Version]
- Kenworthy, K.E.; Reith, P.E.; Prine, G.M.; Blount, A.R.; Quesenberry, K.H. Registration of ‘FL Red’, a later-maturing tetraploid annual ryegrass. J. Plant Reg. 2017, 11, 46–50. [Google Scholar] [CrossRef]
- Smith, K.F.; Simpson, R.J.; Culvenor, R.A.; Humphreys, M.O.; Prud’Homme, M.P.; Oram, R.N. The effects of ploidy and a phenotype conferring a high water-soluble carbohydrate concentration on carbohydrate accumulation, nutritive value and morphology of perennial ryegrass (Lolium perenne L.). J. Agric. Sci. 2001, 136, 65–74. [Google Scholar] [CrossRef]
- Pereira, R.C.; Santos, N.D.S.; Bustamante, F.D.O.; Mittelmann, A.; Techio, V.H. Stability in chromosome number and DNA content in synthetic tetraploids of Lolium multiflorum after two generations of selection. Ciência Rural 2017, 47, 2. [Google Scholar] [CrossRef]
- Ren-Hua, J.I.; Zhong-Ling, C.I.; Lin-Qing, Y.U.; LIU, S.-N.; Chen, S.R. Chromosome doubling technique of forage in vitro and its application progress in breeding. Pratacult. Anim. Husband. 2011, 9, 33–39. [Google Scholar]
- Hegarty, M.; Coate, J.; Sherman-Broyles, S.; Abbott, R.; Hiscock, S.; Doyle, J. Lessons from natural and artificial polyploids in higher plants. Cytogenet. Genome Res. 2013, 140, 204–225. [Google Scholar] [CrossRef] [PubMed]
- Münzbergová, Z. Colchicine application significantly affects plant performance in the second generation of synthetic polyploids and its effects vary between populations. Ann. Bot. 2017, 120, 329–339. [Google Scholar] [CrossRef]
- Innes, L.A.; Denton, M.D.; Dundas, I.S.; Peck, D.M.; Humphries, A.W. The effect of ploidy number on vigor, productivity, and potential adaptation to climate change in annual Medicago species. Crop Sci. 2020, 60. [Google Scholar] [CrossRef]
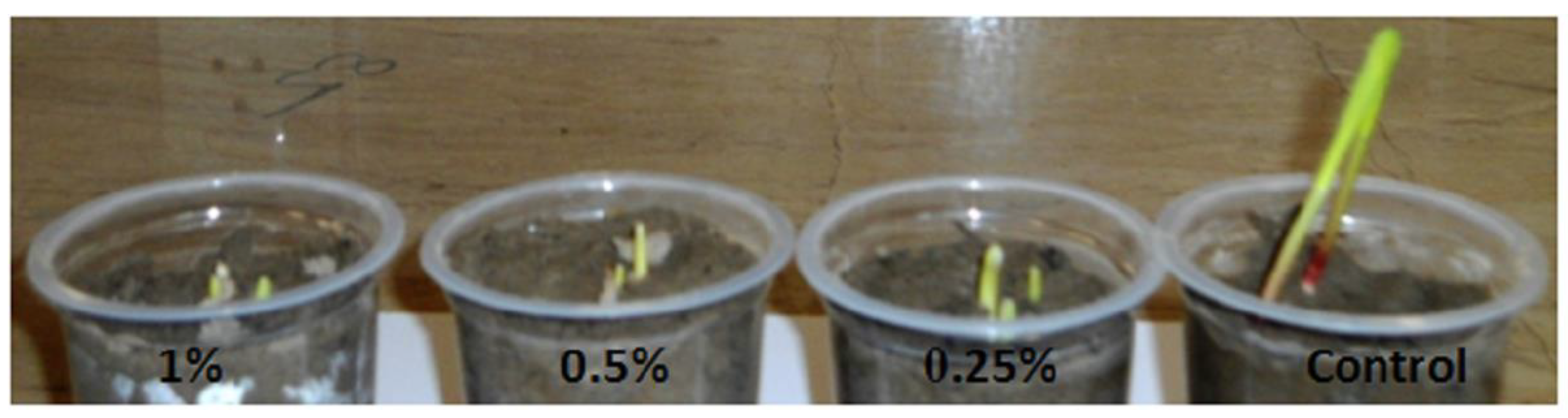
| Species | Ploidy Level | Treatment | Polyploid Frequency | References |
|---|---|---|---|---|
| Vicia faba L. | 2× = 12 | 0.005% for 8 h, seed imbibition | 4× = 50% | [31] |
| Cyamopsis tetragonoloba L. Taub. | 2× = 14 | 0.2% cotton swab for 10 h | 4× = 12–40% | [15] |
| Brachiaria decumbens (Mez) Davidse | 2× = 18, 36, 45 | Colchicine concentration (0.01%, 0.01%, and 0.1%) | 0.1% colchicine for 48 h induces 11% of polyploid | [20] |
| Psathyrostachys juncea (Fisch.) Nevski | 2× = 14 | 100 mg L−1 colchicine and 1.5% DMSO, callus tissue | 4× = 53.58% Stomata size increased by 13.52% while stomata frequency decreased | [30] |
| Lolium multiflorum Lam. | 2× = 14 | Seedling immersion (0.2%) for 3 h and 24 h in colchicine | 4× = 20% | [32] |
| Sorghum bicolor (L.) Mönch | 2× = 20 | MS medium supplemented with 0.1% of colchicine | In vitro media supplemented with 0.1% colchicine has the highest frequency 4× induction | [18] |
| Sorghum bicolor (L.) Mönch | 2× = 20 | 4 colchicine levels and 5 soaking times | 0.2% concentration for 48 h had ploidy induction of 3.8 to 4.2% colchicine | [19] |
| Trifolium pratense L. Trifolium hybridum L. | 2× = 14 2× = 16 | Callus vs. seed germination treatment of colchicine, amiprophos-methyl, trifluralin and oryzalin | Amiprophos-methyl and oryzalin had similar frequency of ploidy induction in T. pratense; colchicine had higher efficiency for induction of tetratploid in T. hybridum | [25] |
| Panicum virgatum L. | 4× = 36 | 0.2% colchicine solution applied to seed | 8× = 19 families | [33] |
| Pennisetum purpureum Schumach. × Pennisetum glaucum (L.) Brown | 3× = 21 | In vitro colchicine (0.1%) solution, seed imbibition | 6× = 17/480, larger stomata size | [34] |
| Brachiaria ruziziensis (Mez) Davidse | 2× = 18 | In vitro 0.0125–0.1% colchicine | 4× = 23/400 (31.3%) 0.1% colchicine for 3 BAP | [35] |
| Brachiaria ruziziensis (Mez) Davidse | 2× = 18 | 0.1% colchicine, seed | 4× = 11.45% | [36] |
| Miscanthus species Andersson | 2× = 38 | Callus treatment with 313–626 µM colchicine | 4× was induced with 40% success | [37] |
| Species | Characteristics | References |
|---|---|---|
| Zea mays L. × Zea mays ssp. mexicana (Schrad.) Kuntze | 4× had higher leaf soluble proteins, oil contents, and total soluble sugars | [16] |
| Sorghum bicolor (L.) Mönch | 4× had higher chlorophyll contents, water soluble carbohydrates, and proteins | [18] |
| Lolium multiflorum Lam., Lolium perenne L. and Festuca pratensis (Huds.) Darbysh. | 4× produced higher dry matter and seed yield when compared with 2× and may be used for the development of new cultivars | [26] |
| Trifolium alexandrinum L. | Induced 4× had taller plants, faster regrowth, increased tillering and branching | [39] |
| Lolium perenne L. | 4× had larger and wider leaves, taller seedlings, and fewer tillers | [47] |
| Sorghum bicolor (L.) Mönch | 4× pollen had lower germination percentage and pollen tube growth than diploid pollens resulting in lower seed setting percentages | [48] |
| Sorghum bicolor (L.) Mönch | 4× had thicker leaves, taller plantslonger leaves, higher antioxidant activity, panicle length and diameter | [49] |
| Trifolium alexandrinum L. | 4× had better pentafoliate trait than 2× | [50] |
| Cynodon dactylon Pers. | Stomatal size increased while stomatal density decreased. Leaf anatomy was used to discriminate 2× vs. 3× accessions | [51] |
| Progenies | Number of | Forage Yield (g plant−1) | Seed Setting (%) | |||
|---|---|---|---|---|---|---|
| Univalents (I) | Bivalents (II) | Trivalents (III) | Quadrivalents (IV) | |||
| S1-2× | - | 10 | - | - | 241.34 b ± 42.64 | 98.13 a ± 2.01 |
| C1-4× | 2.81 a ± 0.39 | 1.52 c ± 0.22 | 3.11 a ± 0.41 | 6.12 d ± 0.37 | 321.55 a ± 36.55 | 26.19 d ± 4.12 |
| C2-4× | 2.21 b ± 0.42 | 1.84 b ± 0.34 | 2.19 b ± 0.29 | 6.88 c ± 0.74 | 334.42 a ± 44.37 | 29.27 d ± 2.06 |
| C3-4× | 1.51 c ± 0.21 | 2.02 a ± 0.42 | 1.14 c ± 0.33 | 7.58 b ± 0.52 | 339.27 a ± 36.34 | 41.23 c ± 5.01 |
| C4-4× | 0.81 d ± 0.17 | 2.06 a ± 0.27 | 0.54 d ± 0.21 | 8.32 a ± 0.43 | 351.21 a ± 40.19 | 71.40 b ± 3.71 |
| Stress | Species | Resistance | Reference |
|---|---|---|---|
| Salinity | Chloris gayana L. (Rhodes grass) | Wild genetic diversity in tetraploid species may be useful for making selection for salinity tolerance | [78] |
| Water stress tolerance | Eight Cenchrus species | Morphological traits such as plant height, leaf rolling and wilting indicated higher water stress tolerance in natural polysomic tetraploid species | [81] |
| Water stress tolerance | Lolium multiflorum Lam. and Lolium perenne L. | Polyploid had better tolerance than 2× species | [82] |
| Aluminum tolerance [16 mM Al2(SO4)3] | 16 polyploid vs. diploid species | Polyploidy was positively related with the stress tolerance index | [83] |
| Salt stress | Trigonella foenumgreaum L. | Induced polyploids showed higher values of forage traits under salt stress | [84] |
| Salinity | Brassica rapa L. | 4× plant species had better germination under salinity stress and maintained higher K+/Na+ ratio in roots and shoots | [85] |
| Endophytic fungi | Lolium multiflorum Lam. and Bromus | 2× species had higher endophytic infection rate than 4× | [86] |
| Cold and drought stress | Perennial ryegrass (Lolium perenne L.) | 4× species had better tolerance to cold and drought tolerance, higher regrowth and high dry matter yield | [87] |
| Drought stress | Perennial ryegrass | Greater biomass yield under drought stress | [88] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rauf, S.; Ortiz, R.; Malinowski, D.P.; Clarindo, W.R.; Kainat, W.; Shehzad, M.; Waheed, U.; Hassan, S.W. Induced Polyploidy: A Tool for Forage Species Improvement. Agriculture 2021, 11, 210. https://doi.org/10.3390/agriculture11030210
Rauf S, Ortiz R, Malinowski DP, Clarindo WR, Kainat W, Shehzad M, Waheed U, Hassan SW. Induced Polyploidy: A Tool for Forage Species Improvement. Agriculture. 2021; 11(3):210. https://doi.org/10.3390/agriculture11030210
Chicago/Turabian StyleRauf, Saeed, Rodomiro Ortiz, Dariusz P. Malinowski, Wellington Ronildo Clarindo, Wardah Kainat, Muhammad Shehzad, Ummara Waheed, and Syed Wasim Hassan. 2021. "Induced Polyploidy: A Tool for Forage Species Improvement" Agriculture 11, no. 3: 210. https://doi.org/10.3390/agriculture11030210
APA StyleRauf, S., Ortiz, R., Malinowski, D. P., Clarindo, W. R., Kainat, W., Shehzad, M., Waheed, U., & Hassan, S. W. (2021). Induced Polyploidy: A Tool for Forage Species Improvement. Agriculture, 11(3), 210. https://doi.org/10.3390/agriculture11030210







