Generation of a New Glutinous Photothermosensitive Genic-Male-Sterile (PTGMS) Line by CRISPR/Cas9-Directed Mutagenesis of Wx in Rice (Oryza sativa L.)
Abstract
:1. Introduction
2. Materials and Methods
2.1. Experimental Material, Rice Material Planting, and Growth Environment
2.2. Construction of pYLCRISPR/Cas9-Wx Vector
2.2.1. Target Site Selection and Vector Construction
2.2.2. Construction of pYLCRISPR/Cas9-Wx Vector
2.3. Genotyping, Phenotyping, and Screening of T-DNA-Free Plants
2.4. Grain Phenotype of Endosperm and Iodine Staining
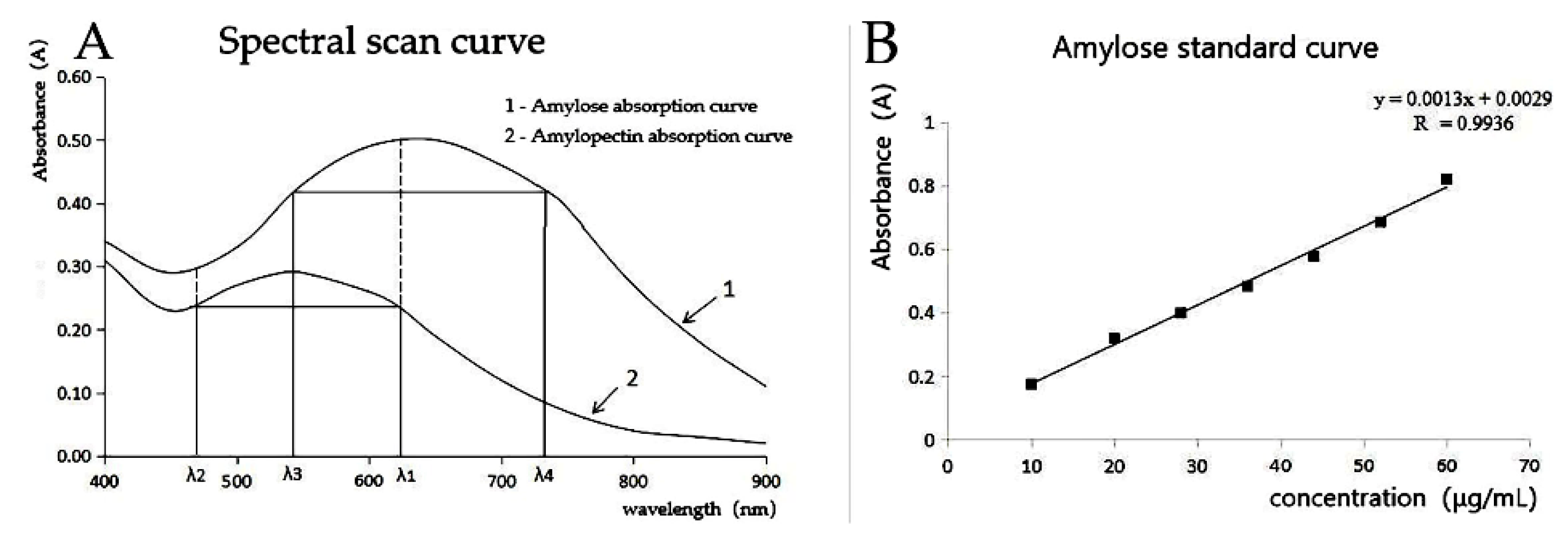
2.5. Determination of Amylose Content by Dual-Wavelength Method
2.6. Pollen Fertility Survey
2.7. Data Analysis
3. Results
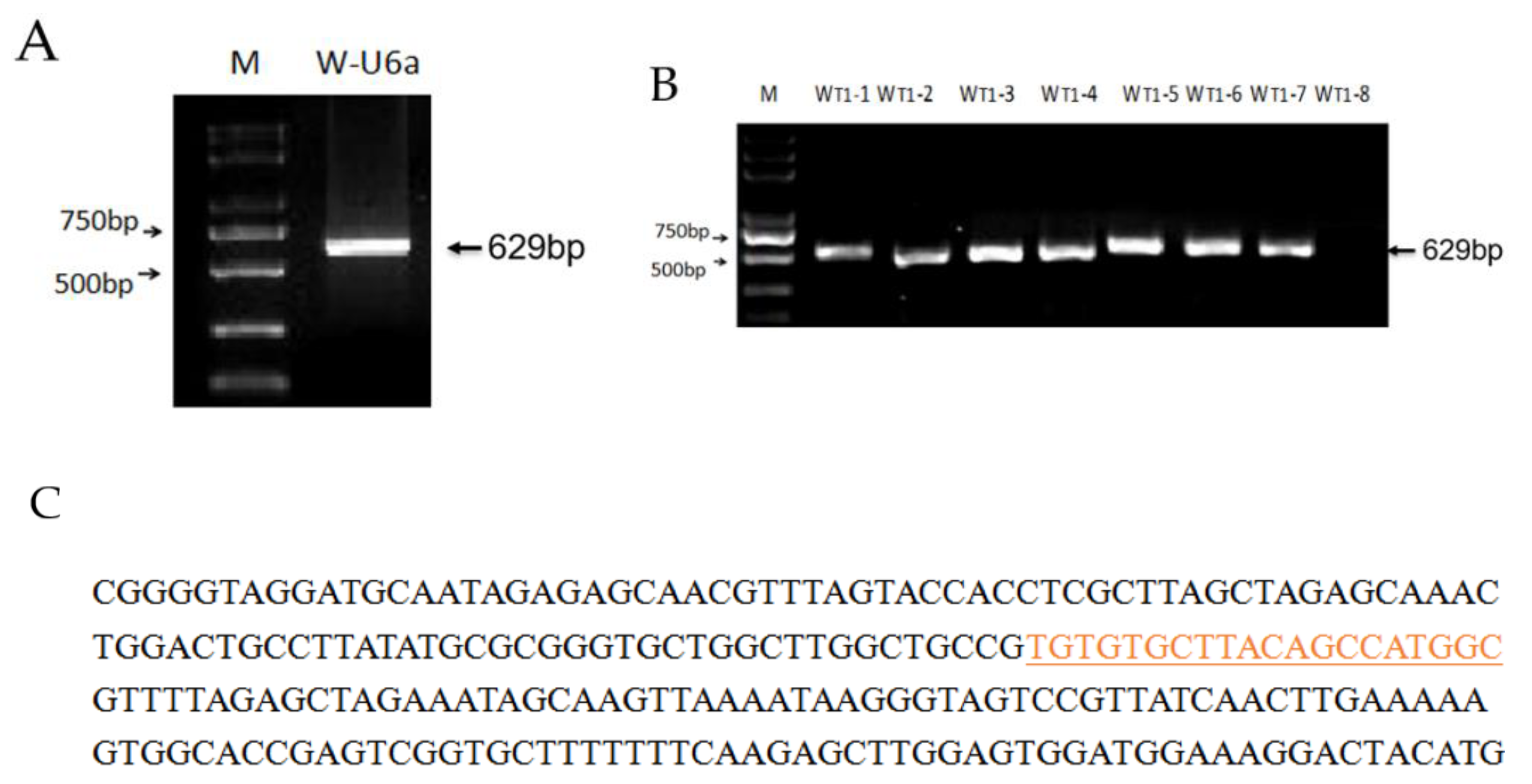
3.1. Vector Construction
3.2. Genotyping and Analysis of Mutation Types in T0 Plants
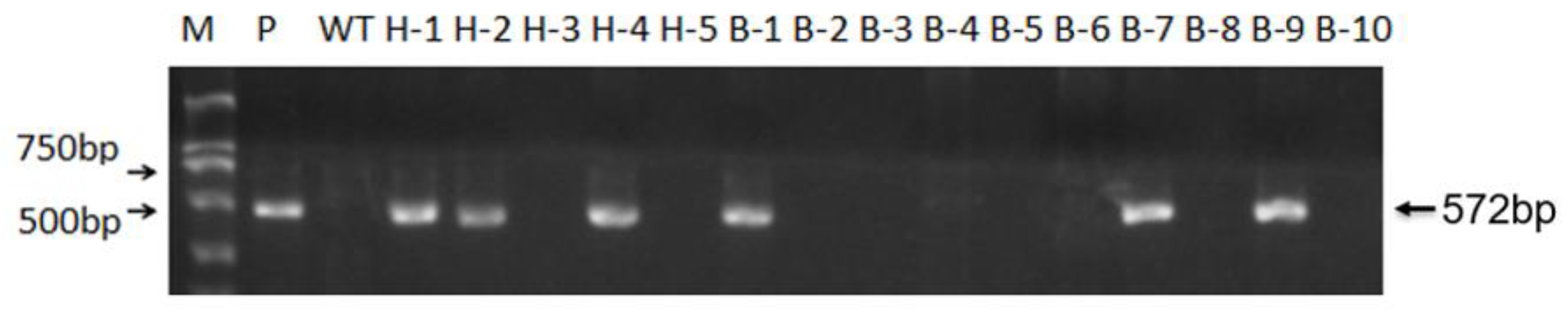
3.3. Screening of T-DNA-Free Plants
3.4. Determination of Amylose Content in T2 Generation
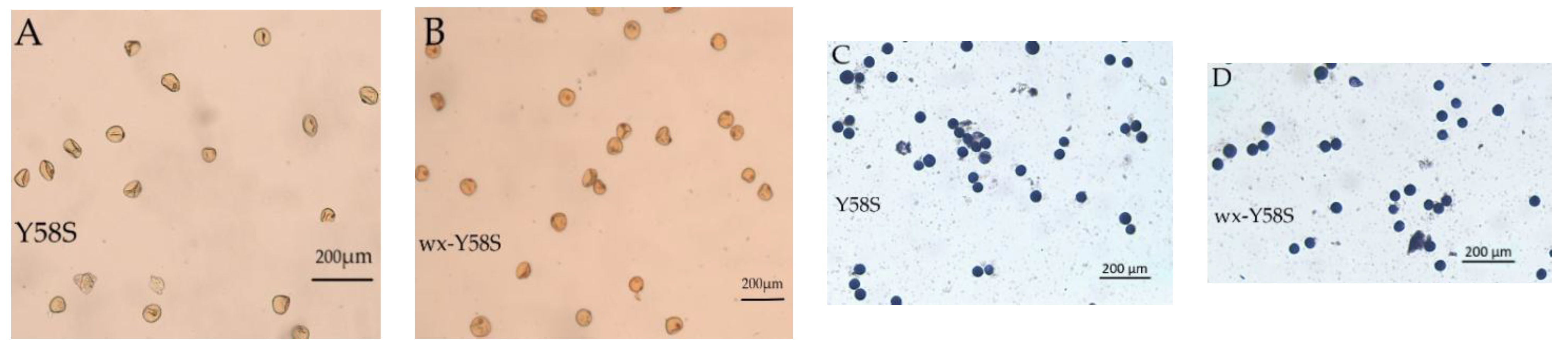
3.5. Plant Type, Fertility Transformation, and Appearance Analysis of Grain and Rice Kernels of T2 Homozygous Mutant Lines
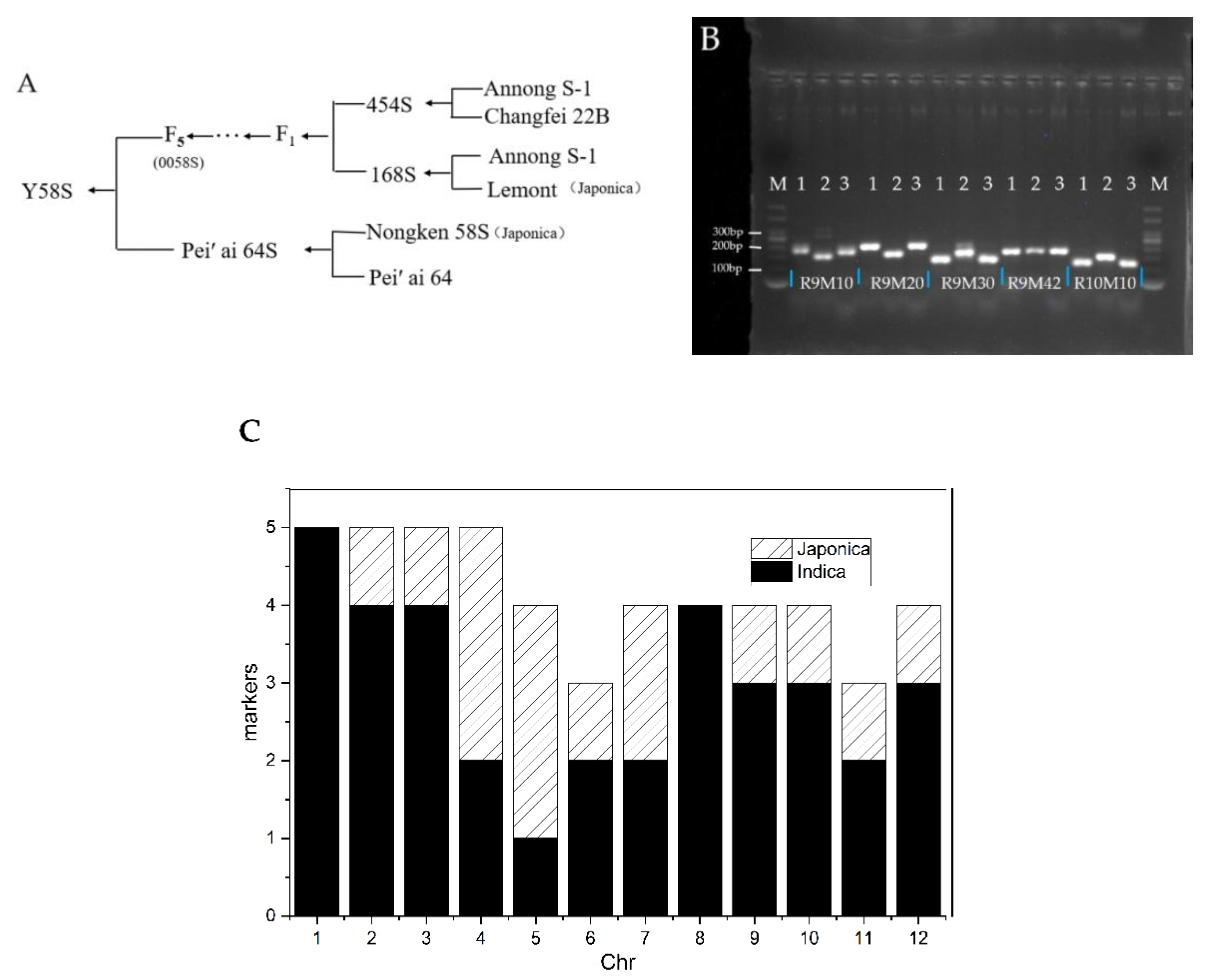
3.6. Analysis of the Genetic Background of Y58S
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
Abbreviations
| CRISPR/Cas9 | Clustered regularly interspaced short palindromic repeats/Cas9 |
| AC | Amylose content |
| PAM | Protospacer adjacent motif |
| QTL | Quantitative trait loci |
| PTGMS | Photothermo-sensitive genic-male-sterile |
| WT | Wild type |
| Wx | Waxy |
| T-DNA | Transfer DNA |
References
- Hsu, Y.-C.; Tseng, M.-C.; Wu, Y.-P.; Lin, M.-Y.; Wei, F.-J.; Hwu, K.-K.; Hsing, Y.-I.; Lin, Y.-R. Genetic factors responsible for eating and cooking qualities of rice grains in a recombinant inbred population of an inter-subspecific cross. Mol. Breed. 2014, 34, 655–673. [Google Scholar] [CrossRef] [Green Version]
- Zeng, D.; Tian, Z.; Rao, Y.; Dong, G.; Yang, Y.; Huang, L.; Leng, Y.; Xu, J.; Sun, C.; Zhang, G.; et al. Rational design of high-yield and superior-quality rice. Nat. Plants 2017, 3, 17031. [Google Scholar] [CrossRef]
- Wani, A.A.; Singh, P.; Shah, M.A.; Schweiggert-Weisz, U.; Gul, K.; Wani, I.A. Rice Starch Diversity: Effects on Structural, Morphological, Thermal, and Physicochemical Properties-A Review. Compr. Rev. Food Sci. Food Saf. 2012, 11, 417–436. [Google Scholar] [CrossRef]
- James, M.G.; Denyer, K.; Myers, A.M. Starch synthesis in the cereal endosperm. Curr. Opin. Plant. Biol. 2003, 6, 215–222. [Google Scholar] [CrossRef]
- Huang, L.; Li, Q.-F.; Zhang, C.; Chu, R.; Gu, Z.; Tan, H.; Zhao, D.; Fan, X.; Liu, Q. Creating novel Wx alleles with fine-tuned amylose levels and improved grain quality in rice by promoter editing using CRISPR/Cas9 system. Plant Biotechnol. J. 2020, 18, 2164–2166. [Google Scholar] [CrossRef]
- Zeng, D.; Liu, T.; Ma, X.; Wang, B.; Zheng, Z.; Zhang, Y.; Xie, X.; Yang, B.; Zhao, Z.; Zhu, Q.; et al. Quantitative regulation of Waxy expression by CRISPR/Cas9-based promoter and 5’UTR-intron editing improves grain quality in rice. Plant Biotechnol. J. 2020, 18, 2385–2387. [Google Scholar] [CrossRef]
- Wang, Z.-Y.; Zheng, F.-Q.; Shen, G.-Z.; Gao, J.-P.; Snustad, D.P.; Li, M.-G.; Zhang, J.-L.; Hong, M.-M. The amylose content in rice endosperm is related to the post-transcriptional regulation of the waxy gene. Plant J. 1995, 7, 613–622. [Google Scholar] [CrossRef] [PubMed]
- Sano, Y. Differential regulation of waxy gene expression in rice endosperm. Theor. Appl. Genet. 1984, 68, 467–473. [Google Scholar] [CrossRef]
- Xu, Y.; Lin, Q.; Li, X.; Wang, F.; Chen, Z.; Wang, J.; Li, W.; Fan, F.; Tao, Y.; Jiang, Y.; et al. Fine-tuning the amylose content of rice by precise base editing of the Wx gene. Plant Biotechnol. J. 2021, 19, 11–13. [Google Scholar] [CrossRef] [PubMed]
- Shu, Q.Y.; Wu, D.X.; Xia, Y.W.; Gao, M.W.; Ayres, N.M.; Larkin, P.D.; Park, W.D. Microsatellites Polymorphism on the Waxy Gene Locus and Their Relationship to Amylose Content in indica and japonica Rice (Oryza sativa L). J. Genet. Genom. 1999, 26, 350–358. [Google Scholar]
- Hirano, H.Y.; Eiguchi, M.; Sano, Y. A single base change altered the regulation of the Waxy gene at the posttranscriptional level during the domestication of rice. Mol. Biol. Evol. 1998, 15, 978–987. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Isshiki, M.; Morino, K.; Nakajima, M.; Okagaki, R.J.; Wessler, S.R.; Izawa, T.; Shimamoto, K. A naturally occurring functional allele of the rice waxy locus has a GT to TT mutation at the 5’ splice site of the first intron. Plant J. 1998, 15, 133–138. [Google Scholar] [CrossRef] [Green Version]
- Chen, Y.L.; Bao, J.S. Progress in structures, functions and interactions of enzymes related to starch synthesis in rice endosperm. J. China Rice Sci. 2017, 31, 1–12. [Google Scholar] [CrossRef]
- Terada, R.; Johzuka-Hisatomi, Y.; Saitoh, M.; Asao, H.; Iida, S. Gene Targeting by Homologous Recombination as a Biotechnological Tool for Rice Functional Genomics. Plant Physiol. 2007, 144, 846–856. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shan, Q.; Wang, Y.; Li, J.; Zhang, Y.; Chen, K.; Liang, Z.; Zhang, K.; Liu, J.; Xi, J.J.; Qiu, J.-L.; et al. Targeted genome modification of crop plants using a CRISPR-Cas system. Nat. Biotechnol. 2013, 31, 686–688. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Li, X.; Zhou, Z.; Wu, P.; Fang, M.; Pan, X.; Lin, Q.; Luo, W.; Wu, G.; Li, H. Reassessment of the Four Yield-related Genes Gn1a, DEP1, GS3, and IPA1 in Rice Using a CRISPR/Cas9 System. Front. Plant Sci. 2016, 7, 377. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Feng, Z.; Zhang, B.; Ding, W.; Liu, X.; Yang, D.-L.; Wei, P.; Cao, F.; Zhu, S.; Zhang, F.; Mao, Y.; et al. Efficient genome editing in plants using a CRISPR/Cas system. Cell Res. 2013, 23, 1229–1232. [Google Scholar] [CrossRef] [Green Version]
- Zhang, H.; Zhang, J.; Wei, P.; Zhang, B.; Gou, F.; Feng, Z.; Mao, Y.; Yang, L.; Zhang, H.; Xu, N.; et al. The CRISPR/Cas9 system produces specific and homozygous targeted gene editing in rice in one generation. Plant Biotechnol. J. 2014, 12, 797–807. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.; Zhang, Q.; Zhu, Q.; Liu, W.; Chen, Y.; Qiu, R.; Wang, B.; Yang, Z.; Li, H.; Lin, Y.; et al. A Robust CRISPR/Cas9 System for Convenient, High-Efficiency Multiplex Genome Editing in Monocot and Dicot Plants. Mol. Plant 2015, 8, 1274–1284. [Google Scholar] [CrossRef]
- Wang, J.-F.; Zheng, C.-M.; Liu, W.; Luo, W.-L.; Wang, H.; Chen, Z.-Q.; Guo, T. Construction of tgw6 Mutants in Rice Based on CRISPR/Cas9 Technology. Acta Agron. Sin. 2016, 42, 1160–1167. [Google Scholar] [CrossRef]
- DiCarlo, J.; Norville, J.; Mali, P.; Rios, X.; Aach, J.; Church, G.M. Genome engineering in Saccharomyces cerevisiae using CRISPR-Cas systems. Nucleic Acids Res. 2013, 41, 4336–4343. [Google Scholar] [CrossRef] [Green Version]
- Zhang, J.; Zhang, H.; Botella, J.R.; Zhu, J.-K. Generation of new glutinous rice by CRISPR/Cas9-targeted mutagenesis of the Waxy gene in elite rice varieties. J. Integr. Plant Biol. 2018, 60, 369–375. [Google Scholar] [CrossRef]
- Yunyan, F.; Jie, Y.; Fangquan, W.; Fangjun, F.; Wenqi, L.; Jun, W.; Yang, X.; Jinyan, Z.; Weigong, Z. Production of Two Elite Glutinous Rice Varieties by Editing Wx Gene. Rice Sci. 2019, 26, 118–124. [Google Scholar] [CrossRef]
- Hiei, Y.; Komari, T.; Kubo, T. Transformation of rice mediated by Agrobacterium tumefaciens. Plant Mol. Biol. 1997, 35, 205–218. [Google Scholar] [CrossRef] [PubMed]
- Milligan, B.G. Purification of chloroplast DNA using hexadecyltrimethylammonium bromide. Plant Mol. Biol. Rep. 1989, 7, 144–149. [Google Scholar] [CrossRef]
- Lin, M.J.; Song, J.F.; Li, D.J. Determination of amylose and amylopectin content in fresh corn by dual-wavelength spectrophotometry. Acta Agric. Jiangxi 2010, 22, 117–119. (In Chinese) [Google Scholar] [CrossRef]
- Huang, J.R.; Wang, Z.J.; Li, L.C. Determination of amylose and amylopectin in mung bean by dual wavelength spectrophotometry. J. Mach. Food 2015, 31, 48–51. (In Chinese) [Google Scholar] [CrossRef]
- Jialiexi, M.; Jing, W.W.; Xuelaiti, Z. Determination of Amylose and Amylopectin in Grain and Bean by Dual-Wavelength Spectrophotometry. J. Xinjiang Agric. Sci. 2010, 47, 564–568. (In Chinese) [Google Scholar]
- Shen, Y.-J.; Jiang, H.; Jin, J.-P.; Zhang, Z.-B.; Xi, B.; He, Y.-Y.; Wang, G.; Wang, C.; Qian, L.; Li, X.; et al. Development of Genome-Wide DNA Polymorphism Database for Map-Based Cloning of Rice Genes. Plant Physiol. 2004, 135, 1198–1205. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gao, C. Genome engineering for crop improvement and future agriculture. Cell 2021, 184, 1621–1635. [Google Scholar] [CrossRef] [PubMed]
- Liao, S.; Qin, X.; Luo, L.; Han, Y.; Wang, X.; Usman, B.; Nawaz, G.; Zhao, N.; Liu, Y.-G.; Li, R. CRISPR/Cas9-Induced Mutagenesis of Semi-Rolled Leaf1,2 Confers Curled Leaf Phenotype and Drought Tolerance by Influencing Protein Expression Patterns and ROS Scavenging in Rice (Oryza sativa L.). Agronomy 2019, 9, 728. [Google Scholar] [CrossRef] [Green Version]
- Han, Y.; Teng, K.; Nawaz, G.; Feng, X.; Usman, B.; Wang, X.; Luo, L.; Zhao, N.; Liu, Y.; Li, R. Generation of semi-dwarf rice (Oryza sativa L.) lines by CRISPR/Cas9-directed mutagenesis of OsGA20ox2 and proteomic analysis of unveiled changes caused by mutations. 3 Biotech. 2019, 9, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Han, Y.; Luo, D.; Usman, B.; Nawaz, G.; Zhao, N.; Liu, F.; Li, R. Development of High Yielding Glutinous Cytoplasmic Male Sterile Rice (Oryza sativa L.) Lines through CRISPR/Cas9 Based Mutagenesis of Wx and TGW6 and Proteomic Analysis of Anther. Agronomy 2018, 8, 290. [Google Scholar] [CrossRef] [Green Version]
- Deng, H.B. Research quality of status and evolution of waxy rice. J. Crop Res. 2000, 14, 45–48. [Google Scholar]
- Zhang, H.X.; Zhang, B.Q.; Kong, X.D. Breeding and application of indica glutinous hybrid rice. J. Hybrid Rice 1989, 4, 39–42. (In Chinese) [Google Scholar]
- Deng, X.L. Selection of two-line hybrid glutinous rice. J. Hybrid Rice 1992, 7, 33–34. (In Chinese) [Google Scholar] [CrossRef]
- Deng, S.D.; Guang, H.R.; Deng, W.M. Hybrid glutinous rice Nuoyou No.1. J. Hybrid Rice 1996, 11, 34. (In Chinese) [Google Scholar] [CrossRef]
- Si, H.-M.; Fu, Y.-P.; Liu, W.-Z.; Sun, Z.-X.; Hu, G.-C. Pedigree Analysis of Photoperiod-thermo Sensitive Genic Male Sterile Rice. Acta Agron. Sin. 2012, 38, 394. [Google Scholar] [CrossRef]
- Deng, Q.Y. Selection and breeding of a wide adaptability rice photo thermo sensitive male sterile line Y58S. J. Hybrid Rice 2005, 2, 18–21. (In Chinese) [Google Scholar] [CrossRef]
- Luo, X.H.; Qiu, Z.Z.; Li, R.H. Pei’ai 64S, a dual-purpose sterile line with low critical sterile temperature. J. Hybrid Rice 1992, 1, 27–29. (In Chinese) [Google Scholar] [CrossRef]





| Primer Name | Primer Sequence 5’-3’ |
|---|---|
| Wx-text-F | TCCGCCACGGGTTCCAG |
| Wx-text-R | CTCCTACCTCAGCCACAACG |
| U-F | CTCCGTTTTACCTGTGGAATCG |
| gR-R | CGGAGGAAAATTCCATCCAC |
| Wx-U6a-1F | TGTGTGCTTACAGCCATGGCGTTTTAGAGCTAGAAAT |
| Wx-U6a-1R | GCCATGGCTGTAAGCACACACGGCAGCCAAGCCAGCA |
| Pps-GGL | TTCAGAGGTCTCTCTCGACTAGTATGGAATCGGCAGCAAAGG |
| Pgs-GGR | AGCGTGGGTCTCGACCGACGCGTATCCATCCACTCCAAGCTC |
| PB-R | GCGCGCGGTCTCTACCGACGCGTATCC |
| PB-L | GCGCGCgGTCTCGCTCGACTAGTATGG |
| HPT-F | ATTTGTGTACGCCCGACAGT |
| HPT-R | GTGCTTGACATTGGGGAGTT |
| CAS9-F | CTGACGCTAACCTCGACAAG |
| CAS-R | CCGATCTAGTAACATAGATGACACC |
| Types | Heterozygous | Bi-Allelic | Homozygous | Total |
|---|---|---|---|---|
| Number | 5 | 7 | 6 | 18 |
| Transformation Material | Mutation | Type |
|---|---|---|
| W-T | GCCA_TGGCTGTAAGCACACA | |
| W-1 | GCCTCTGGATGGAAGCACACA | +1/+3substitution |
| W-4 | GCCAATGGCTGTAAGCACACA | +1 |
| W-6 | GCCA_TGGGCTGTAAGCACACA | +1 |
| T2 Line | AC (%) |
|---|---|
| W-4-H-3 | 0.8 |
| W-4-H-5 | 1.0 |
| W-1-B-2 | 1.8 |
| W-1-B-5 | 0.6 |
| W-1-B-10 | 0.9 |
| Y58S (control) | 13.8 |
| T2 Homozygous Mutation Lines | Plant Height (cm) | No. of Panicle/Plant | Flag Leaf Length (cm) | Flag Leaf Width (cm) | Length of Panicle (cm) | Grain no. per Spike | Grain Set Rate (%) |
|---|---|---|---|---|---|---|---|
| W-4-H-3 | 83.3a | 6 | 45.3 a | 1.5 a | 25.0 a | 183 a | 84.6 a |
| W-4-H-5 | 83.5 a | 6 | 45.5 a | 1.5 a | 26.2 a | 184 a | 85.0 a |
| W-1-B-2 | 82.7 a | 6 | 46.2 a | 1.6 a | 25.8 a | 181 a | 85.2 a |
| W-1-B-5 | 81.9 a | 7 | 46.5 a | 1.5 a | 25.8 a | 180 a | 85.0 a |
| Y58S (CK) | 82.7 a | 7 | 46.8 a | 1.6 a | 26.2 a | 186 a | 85.5 a |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Teng, K.; Wang, X.; Guo, X.; Liu, Y.; Li, R. Generation of a New Glutinous Photothermosensitive Genic-Male-Sterile (PTGMS) Line by CRISPR/Cas9-Directed Mutagenesis of Wx in Rice (Oryza sativa L.). Agriculture 2021, 11, 1044. https://doi.org/10.3390/agriculture11111044
Teng K, Wang X, Guo X, Liu Y, Li R. Generation of a New Glutinous Photothermosensitive Genic-Male-Sterile (PTGMS) Line by CRISPR/Cas9-Directed Mutagenesis of Wx in Rice (Oryza sativa L.). Agriculture. 2021; 11(11):1044. https://doi.org/10.3390/agriculture11111044
Chicago/Turabian StyleTeng, Kaichong, Xin Wang, Xinying Guo, Yaoguang Liu, and Rongbai Li. 2021. "Generation of a New Glutinous Photothermosensitive Genic-Male-Sterile (PTGMS) Line by CRISPR/Cas9-Directed Mutagenesis of Wx in Rice (Oryza sativa L.)" Agriculture 11, no. 11: 1044. https://doi.org/10.3390/agriculture11111044
APA StyleTeng, K., Wang, X., Guo, X., Liu, Y., & Li, R. (2021). Generation of a New Glutinous Photothermosensitive Genic-Male-Sterile (PTGMS) Line by CRISPR/Cas9-Directed Mutagenesis of Wx in Rice (Oryza sativa L.). Agriculture, 11(11), 1044. https://doi.org/10.3390/agriculture11111044







