Selection of Lactic Acid Bacteria from Alfalfa Silage and Its Effects as Inoculant on Silage Fermentation
Abstract
1. Introduction
2. Materials and Methods
2.1. Experimental Area
2.2. Part 1—Isolation, Characterization and Identification of LAB from Alfalfa Forage and Its Silage
2.2.1. Ensiling and LAB Isolation
2.2.2. Physiological Tests
2.2.3. Determination of Antimicrobial Activity
2.2.4. Extraction of LAB Genomic DNA
2.2.5. Species Identification by 16S rRNA Gene Sequencing
2.2.6. Selection of LAB Strains for Alfalfa Silage Production
2.3. Part 2—Fermentation of Alfalfa Silage Inoculated with Wild LAB Strains
2.3.1. Experimental Design and Silage Preparation
2.3.2. Preparation of the Isolated Strains for Use as Inoculant
2.3.3. Chemical Analysis
2.3.4. Quantification of Microbial Populations
2.4. Statistical Analysis
3. Results
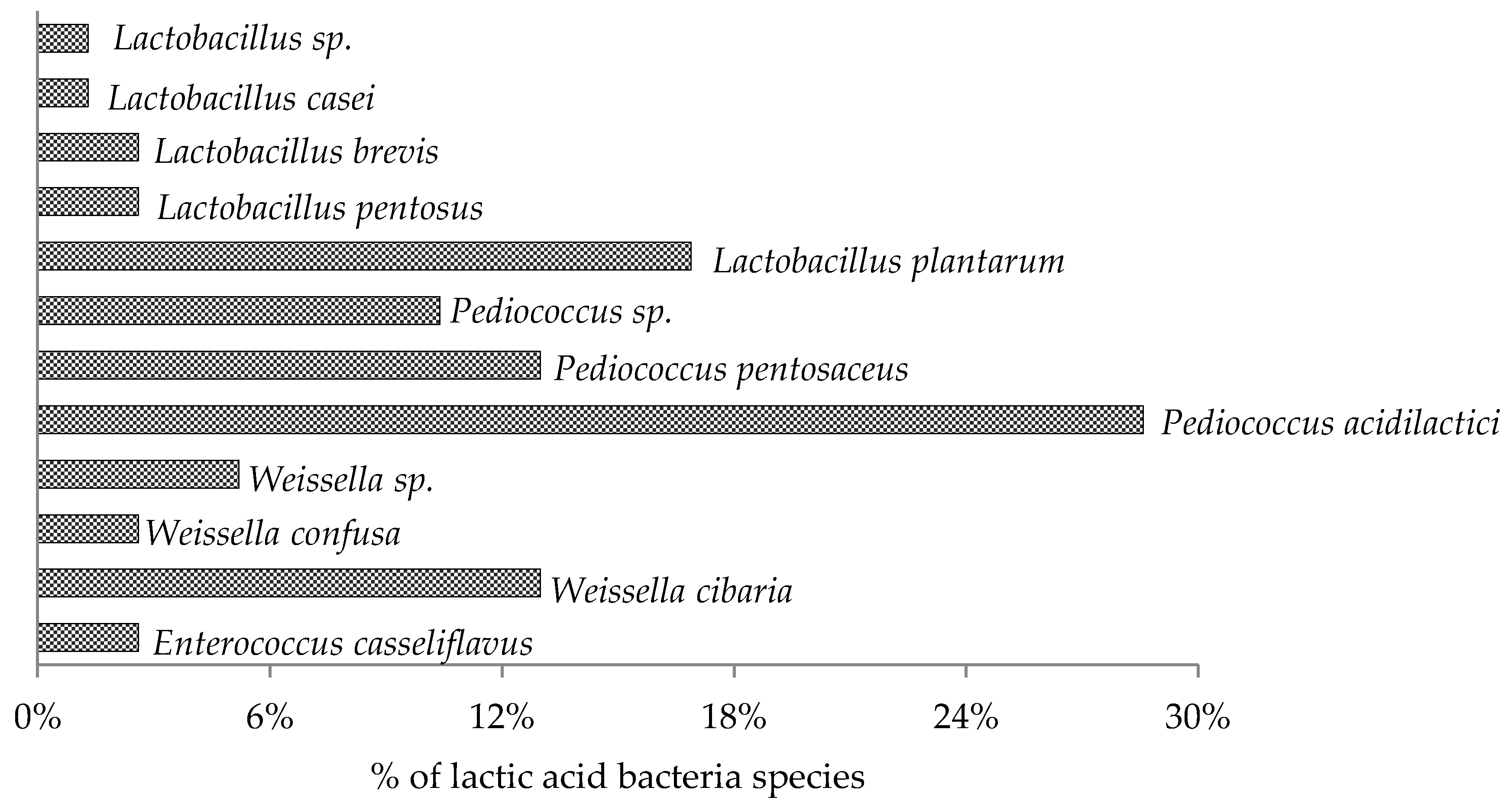
3.1. Screening and Identification of LAB from Alfalfa Forage and Its Silage (Part 1)
3.2. Chemical Composition (Part 2)
3.3. Fermentation Characteristics and Microbial Population (Part 2)
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Muck, R.R.; Nadeau, T.A.; McAllister, T.A.; Contreras-Govea, F.E.; Santos, M.C.; Kung, L. Silage review: Recent advances and future uses of silage additives. J. Dairy Sci. 2018, 101, 3980–4000. [Google Scholar] [CrossRef] [PubMed]
- Fabiszewska, A.U.; Zielińska, K.J.; Wróbel, B. Trends in designing microbial silage quality by biotechnological methods using lactic acid bacteria inoculants: A minireview. World J. Microbiol. Biotechnol. 2019, 35, 76. [Google Scholar] [CrossRef] [PubMed]
- Li, P.; Zhang, Y.; Gou, W.; Cheng, Q.; Bai, S.; Cai, Y. Silage fermentation and bacterial community of bur clover, annual ryegrass and their mixtures prepared with microbial inoculant and chemical additive. Anim. Feed Sci. Technol. 2019, 247, 285–293. [Google Scholar] [CrossRef]
- Yan, Y.; Li, X.; Guan, H.; Huang, L.; Ma, X.; Peng, Y.; Li, Z.; Nie, G.; Zhou, J.; Yang, W.; et al. Microbial community and fermentation characteristic of Italian ryegrass silage prepared with corn stover and lactic acid bacteria. Bioresour. Technol. 2019, 279, 166–173. [Google Scholar] [CrossRef] [PubMed]
- Oliveira, A.S.; Weinberg, Z.G.; Ogunade, I.M.; Cervantes, A.A.P.; Arriola, K.G.; Jiang, Y.; Kim, D.; Li, X.; Gonçalves, M.C.M.; Vyas, D.; et al. Meta-analysis of effects of inoculation with homofermentative and facultative heterofermentative lactic acid bacteria on silage fermentation, aerobic stability, and the performance of dairy cows. J. Dairy Sci. 2017, 100, 4587–4603. [Google Scholar] [CrossRef]
- Khota, W.; Pholsen, S.; Higgs, D.; Cai, Y. Natural lactic acid bacteria population of tropical grasses and their fermentation factor analysis of silage prepared with cellulase and inoculant. J. Dairy Sci. 2016, 99, 9768–9781. [Google Scholar] [CrossRef]
- Chen, M.M.; Liu, Q.H.; Xin, G.R.; Zhang, J.G. Characteristics of lactic acid bacteria isolates and their inoculating effects on the silage fermentation at high temperature. Lett. Appl. Microbiol. 2013, 56, 71–78. [Google Scholar] [CrossRef]
- Sifeeldein, A.; Wang, S.; Li, J.; Dong, Z.; Chen, L.; Kaka, N.A.; Shao, T. Phylogenetic identification of lactic acid bacteria isolates and their effects on the fermentation quality of sweet sorghum (Sorghum bicolor) silage. J. Appl. Microbiol. 2019, 126, 718–729. [Google Scholar] [CrossRef]
- Zhang, Q.; Yu, Z.; Wang, X. Isolating and evaluating lactic acid bacteria strains with or without sucrose for effectiveness of silage fermentation. Grassl. Sci. 2015, 61, 167–176. [Google Scholar] [CrossRef]
- Pang, H.; Qin, G.; Tan, Z.; Li, Z.; Wang, Y.; Cai, Y. Natural populations of lactic acid bacteria associated with silage fermentation as determined by phenotype, 16S ribosomal RNA and recA gene analysis. Syst. Appl. Microbiol. 2011, 34, 235–241. [Google Scholar] [CrossRef]
- Agarussi, M.C.N.; Pereira, O.G.; Silva, V.P.; Leandro, E.S.; Ribeiro, K.G.; Santos, S.A. Fermentative profile and lactic acid bacterial dynamics in non- wilted and wilted alfalfa silage in tropical conditions. Mol. Biol. Rep. 2019, 46, 451–460. [Google Scholar] [CrossRef] [PubMed]
- Leandro, E.S.; Oliveira, M.N.V.; Rufino, L.D.; Ribeiro, K.G.; Pereira, O.G. Characterization of Arachis pintoi and its silages at different fermentation periods. Mol. Biol. Rep. 2019, 46, 5019–5024. [Google Scholar] [CrossRef] [PubMed]
- Grev, A.M.; Wells, M.C.; Samac, D.A.; Martinson, K.L.; Sheaffer, C.C. Forage accumulation and nutritive value of reduced lignin and reference alfalfa cultivars. Agron. J. 2017, 109, 2749–2761. [Google Scholar] [CrossRef]
- Zhang, T.; Kang, J.; Guo, W.; Zhao, Z.; Xu, Y.; Yan, X.; Yang, Q. Yield evaluation of twenty-eight alfalfa cultivars in Hebei Province of China. J. Integr. Agric. 2014, 13, 2260–2267. [Google Scholar] [CrossRef]
- Kopp, M.M.; Pereira, A.V.; Ferreira, R.P. Alfalfa cultivars in Brazil (“Cultivares de alfafa no Brasil”). In Genetic Improvement of Alfalfa (“Melhoramento Genético Da Alfafa”); Ferreira, R.P., Basigalup, D.H., Gieco, J.O., Eds.; Embrapa Pecuaária Sudeste: São Carlos, Brazil, 2011; pp. 309–331. [Google Scholar]
- Dewhurst, R.J.; Fisher, W.J.; Tweed, J.K.S.; Wilkins, R.J. Comparison of grass and legume silages for milk production. Production responses with different levels of concentrate. J. Dairy Sci. 2003, 86, 2598–2611. [Google Scholar] [CrossRef]
- McDonald, P.; Henderson, A.R. Buffering capacity of herbage samples as a factor in ensilage. J. Sci. Food Agric. 1962, 13, 395–400. [Google Scholar] [CrossRef]
- Ertekin, I.; Kızılşimşek, M. Effects of lactic acid bacteria inoculation in pre-harvesting period on fermentation and feed quality properties of alfalfa silage. Asian-Australas. J. Anim. Sci. 2019, 33, 245–253. [Google Scholar] [CrossRef]
- Zhao, S.S.; Wang, Y.P.; Yang, F.Y.; Wang, Y.; Zhang, H. Screening a Lactobacillus plantarum strain for good adaption in alfalfa ensiling and demonstrating its improvement of alfalfa silage quality. J. Appl. Microbiol. 2020, 129, 233–242. [Google Scholar] [CrossRef]
- Contreras-Govea, F.E.; Muck, R.E.; Weimer, P.J.; Hymes-Fecht, U.C. In vitro ruminal fermentation of treated alfalfa silage using ruminal inocula from high and low feed efficient lactating cows. J. Appl. Microbiol. 2016, 121, 333–340. [Google Scholar] [CrossRef]
- Holt, J.G.; Krieg, N.R.; Sneath, P.H.A.; Staley, J.T.; Williams, S.T. Bergey’s Manual of Determinative Bacteriology, 9th ed.; Williams & Wilkins: Baltimore, MD, USA, 1994. [Google Scholar]
- Liu, Q.; Chen, M.; Zhang, J.; Shi, S.; Cai, Y. Characteristics of isolated lactic acid bacteria and their effectiveness to improve stylo (Stylosanthes guianensis Sw.) silage quality at various temperatures. Anim. Sci. J. 2012, 83, 128–135. [Google Scholar] [CrossRef]
- Tagg, J.R.; Dajani, A.S.; Wannamaker, L.W. Bacteriocins of Gram-positive bacteria. Bacteriol. Rev. 1976, 40, 722–756. [Google Scholar] [CrossRef] [PubMed]
- Heuer, H.; Krsek, M.; Baker, P.; Smalla, K.; Wellington, E.M.H. Analysis of actinomycete communities by specific amplification of genes encoding 16S rRNA and gel-electrophoretic separation in denaturing gradients. Appl. Environ. Microbiol. 1997, 63, 3233–3241. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Jobim, C.C.; Nussio, L.G.; Reis, R.A.; Schmidt, P. Methodological advances in evaluation of preserved forage quality. R. Bras. Zootec. 2007, 36, 101–119. [Google Scholar] [CrossRef]
- Association of Official Analytical Chemistry (AOAC). Official Methods of Analysis, 15th ed.; AOAC International: Arlington, TX, USA, 1990. [Google Scholar]
- Mertens, D.R. Gravimetric determination of amylase treated neutral detergent fiber in feeds with refluxing in beaker or crucibles: Collaborative study. J. AOAC Int. 2002, 85, 1217–1240. [Google Scholar] [PubMed]
- Licitra, G.; Hernandez, T.M.; Van Soest, P.J. Standardization of procedures for nitrogen fractionation of ruminant feeds. Anim. Feed Sci. Technol. 1996, 57, 347–358. [Google Scholar] [CrossRef]
- Weatherburn, M.W. Phenol-hypochlorite reaction for determination of ammonia. Anal. Chem. 1967, 39, 971–974. [Google Scholar] [CrossRef]
- Nelson, N. A photometric adaptation of the Somogyi method for the determination of glucose. J. Biol. Chem. 1944, 153, 375–380. [Google Scholar]
- Siegrifield, V.R.; Ruckemann, H.; Stumpf, G. Method for the determination of organic acids in silage by high performance liquid chromatography. Landwirtsch. Forsch. 1984, 37, 298–304. [Google Scholar]
- Zhang, Q.; Li, X.J.; Zho, M.M.; YU, Z. Isolating and evaluating lactic acid bacteria strains for effectiveness of Leymus chinensis silage fermentation. Lett. Appl. Microbiol. 2014, 59, 391–397. [Google Scholar] [CrossRef]
- Li, D.; Ni, K.; Pang, H.; Wang, Y.; Cai, Y.; Jin, Q. Identification and antimicrobial activity detection of lactic acid bacteria isolated from corn stover silage. Asian-Australas. J. Anim. Sci. 2015, 28, 620–631. [Google Scholar] [CrossRef] [PubMed]
- Amaral, R.C.; Carvalho, B.F.; Costa, D.M.; Morenz, M.J.F.; Schwan, R.F.; Ávila, C.L.S. Novel lactic acid bacteria strains enhance the conservation of elephant grass silage cv. BRS Capiaçu. Anim. Feed Sci. Technol. 2020, 264, 114472. [Google Scholar] [CrossRef]
- Santos, A.O.; Ávila, C.L.S.; Soares, C.; Carvalho, B.F.; Schwan, R.F.; Lima, N. Lactic acid bacteria diversity in corn silage produced in Minas Gerais (Brazil). Ann. Microbiol. 2019, 69, 1445–1459. [Google Scholar] [CrossRef]
- Li, D.; Wang, Y.; Zhang, Y.; Lin, Y.; Yang, F. Evaluation of lactic acid bacteria isolated from alfalfa for silage fermentation. Grassl. Sci. 2018, 64, 190–198. [Google Scholar] [CrossRef]
- Dworkin, M.; Falkow, S.; Rosenberg, E.; Schleifer, K.H.; Stackebrandt, E. (Eds.) The Prokaryotes: A Handbook on the Biology of Bacteria: Firmicutes and Cyanobacteria, 3rd ed.; Springer: New York, NY, USA, 2006. [Google Scholar]
- Pholsen, S.; Khota, W.; Pang, H.; Higgs, D.; Cai, Y. Characterization and application of lactic acid bacteria for tropical silage preparation. Anim. Sci. J. 2016, 87, 1202–1211. [Google Scholar] [CrossRef]
- Silva, V.P.; Pereira, O.G.; Leandro, E.S.; Silva, T.C.; Ribeiro, K.G.; Mantovani, H.C.; Santos, S.A. Effects of lactic acid bacteria with bacteriocinogenic potential on the fermentation profile and chemical composition of alfalfa silage in tropical conditions. J. Dairy Sci. 2016, 99, 1895–1902. [Google Scholar] [CrossRef] [PubMed]
- Kung, L.; Shaver, R.D.; Grant, R.J.; Schmidt, R.J. Silage review: Interpretation of chemical, microbial, and organoleptic components of silages. J. Dairy Sci. 2018, 101, 4020–4033. [Google Scholar] [CrossRef]
- Whiter, A.G.; Kung, L. The effect of a dry or liquid application of Lactobacillus plantarum MTD1 on the fermentation of alfalfa silage. J. Dairy Sci. 2001, 84, 2195–2202. [Google Scholar] [CrossRef]
- Jones, B.A.; Satter, L.D.; Muck, R.E. Influence of bacterial inoculant and substrate addition to lucerne ensiled at different dry matter contents. Grass Forage Sci. 1992, 47, 19–27. [Google Scholar] [CrossRef]
- National Institute of Meteorology. Available online: https://portal.inmet.gov.br/ (accessed on 15 March 2018).
- Tschaplinski, T.J.; Blake, T.J. Photosynthetic reinvigoration of leaves following shoot decapitation and accelerated growth of coppice shoots. Physiol. Plant. 1989, 75, 157–165. [Google Scholar] [CrossRef]
- Guo, L.; Yao, D.; Li, D.; Lin, Y.; Bureenok, S.; Ni, K.; Yang, F. Effects of lactic acid bacteria isolated from rumen fluid and feces of dairy cows on fermentation quality, microbial community, and in vitro digestibility of alfalfa silage. Front. Microbiol. 2020, 10, 2998. [Google Scholar] [CrossRef] [PubMed]
- Mugabe, W.; Yuan, X.; Li, J.; Dong, Z.; Shao, T. Effects of hexanoic acid, Lactobacillus plantarum and their combination on the fermentation characteristics of Napier grass. Anim. Feed Sci. Technol. 2019, 253, 135–140. [Google Scholar] [CrossRef]
- Zhao, J.; Dong, Z.; Li, J.; Chen, L.; Bai, Y.; Jia, Y.; Shao, T. Evaluation of Lactobacillus plantarum MTD1 and waste molasses as fermentation modifier to increase silage quality and reduce ruminal greenhouse gas emissions of rice straw. Sci. Total Environ. 2019, 688, 143–152. [Google Scholar] [CrossRef]
- Blajman, J.E.; Páez, R.B.; Vinderola, C.G.; Lingua, M.S.; Signorini, M.L. A meta-analysis on the effectiveness of homofermentativeand heterofermentative lactic acid bacteria for corn silage. J. Appl. Microbiol. 2018, 125, 1655–1669. [Google Scholar] [CrossRef] [PubMed]
- McDonald, P.; Henderson, A.R.; Heron, S.J.E. Biochemistry of Silage, 2nd ed.; Marlow: Chacombe, UK, 1991. [Google Scholar]

| Item | Harvest 1 | |
|---|---|---|
| 1 | 2 | |
| Dry matter, g/kg | 331.4 | 435.2 |
| Crude protein | 146.7 | 149.5 |
| Neutral detergent fiber | 532.3 | 458.6 |
| Acid detergent fiber | 348.1 | 355.6 |
| Water-soluble carbohydrates | 17.3 | 34 |
| pH | 6.12 | 6.24 |
| Microbial population, log CFU/g of fresh matter | ||
| Lactic acid bacteria | 6.72 | 6.14 |
| Enterobacteria | 7.15 | 6.42 |
| Mold | 4.58 | 4.90 |
| Yeast | 5.21 | 5.03 |
| Item | Strain | ||
|---|---|---|---|
| AV14.2 | AV14.17 | AV56.13 | |
| Species | Lactobacillus brevis | Lactobacillus pentosus | Pediococcus acidilactici |
| Access code 1 | MK713790 | MK713801 | KY613548 |
| Growth at Ph 2 | |||
| 3.5 | ++ | +++ | ++ |
| 4 | +++ | +++ | +++ |
| 4.5 | +++ | +++ | +++ |
| 8.5 | +++ | +++ | +++ |
| Growth at temperature (°C) 2 | |||
| 15 | ++ | ++ | + |
| 45 | +++ | ++ | +++ |
| Growth in NaCl (g/L) 2 | |||
| 40 | +++ | +++ | +++ |
| 65 | +++ | ++ | +++ |
| Radius of inhibition 3 | |||
| Indicator microorganism | |||
| Listeria monocytogenes | ++ | +++ | ++ |
| Escherichia coli | + | +++ | ++ |
| Staphylococcus aureus | + | + | + |
| Bacillus cereus | ++ | +++ | +++ |
| Harvest 1 | Inoculant 2 | Average | SEM 3 | p-Value 4 | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Control | AV14.17 | Combo | CI | I | H | I×H | ||||||
| Dry matter, g/kg of Fresh Matter | ||||||||||||
| 1 | 325 | 329 | 328 | 331 | 328 B | 0.84 | <0.01 | <0.01 | 0.41 | |||
| 2 | 417 | 423 | 423 | 425 | 422 A | |||||||
| Average | 371 b | 376 a | 376 a | 378 a | ||||||||
| Crude protein, g/kg of DM | ||||||||||||
| 1 | 145 | 146 | 149 | 146 | 146 B | 0.07 | 0.16 | <0.01 | 0.92 | |||
| 2 | 149 | 151 | 153 | 149 | 150 A | |||||||
| Average | 147 | 149 | 151 | 147 | ||||||||
| Neutral detergent fiber, g/kg of DM | ||||||||||||
| 1 | 519 Aa | 516 Aa | 490 Aab | 473 Ab | 499 | 0.87 | <0.01 | <0.01 | 0.03 | |||
| 2 | 420 Ba | 405 Ba | 409 Ba | 408 Ba | 410 | |||||||
| Average | 470 | 460 | 449 | 440 | ||||||||
| Acid detergent fiber, g/kg of DM | ||||||||||||
| 1 | 422 Aa | 422 Aa | 369 Ab | 407 Aab | 405 | 0.65 | 0.01 | <0.01 | 0.02 | |||
| 2 | 343 Ba | 356 Ba | 347 Aa | 343 Ba | 347 | |||||||
| Average | 383 | 389 | 358 | 375 | ||||||||
| ADIN 5, g/kg of DM | ||||||||||||
| 1 | 195 | 149 | 154 | 210 | 177 | 0.70 | 0.67 | 0.70 | 0.09 | |||
| 2 | 168 | 174 | 189 | 152 | 171 | |||||||
| Average | 182 | 162 | 172 | 181 | ||||||||
| Harvest 1 | Inoculant 2 | Average | SEM 3 | p-Value 4 | |||||
|---|---|---|---|---|---|---|---|---|---|
| Control | AV14.17 | Combo | CI | I | H | I×H | |||
| pH | |||||||||
| 1 | 4.56 | 4.51 | 4.55 | 4.59 | 4.55 A | 0.02 | 0.04 | <0.01 | 0.73 |
| 2 | 4.36 | 4.31 | 4.35 | 4.35 | 4.34 B | ||||
| Average | 4.46 a | 4.41 b | 4.45 ab | 4.47 a | |||||
| Ammonia nitrogen, g/kg of total nitrogen | |||||||||
| 1 | 142 Aa | 113 Ab | 105 Ab | 104 Ab | 116 | 0.58 | 0.01 | <0.01 | 0.06 |
| 2 | 64.0 Ba | 56.0 Ba | 55.2 Ba | 64.2 Ba | 59.9 | ||||
| Average | 103 | 84.3 | 79.9 | 83.9 | |||||
| Lactic acid, g/kg of DM | |||||||||
| 1 | 12.5 | 15.0 | 15.1 | 17.6 | 15.1 B | 0.10 | 0.73 | <0.01 | 0.55 |
| 2 | 23.6 | 21.2 | 24.4 | 22.6 | 23.0 A | ||||
| Average | 18.1 | 18.1 | 19.8 | 20.1 | |||||
| Acetic acid, g/kg of DM | |||||||||
| 1 | 26.6 | 26.9 | 17.8 | 28.4 | 24.9 A | 0.19 | 0.33 | <0.01 | 0.37 |
| 2 | 10.2 | 6.10 | 8.25 | 9.2 | 8.44 B | ||||
| Average | 18.4 | 16.5 | 13.0 | 18.1 | |||||
| Water-soluble carbohydrate, g/kg of DM | |||||||||
| 1 | 2.68 Ba | 2.76 Ba | 2.53 Ba | 2.53 Ba | 2.63 | 0.04 | 0.02 | <0.01 | 0.04 |
| 2 | 6.32 Ab | 8.45 Aa | 7.39 Aab | 6.10 Ab | 7.07 | ||||
| Average | 4.50 | 5.61 | 4.96 | 4.32 | |||||
| Dry matter recovery, g/kg of DM | |||||||||
| 1 | 974 Aa | 982 Aa | 976 Aa | 960 Aa | 973 | 0.23 | 0.12 | 0.01 | 0.02 |
| 2 | 950 Ba | 966 Aa | 966 Aa | 970 Aa | 963 | ||||
| Average | 962 | 974 | 971 | 965 | |||||
| Harvest 1 | Inoculant 2 | Average | SEM 3 | p-Value 4 | |||||
|---|---|---|---|---|---|---|---|---|---|
| Control | AV14.17 | Combo | CI | I | H | I×H | |||
| Lactic Acid Bacteria | |||||||||
| 1 | 8.26 | 8.35 | 8.35 | 8.28 | 8.31 A | 0.11 | 0.54 | <0.01 | 0.44 |
| 2 | 7.30 | 6.90 | 7.19 | 6.89 | 7.07 B | ||||
| Average | 7.78 | 7.62 | 7.77 | 7.58 | |||||
| Mold | |||||||||
| 1 | 2.33 | 2.19 | 2.07 | 1.72 | 2.07 B | 0.22 | 0.70 | <0.01 | 0.56 |
| 2 | 3.36 | 2.76 | 3.33 | 3.29 | 2.63 A | ||||
| Average | 2.84 | 2.47 | 2.70 | 2.51 | |||||
| Yeast | |||||||||
| 1 | 2.34 | 2.21 | 2.26 | 2.45 | 2.32 | 0.19 | 0.12 | 0.02 | 0.07 |
| 2 | 2.48 | 2.20 | 1.05 | 0.94 | 1.67 | ||||
| Average | 2.41 | 2.20 | 1.65 | 1.70 | |||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Silva, V.P.; Pereira, O.G.; Leandro, E.S.; Paula, R.A.; Agarussi, M.C.N.; Ribeiro, K.G. Selection of Lactic Acid Bacteria from Alfalfa Silage and Its Effects as Inoculant on Silage Fermentation. Agriculture 2020, 10, 518. https://doi.org/10.3390/agriculture10110518
Silva VP, Pereira OG, Leandro ES, Paula RA, Agarussi MCN, Ribeiro KG. Selection of Lactic Acid Bacteria from Alfalfa Silage and Its Effects as Inoculant on Silage Fermentation. Agriculture. 2020; 10(11):518. https://doi.org/10.3390/agriculture10110518
Chicago/Turabian StyleSilva, Vanessa P., Odilon G. Pereira, Eliana S. Leandro, Rosinea A. Paula, Mariele C. N. Agarussi, and Karina G. Ribeiro. 2020. "Selection of Lactic Acid Bacteria from Alfalfa Silage and Its Effects as Inoculant on Silage Fermentation" Agriculture 10, no. 11: 518. https://doi.org/10.3390/agriculture10110518
APA StyleSilva, V. P., Pereira, O. G., Leandro, E. S., Paula, R. A., Agarussi, M. C. N., & Ribeiro, K. G. (2020). Selection of Lactic Acid Bacteria from Alfalfa Silage and Its Effects as Inoculant on Silage Fermentation. Agriculture, 10(11), 518. https://doi.org/10.3390/agriculture10110518






