Molecular Markers and a Quality Trait Evaluation for Assessing the Genetic Diversity of Avocado Landraces from China
Abstract
1. Introduction
2. Materials and Methods
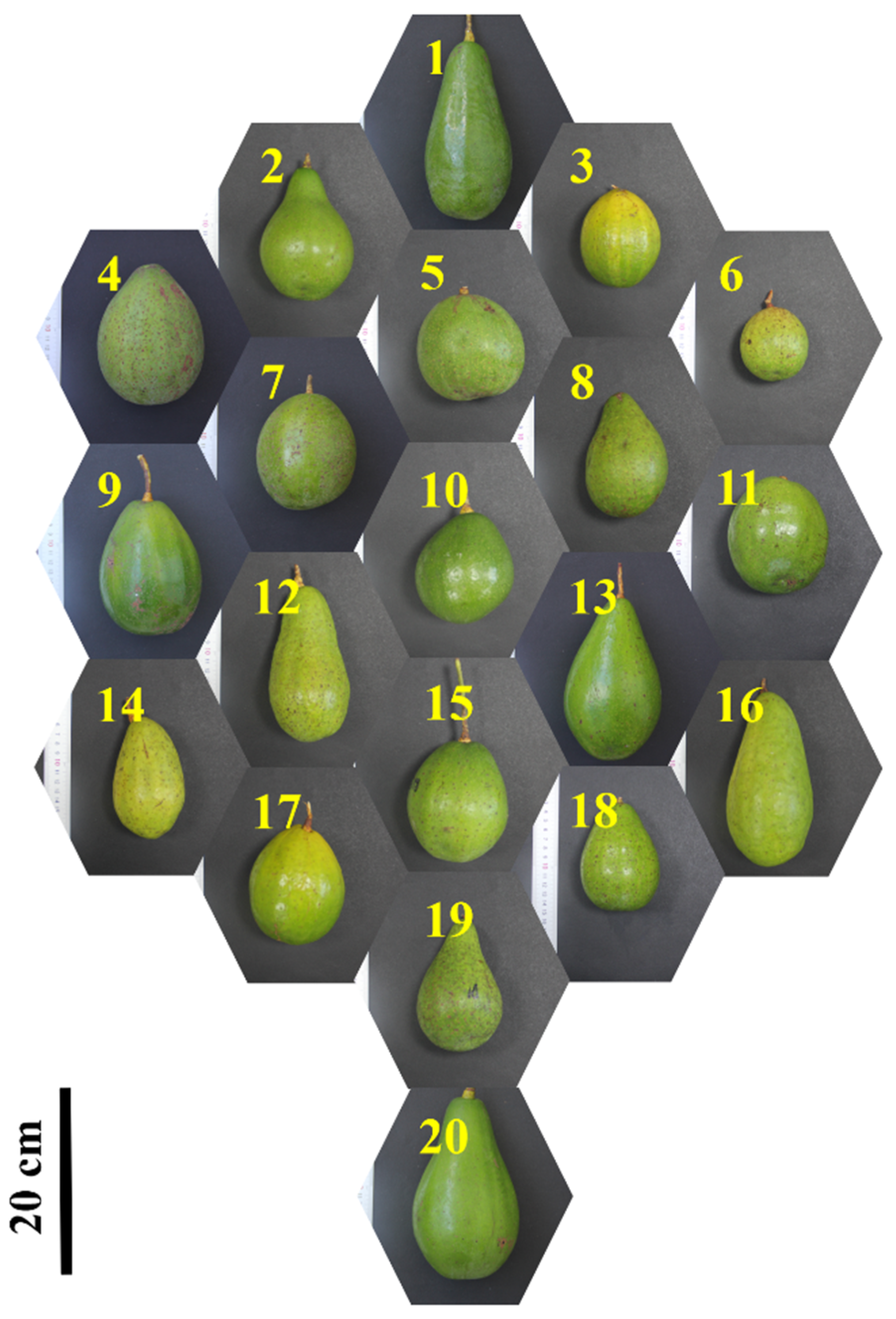
2.1. Sample Collection
2.2. DNA Extraction and Analysis of Avocado Accessions with an Unknown Race
2.3. EST-SSR Marker Analysis
2.4. Quantification of Quality Traits
2.4.1. Moisture Assay
2.4.2. Oil Content Assay
2.4.3. Soluble Sugar Assay
2.4.4. Soluble Protein Assay
2.4.5. Ascorbic Acid Assay
2.4.6. Total Phenol Assay
2.4.7. Total Flavonoid Assay
2.5. Data Analysis
3. Results
3.1. Assignment of Avocado Races
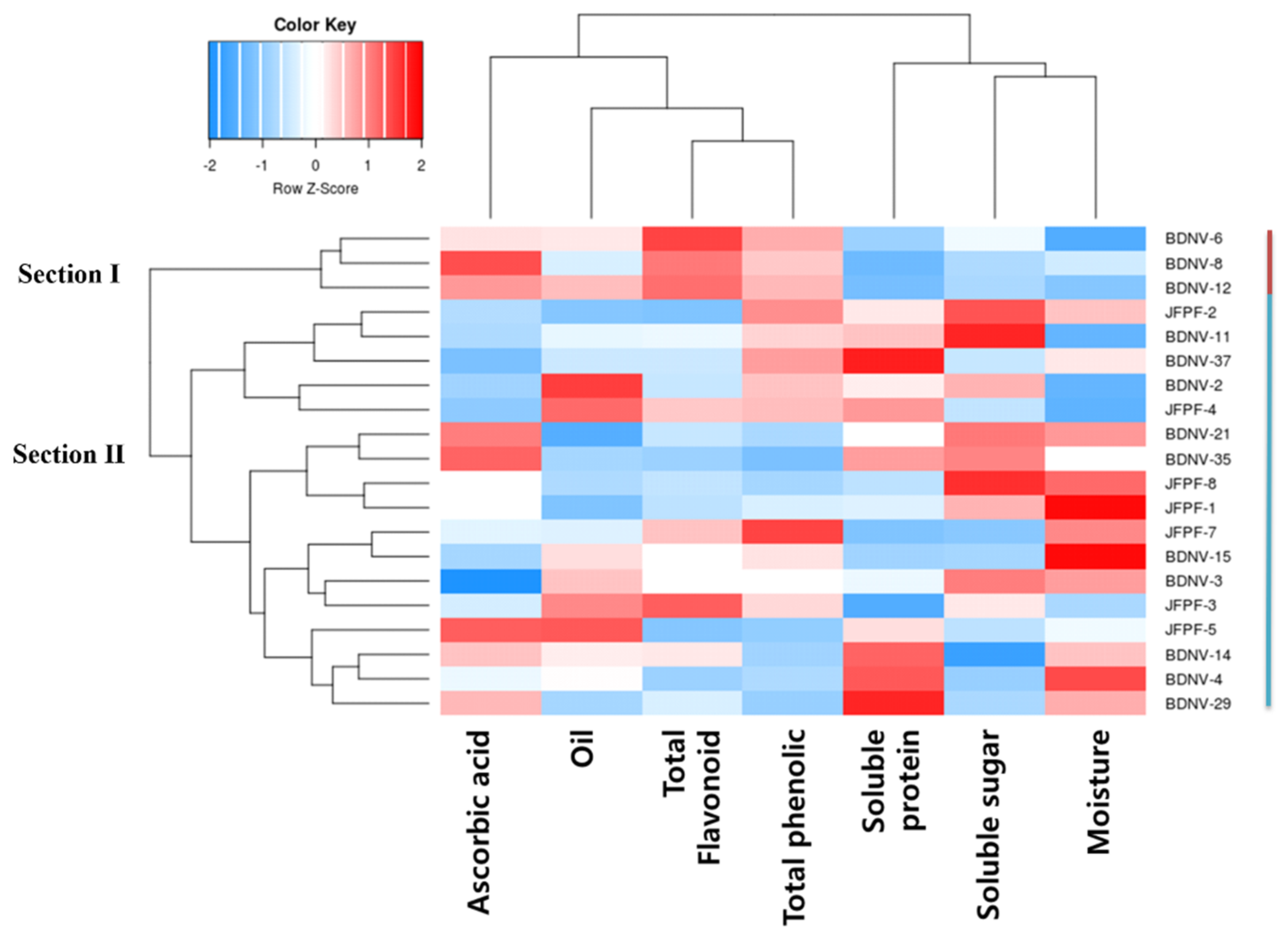
3.2. Characterization of Quality Traits
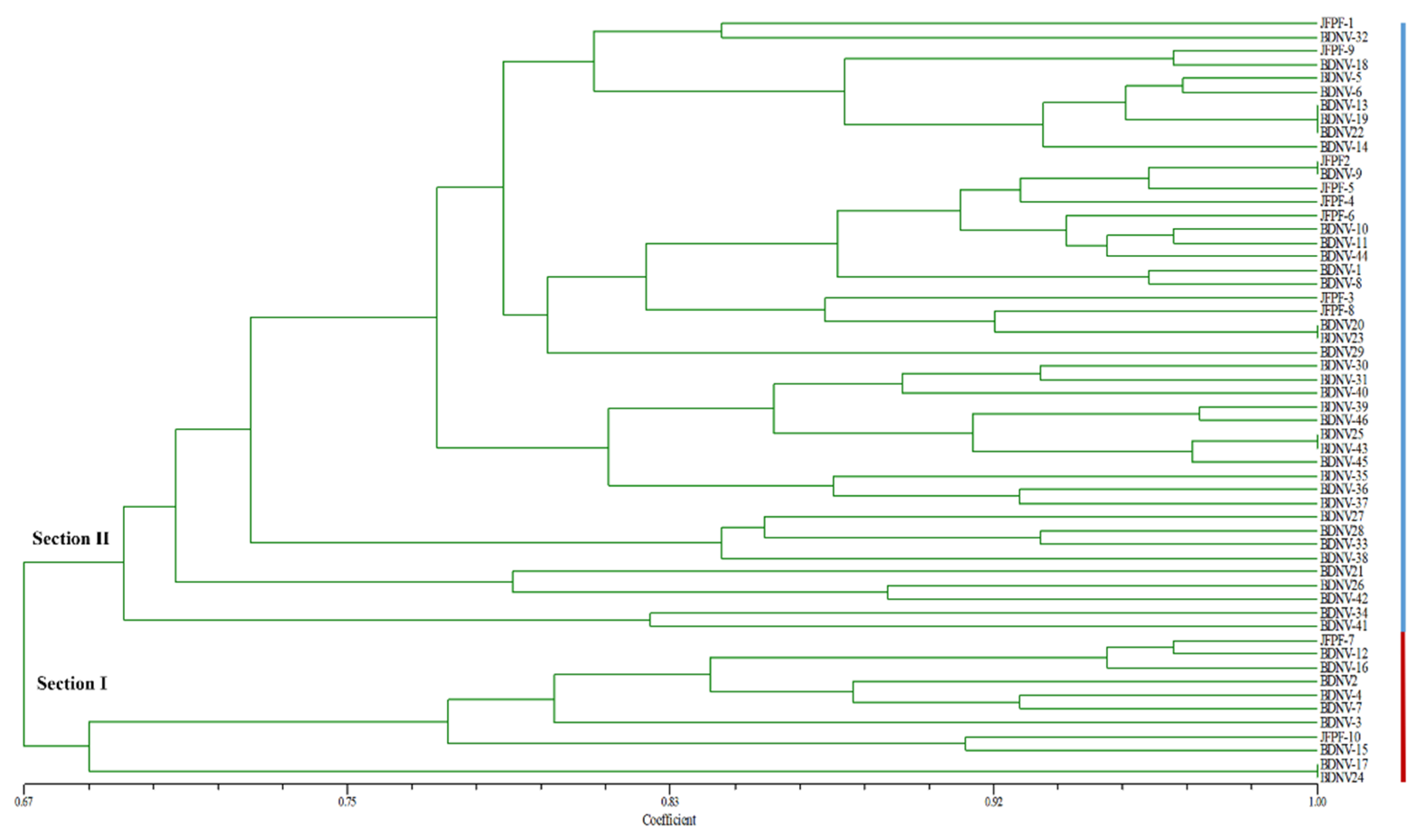
3.3. Molecular Characterization
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Kuhn, D.N.; Groh, A.; Rahaman, J.; Freeman, B.; Arpaia, M.L.; Van den Berg, N.; Abeysekara, N.; Manosalva, P.; Chambers, A.H. Creation of an avocado unambiguous genotype SNP database for germplasm curation and as an aid to breeders. Tree Genet. Genomes 2019, 15, 71. [Google Scholar] [CrossRef]
- Rendón-Anaya, M.; Ibarra-Laclette, E.; Méndez-Bravo, A.; Lan, T.Y.; Zheng, C.F.; Carretero-Paulet, L.; Perez-Torres, C.A.; Chacón-López, A.; Hernandez-Guzmán, G.; Chang, T.H. The avocado genome informs deep angiosperm phylogeny, highlights introgressive hybridization, and reveals pathogen-influenced gene space adaptation. Proc. Natl. Acad. Sci. USA 2019, 116, 17081–17089. [Google Scholar] [CrossRef] [PubMed]
- Ge, Y.; Cheng, Z.H.; Si, X.Y.; Ma, W.H.; Tan, L.; Zang, X.P.; Wu, B.; Xu, Z.N.; Wang, N.; Zhou, Z.X.; et al. Transcriptome profiling provides insight into the genes in carotenoid biosynthesis during the mesocarp and seed developmental stages of avocado (Persea americana). Int. J. Mol. Sci. 2019, 20, 4117. [Google Scholar] [CrossRef] [PubMed]
- Ge, Y.; Dong, X.S.; Wu, B.; Wang, N.; Chen, D.; Chen, H.H.; Zou, M.H.; Xu, Z.N.; Tan, L.; Zhan, R.L. Evolutionary analysis of six chloroplast genomes from three Persea americana ecological races: Insights into sequence divergences and phylogenetic relationships. PLoS ONE 2019, 14, e0221827. [Google Scholar] [CrossRef]
- Galindo-Tovar, M.E.; Ogata-Aguilar, N.; Arzate-Fernandez, A.M. Some aspects of avocado (Persea americana Mill.) diversity and domestication in Mesoamerica. Genet. Resour. Crop Evol. 2008, 55, 441–450. [Google Scholar] [CrossRef]
- Talavera, A.; Soorni, A.; Bombarely, A.; Matas, A.J.; Hormaza, J.I. Genome-Wide SNP discovery and genomic characterization in avocado (Persea americana Mill.). Sci. Rep. 2019, 9, 20137. [Google Scholar] [CrossRef]
- Schaffer, B.; Wolstenholme, B.N.; Whiley, A.W. The Avocado: Botany, Production and Uses, 2nd ed.; CPI Group (UK) Ltd: Croydon, UK, 2012. [Google Scholar]
- Gross-German, E.; Viruel, M.A. Molecular characterization of avocado germplasm with a new set of SSR and EST-SSR markers: Genetic diversity, population structure, and identification of race-specific markers in a group of cultivated genotypes. Tree Genet. Genomes 2013, 9, 539–555. [Google Scholar] [CrossRef]
- Ge, Y.; Hu, F.C.; Tan, L.; Wu, B.; Wang, T.; Zhang, T.; Ma, F.N.; Cao, J.Q.; Xu, Z.N.; Zhan, R.L. Molecular diversity in a germplasm collection of avocado accessions from the tropical and subtropical regions of China. Crop Breed. Appl. Biot. 2019, 19, 153–160. [Google Scholar] [CrossRef]
- Zhang, L.; Zhang, D.S.; Liu, K.D. Environmental analysis and countermeasures for industrial development of Hainan avocado. Chin. J. Agric. Resou. Reg. Plan. 2015, 36, 78–84. [Google Scholar]
- Ge, Y.; Si, X.Y.; Lin, X.E.; Wang, J.S.; Zang, X.P.; Ma, W.H. Advances in avocado (Persea americana Mill.). South China Fruit 2017, 46, 63–70. [Google Scholar] [CrossRef]
- Ge, Y.; Zang, X.P.; Tan, L.; Wang, J.S.; Liu, Y.Z.; Li, Y.X.; Wang, N.; Chen, D.; Zhan, R.L.; Ma, W.H. Single-molecule long-read sequencing of avocado generates microsatellite markers for analyzing the genetic diversity in avocado germplasm. Agronomy 2019, 9, 512. [Google Scholar] [CrossRef]
- Fiedler, J.; Bufler, G.; Bangerth, F. Genetic relationships of avocado (Persea americana Mill.) using RAPD markers. Euphytica 1998, 101, 249–255. [Google Scholar] [CrossRef]
- Furnier, G.R.; Cummings, M.P.; Clegg, M.T. Evolution of the avocados as revealed by DNA restriction site variation. J. Hered. 1990, 81, 183–188. [Google Scholar] [CrossRef]
- Ashworth, V.E.T.M.; Clegg, M.T. Microsatellite markers in avocado (Persea americana Mill.) genealogical relationships among cultivated avocado genotypes. J. Hered. 2003, 94, 407–415. [Google Scholar] [CrossRef]
- Schnell, R.J.; Brown, J.S.; Olano, C.T.; Power, E.J.; Krol, C.A.; Kuhn, D.N.; Motamayor, J.C. Evaluation of avocado germplasm using microsatellite markers. J. Amer. Soc. Hort. Sci. 2003, 128, 881–889. [Google Scholar] [CrossRef]
- Chen, H.; Morrel, P.L.; Ashwoth, V.E.T.M.; De la Cruz, M.; Clegg, M.T. Nucleotide diversity and linkage disequilibrium in wild avocado (Persea americana Mill.). J. Hered. 2008, 99, 382–389. [Google Scholar] [CrossRef]
- Chen, H.; Morrel, P.L.; Ashwoth, V.E.T.M.; De la Cruz, M.; Clegg, M.T. Tracing the geographic origins of mayor avocado cultivars. J. Hered. 2009, 100, 56–65. [Google Scholar] [CrossRef]
- Liu, C.; Ge, Y.; Wang, D.J.; Li, X.; Yang, X.X.; Cui, C.S.; Qu, S.P. Morphological and molecular diversity in a germplasm collection of seed pumpkin. Sci. Hortic. 2013, 154, 8–16. [Google Scholar] [CrossRef]
- Ramírez, I.M.; Fuentes, J.L.; Rodríguez, N.N.; Coto, O.; Cueto, J.; Becker, D.; Rohde, W. Diversity analysis of Cuban avocado varieties based on agro-morphological traits and DNA polymorphisms. J. Genet. Breed. 2005, 59, 241–252. [Google Scholar]
- Ge, Y.; Zhang, T.; Wu, B.; Tan, L.; Ma, F.; Zou, M.; Chen, H.; Pei, J.; Liu, Y.; Chen, Z.; et al. Genome-wide assessment of avocado germplasm determined from specific length amplified fragment sequencing and transcriptomes: Population structure, genetic diversity, identification, and application of race-specific markers. Genes 2019, 10, 215. [Google Scholar] [CrossRef]
- Ge, Y.; Tan, L.; Wu, B.; Wang, T.; Zhang, T.; Chen, H.; Zou, M.; Ma, F.; Xu, Z.; Zhan, R. Transcriptome sequencing of difference avocado ecotypes: De novo transcriptome assembly, annotation, identification and validation of EST-SSR markers. Forests 2019, 10, 411. [Google Scholar] [CrossRef]
- Ge, Y.; Ramchiary, N.; Wang, T.; Liang, C.; Wang, N.; Wang, Z.; Choi, S.R.; Lim, Y.P.; Piao, Z.Y. Development and linkage mapping of unigene-derived microsatellite markers in Brassica rapaL. Breed. Sci. 2011, 61, 160–167. [Google Scholar] [CrossRef]
- Ge, Y.; Si, X.Y.; Cao, J.Q.; Zhou, Z.X.; Wang, W.L.; Ma, W.H. Morphological characteristics, nutritional quality, and bioactive constituents in fruits of two avocado (Persea americana) varieties from hainan province, China. J. Agric. Sci. 2017, 9, 8–17. [Google Scholar] [CrossRef]
- Krawczak, M.; Nikolaus, S.; von Eberstein, H.; Croucher, P.J.; El Mokhtari, N.E.; Schreiber, S. PopGen: Population based recruitment of patients and controls for the analysis of complex genotype-phenotype relationships. Community Genet. 2006, 9, 55–61. [Google Scholar] [CrossRef]
- Rohlf, F.J. NTSYS pc: Numerical Taxonomy and Multivariate Analysis System; Version 2.1; Exeter Software: New York, NY, USA, 2000. [Google Scholar]
- Alcaraz, M.L.; Hormaza, J.I. Molecular characterization and genetic diversity in an avocado collection of cultivars and local Spanish genotypes using SSRs. Hereditas 2007, 144, 244–253. [Google Scholar] [CrossRef]
- Ge, Y.; Si, X.Y.; Cao, J.Q.; Hu, F.C.; Wu, K.; Xu, H.M.; Tan, H.R.; Zang, X.P.; Ma, W.H. The analysis on the nutrition and biological activity of avocado fruit. South China Fruits 2017, 46, 50–56. [Google Scholar] [CrossRef]
- Donetti, M.; Terry, L.A. Biochemical markers defining growing area and ripening stage of imported avocado fruit cv. Hass. J. Food Compos. Anal. 2014, 34, 90–98. [Google Scholar] [CrossRef]
| Quality Traits | Maximum | Minimum | Mean | Standard Deviation | Coefficient of Variation (%) |
|---|---|---|---|---|---|
| Moisture (g/100g) | 86.33 | 77.00 | 82.50 | 2.63 | 3.19 |
| Oil (g/100g) | 11.64 | 5.44 | 8.01 | 1.56 | 19.53 |
| Soluble sugar (g/100g) | 1.44 | 0.76 | 1.12 | 0.22 | 19.77 |
| Soluble protein (mg/100g) | 3.56 | 1.11 | 2.19 | 0.61 | 28.08 |
| Ascorbic acid (mg/100g) | 66.70 | 12.75 | 40.83 | 13.75 | 33.68 |
| Total phenol (mg/100g) | 8.92 | 3.27 | 6.21 | 1.87 | 30.05 |
| Total flavonoid (mg/100g) | 9.12 | 1.29 | 3.91 | 2.15 | 54.94 |
| Marker Name | Na 1 | Ne 2 | Ho 3 | He 4 | I 5 |
|---|---|---|---|---|---|
| Pa-eSSR-1 | 2 | 1.26 | 0.23 | 0.21 | 0.36 |
| Pa-eSSR-2 | 3 | 2.64 | 0.68 | 0.62 | 1.03 |
| Pa-eSSR-4 | 3 | 1.46 | 0.35 | 0.31 | 0.58 |
| Pa-eSSR-5 | 4 | 2.04 | 0.68 | 0.51 | 0.90 |
| Pa-eSSR-7 | 3 | 1.90 | 0.24 | 0.47 | 0.81 |
| Pa-eSSR-9 | 4 | 1.46 | 0.27 | 0.31 | 0.66 |
| Pa-eSSR-12 | 2 | 1.13 | 0.13 | 0.12 | 0.23 |
| Pa-eSSR-13 | 4 | 1.96 | 0.63 | 0.49 | 0.92 |
| Pa-eSSR-14 | 3 | 1.14 | 0.13 | 0.12 | 0.27 |
| Pa-eSSR-15 | 3 | 2.03 | 0.59 | 0.51 | 0.74 |
| Total | 31 | ||||
| Mean | 3.10 | 1.70 | 0.39 | 0.36 | 0.65 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, Y.; Ge, Y.; Zhan, R.; Lin, X.; Zang, X.; Li, Y.; Yang, Y.; Ma, W. Molecular Markers and a Quality Trait Evaluation for Assessing the Genetic Diversity of Avocado Landraces from China. Agriculture 2020, 10, 102. https://doi.org/10.3390/agriculture10040102
Liu Y, Ge Y, Zhan R, Lin X, Zang X, Li Y, Yang Y, Ma W. Molecular Markers and a Quality Trait Evaluation for Assessing the Genetic Diversity of Avocado Landraces from China. Agriculture. 2020; 10(4):102. https://doi.org/10.3390/agriculture10040102
Chicago/Turabian StyleLiu, Yuanzheng, Yu Ge, Rulin Zhan, Xinge Lin, Xiaoping Zang, Yanxia Li, Ying Yang, and Weihong Ma. 2020. "Molecular Markers and a Quality Trait Evaluation for Assessing the Genetic Diversity of Avocado Landraces from China" Agriculture 10, no. 4: 102. https://doi.org/10.3390/agriculture10040102
APA StyleLiu, Y., Ge, Y., Zhan, R., Lin, X., Zang, X., Li, Y., Yang, Y., & Ma, W. (2020). Molecular Markers and a Quality Trait Evaluation for Assessing the Genetic Diversity of Avocado Landraces from China. Agriculture, 10(4), 102. https://doi.org/10.3390/agriculture10040102





