Metastatic Burden Defines Clinically and Biologically Distinct Subgroups of Stage 4 High-Risk Neuroblastoma
Abstract
:1. Introduction
2. Methods
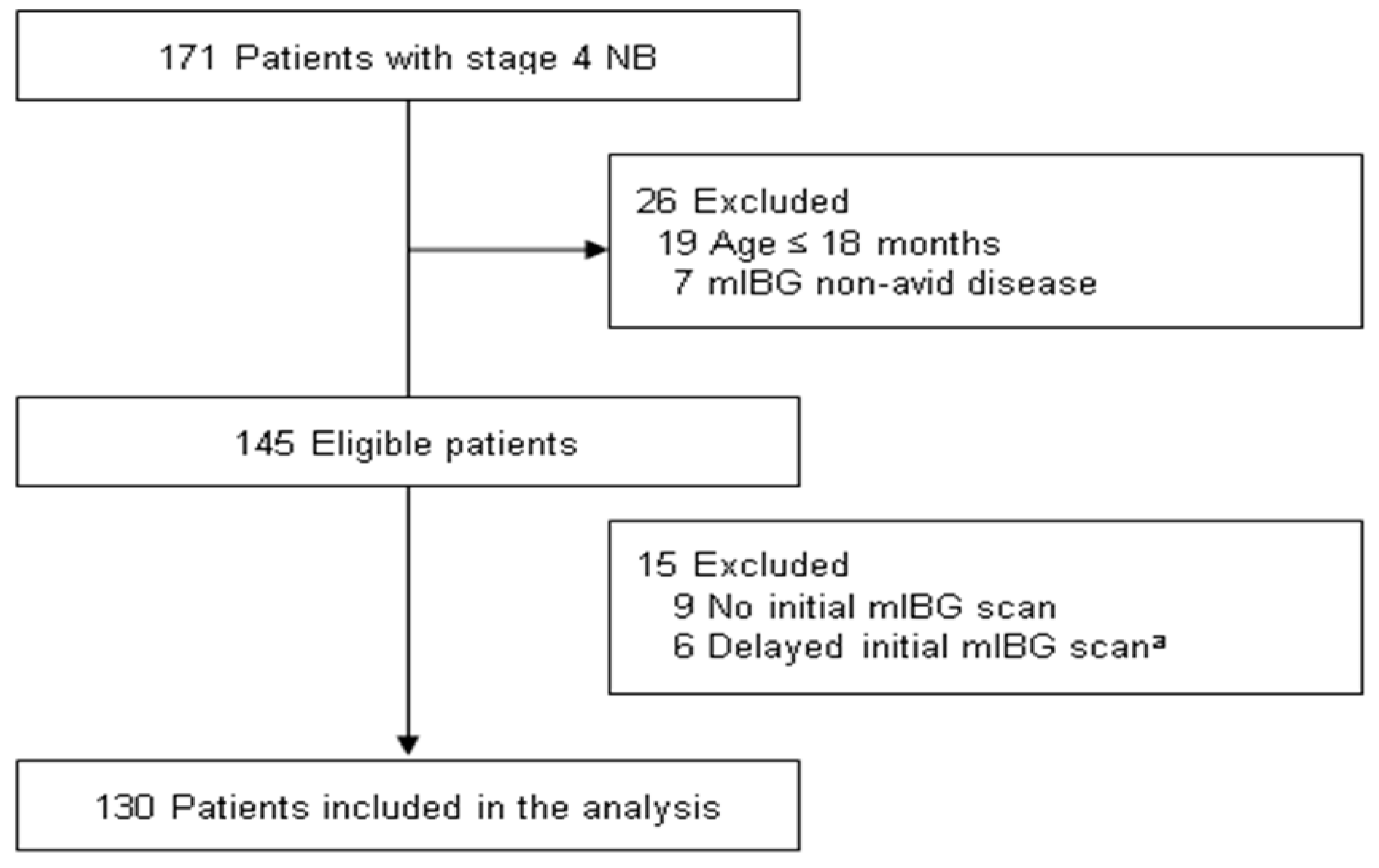
2.1. Patient Cohort and Data Collection
2.2. Treatment
2.3. Assessment of Metastasis
2.4. Response Criteria
2.5. RNA Isolation
2.6. RNA Sequencing (RNA-Seq) and Data Analysis
2.7. Gene Set Variation Analysis (GSVA)
2.8. Statistical Methods
3. Results
3.1. Characteristics and Outcomes of the Study Population
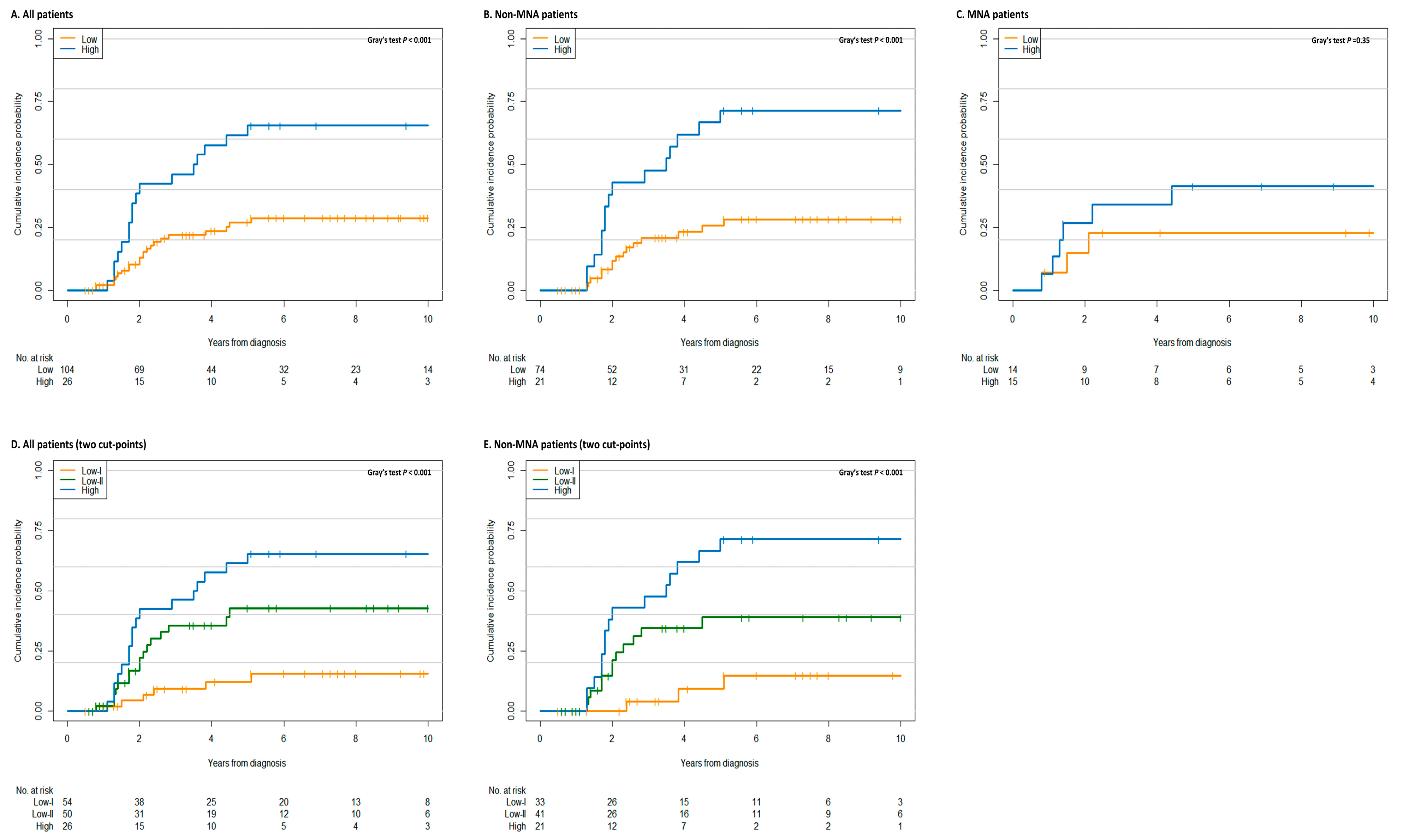
3.2. Prognostic Significance of Metastatic Burden
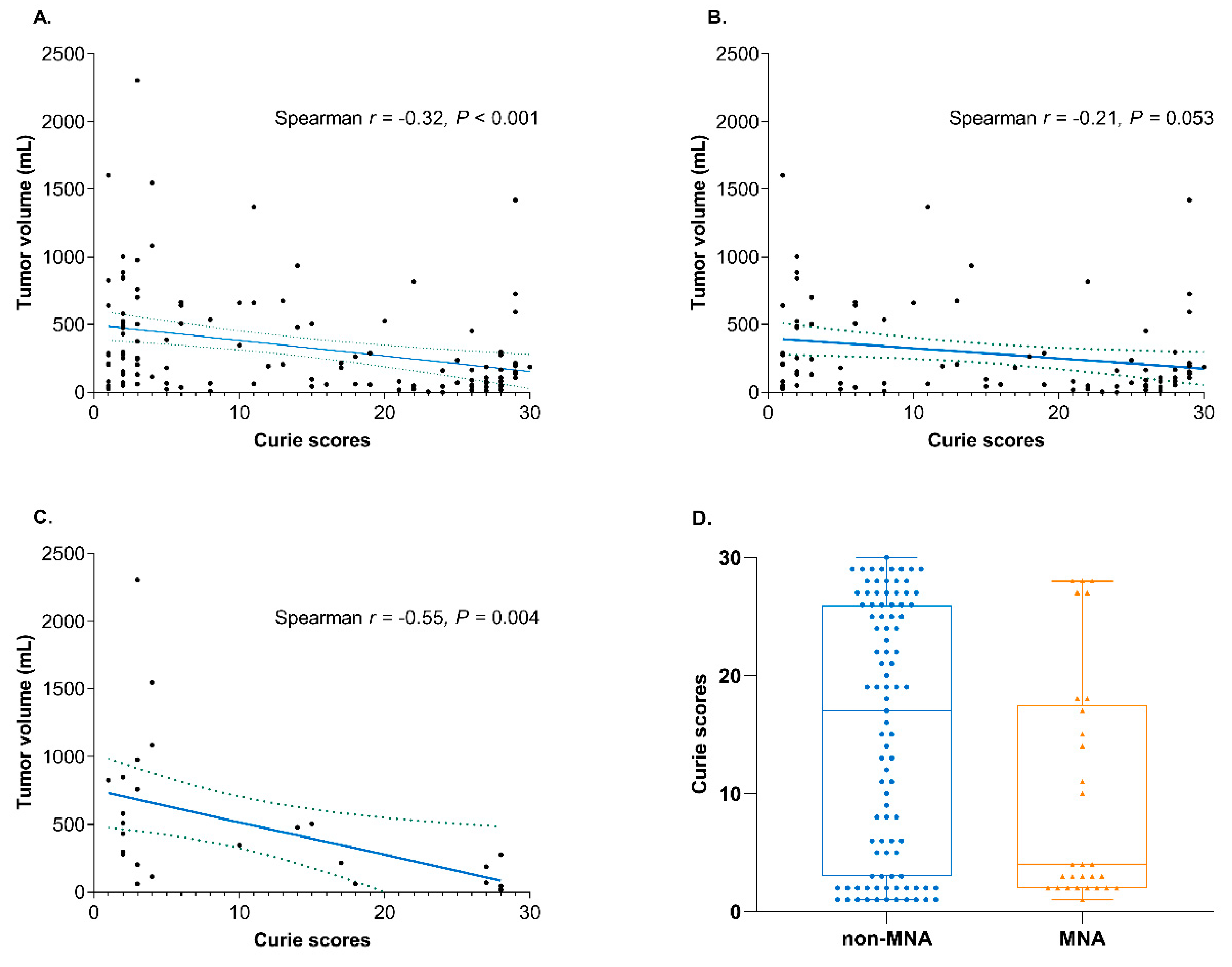
3.3. Distinct Clinical Characteristics Based on the Metastatic Burden
3.4. Identification of DEGs and Gene Sets Based on the Metastatic Burden
4. Discussion
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Maris, J.M.; Hogarty, M.D.; Bagatell, R.; Cohn, S.L. Neuroblastoma. Lancet 2007, 369, 2106–2120. [Google Scholar] [CrossRef]
- Maris, J.M. Recent advances in neuroblastoma. N. Engl. J. Med. 2010, 362, 2202–2211. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pinto, N.R.; Applebaum, M.A.; Volchenboum, S.L.; Matthay, K.K.; London, W.B.; Ambros, P.F.; Nakagawara, A.; Berthold, F.; Schleiermacher, G.; Park, J.R.; et al. Advances in Risk Classification and Treatment Strategies for Neuroblastoma. J. Clin. Oncol. 2015, 33, 3008–3017. [Google Scholar] [CrossRef] [PubMed]
- Morgenstern, D.A.; Bagatell, R.; Cohn, S.L.; Hogarty, M.D.; Maris, J.M.; Moreno, L.; Park, J.R.; Pearson, A.D.; Schleiermacher, G.; Valteau-Couanet, D.; et al. The challenge of defining “ultra-high-risk” neuroblastoma. Pediatr. Blood Cancer 2019, 66, e27556. [Google Scholar] [CrossRef] [PubMed]
- Yanik, G.A.; Parisi, M.T.; Shulkin, B.L.; Naranjo, A.; Kreissman, S.G.; London, W.B.; Villablanca, J.G.; Maris, J.M.; Park, J.R.; Cohn, S.L.; et al. Semiquantitative mIBG scoring as a prognostic indicator in patients with stage 4 neuroblastoma: A report from the Children’s oncology group. J. Nucl. Med. 2013, 54, 541–548. [Google Scholar] [CrossRef] [Green Version]
- Morgenstern, D.A.; London, W.B.; Stephens, D.; Volchenboum, S.L.; Simon, T.; Nakagawara, A.; Shimada, H.; Schleiermacher, G.; Matthay, K.K.; Cohn, S.L.; et al. Prognostic significance of pattern and burden of metastatic disease in patients with stage 4 neuroblastoma: A study from the International Neuroblastoma Risk Group database. Eur. J. Cancer 2016, 65, 1–10. [Google Scholar] [CrossRef]
- Matthay, K.K.; Edeline, V.; Lumbroso, J.; Tanguy, M.L.; Asselain, B.; Zucker, J.M.; Valteau-Couanet, D.; Hartmann, O.; Michon, J. Correlation of early metastatic response by 123I-metaiodobenzylguanidine scintigraphy with overall response and event-free survival in stage IV neuroblastoma. J. Clin. Oncol. 2003, 21, 2486–2491. [Google Scholar] [CrossRef]
- Decarolis, B.; Schneider, C.; Hero, B.; Simon, T.; Volland, R.; Roels, F.; Dietlein, M.; Berthold, F.; Schmidt, M. Iodine-123 metaiodobenzylguanidine scintigraphy scoring allows prediction of outcome in patients with stage 4 neuroblastoma: Results of the Cologne interscore comparison study. J. Clin. Oncol. 2013, 31, 944–951. [Google Scholar] [CrossRef]
- Riaz, S.; Bashir, H.; Khan, S.J.; Qazi, A. I-131 mIBG Scintigraphy Curie Versus SIOPEN Scoring: Prognostic Value in Stage 4 Neuroblastoma. Mol. Imaging Radionucl. Ther. 2018, 27, 121–125. [Google Scholar] [CrossRef]
- Lee, J.W.; Lee, S.; Cho, H.W.; Ma, Y.; Yoo, K.H.; Sung, K.W.; Koo, H.H.; Cho, E.J.; Lee, S.K.; Lim, D.H. Incorporation of high-dose (131)I-metaiodobenzylguanidine treatment into tandem high-dose chemotherapy and autologous stem cell transplantation for high-risk neuroblastoma: Results of the SMC NB-2009 study. J. Hematol. Oncol. 2017, 10, 108. [Google Scholar] [CrossRef]
- Sung, K.W.; Son, M.H.; Lee, S.H.; Yoo, K.H.; Koo, H.H.; Kim, J.Y.; Cho, E.J.; Lee, S.K.; Choi, Y.S.; Lim, D.H.; et al. Tandem high-dose chemotherapy and autologous stem cell transplantation in patients with high-risk neuroblastoma: Results of SMC NB-2004 study. Bone Marrow Transpl. 2013, 48, 68–73. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sung, K.W.; Lee, S.H.; Yoo, K.H.; Jung, H.L.; Cho, E.J.; Koo, H.H.; Lee, S.K.; Kim, J.; Lim, D.H.; Suh, Y.L.; et al. Tandem high-dose chemotherapy and autologous stem cell rescue in patients over 1 year of age with stage 4 neuroblastoma. Bone Marrow Transpl. 2007, 40, 37–45. [Google Scholar] [CrossRef] [PubMed]
- Brodeur, G.M.; Pritchard, J.; Berthold, F.; Carlsen, N.L.; Castel, V.; Castelberry, R.P.; De Bernardi, B.; Evans, A.E.; Favrot, M.; Hedborg, F.; et al. Revisions of the international criteria for neuroblastoma diagnosis, staging, and response to treatment. J. Clin. Oncol. 1993, 11, 1466–1477. [Google Scholar] [CrossRef] [PubMed]
- Bagatell, R.; McHugh, K.; Naranjo, A.; Van Ryn, C.; Kirby, C.; Brock, P.; Lyons, K.A.; States, L.J.; Rojas, Y.; Miller, A.; et al. Assessment of Primary Site Response in Children With High-Risk Neuroblastoma: An International Multicenter Study. J. Clin. Oncol. 2016, 34, 740–746. [Google Scholar] [CrossRef]
- Matthay, K.K.; Shulkin, B.; Ladenstein, R.; Michon, J.; Giammarile, F.; Lewington, V.; Pearson, A.D.; Cohn, S.L. Criteria for evaluation of disease extent by (123)I-metaiodobenzylguanidine scans in neuroblastoma: A report for the International Neuroblastoma Risk Group (INRG) Task Force. Br. J. Cancer 2010, 102, 1319–1326. [Google Scholar] [CrossRef]
- Kim, D.; Salzberg, S.L. TopHat-Fusion: An algorithm for discovery of novel fusion transcripts. Genome Biol. 2011, 12, R72. [Google Scholar] [CrossRef] [Green Version]
- Trapnell, C.; Williams, B.A.; Pertea, G.; Mortazavi, A.; Kwan, G.; van Baren, M.J.; Salzberg, S.L.; Wold, B.J.; Pachter, L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 2010, 28, 511–515. [Google Scholar] [CrossRef] [Green Version]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R.; Genome Project Data Processing, S. The Sequence Alignment/Map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef] [Green Version]
- Li, B.; Dewey, C.N. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. Bioinformatics 2011, 12, 323. [Google Scholar]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef] [Green Version]
- Robinson, M.D.; Oshlack, A. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol 2010, 11, R25. [Google Scholar] [CrossRef] [Green Version]
- Law, C.W.; Chen, Y.; Shi, W.; Smyth, G.K. voom: Precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol. 2014, 15, R29. [Google Scholar] [CrossRef] [Green Version]
- Hanzelmann, S.; Castelo, R.; Guinney, J. GSVA: Gene set variation analysis for microarray and RNA-seq data. Bioinformatics 2013, 14, 7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef] [Green Version]
- Laska, E.; Meisner, M.; Wanderling, J. A maximally selected test of symmetry about zero. Stat. Med. 2012, 31, 3178–3191. [Google Scholar] [CrossRef] [PubMed]
- Morgenstern, D.A.; Potschger, U.; Moreno, L.; Papadakis, V.; Owens, C.; Ash, S.; Pasqualini, C.; Luksch, R.; Garaventa, A.; Canete, A.; et al. Risk stratification of high-risk metastatic neuroblastoma: A report from the HR-NBL-1/SIOPEN study. Pediatr. Blood Cancer 2018, 65, e27363. [Google Scholar] [CrossRef]
- Li, D.; Hu, B.; Zhou, Y.; Wan, T.; Si, X. Impact of tumor size on survival of patients with resected pancreatic ductal adenocarcinoma: A systematic review and meta-analysis. BMC Cancer 2018, 18, 985. [Google Scholar] [CrossRef] [Green Version]
- Okada, M.; Nishio, W.; Sakamoto, T.; Uchino, K.; Yuki, T.; Nakagawa, A.; Tsubota, N. Effect of tumor size on prognosis in patients with non-small cell lung cancer: The role of segmentectomy as a type of lesser resection. J. Thorac. Cardiovasc. Surg. 2005, 129, 87–93. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yoo, S.Y.; Kim, J.S.; Sung, K.W.; Jeon, T.Y.; Choi, J.Y.; Moon, S.H.; Son, M.H.; Lee, S.H.; Yoo, K.H.; Koo, H.H. The degree of tumor volume reduction during the early phase of induction chemotherapy is an independent prognostic factor in patients with high-risk neuroblastoma. Cancer 2013, 119, 656–664. [Google Scholar] [CrossRef]
- Xia, H.; Chen, J.; Shi, M.; Gao, H.; Sekar, K.; Seshachalam, V.P.; Ooi, L.L.; Hui, K.M. EDIL3 is a novel regulator of epithelial-mesenchymal transition controlling early recurrence of hepatocellular carcinoma. J. Hepatol. 2015, 63, 863–873. [Google Scholar] [CrossRef]
- Beckham, C.J.; Olsen, J.; Yin, P.N.; Wu, C.H.; Ting, H.J.; Hagen, F.K.; Scosyrev, E.; Messing, E.M.; Lee, Y.F. Bladder cancer exosomes contain EDIL-3/Del1 and facilitate cancer progression. J. Urol. 2014, 192, 583–592. [Google Scholar] [CrossRef] [PubMed]
- Mathieu, M.G.; Linley, A.J.; Reeder, S.P.; Badoual, C.; Tartour, E.; Rees, R.C.; McArdle, S.E. HAGE, a cancer/testis antigen expressed at the protein level in a variety of cancers. Cancer Immun. 2010, 10, 2. [Google Scholar]
- Abdel-Fatah, T.M.; McArdle, S.E.; Johnson, C.; Moseley, P.M.; Ball, G.R.; Pockley, A.G.; Ellis, I.O.; Rees, R.C.; Chan, S.Y. HAGE (DDX43) is a biomarker for poor prognosis and a predictor of chemotherapy response in breast cancer. Br. J. Cancer 2014, 110, 2450–2461. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Singh, N.; Kulikovskaya, I.; Barrett, D.M.; Binder-Scholl, G.; Jakobsen, B.; Martinez, D.; Pawel, B.; June, C.H.; Kalos, M.D.; Grupp, S.A. T cells targeting NY-ESO-1 demonstrate efficacy against disseminated neuroblastoma. Oncoimmunology 2016, 5, e1040216. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bao, L.; Dunham, K.; Lucas, K. MAGE-A1, MAGE-A3, and NY-ESO-1 can be upregulated on neuroblastoma cells to facilitate cytotoxic T lymphocyte-mediated tumor cell killing. Cancer Immunol. Immunother. 2011, 60, 1299–1307. [Google Scholar] [CrossRef] [PubMed]
- Oliynyk, G.; Ruiz-Perez, M.V.; Sainero-Alcolado, L.; Dzieran, J.; Zirath, H.; Gallart-Ayala, H.; Wheelock, C.E.; Johansson, H.J.; Nilsson, R.; Lehtio, J.; et al. MYCN-enhanced Oxidative and Glycolytic Metabolism Reveals Vulnerabilities for Targeting Neuroblastoma. iScience 2019, 21, 188–204. [Google Scholar] [CrossRef] [Green Version]
- Hald, O.H.; Olsen, L.; Gallo-Oller, G.; Elfman, L.H.M.; Lokke, C.; Kogner, P.; Sveinbjornsson, B.; Flaegstad, T.; Johnsen, J.I.; Einvik, C. Inhibitors of ribosome biogenesis repress the growth of MYCN-amplified neuroblastoma. Oncogene 2019, 38, 2800–2813. [Google Scholar] [CrossRef]
- Lee, J.W.; Son, M.H.; Cho, H.W.; Ma, Y.E.; Yoo, K.H.; Sung, K.W.; Koo, H.H. Clinical significance of MYCN amplification in patients with high-risk neuroblastoma. Pediatr Blood Cancer 2018, 65, e27257. [Google Scholar] [CrossRef]

| Characteristics | Number (%) a |
|---|---|
| Sex | |
| Male | 77 (59) |
| Female | 53 (41) |
| Age at diagnosis, mean ± Std | 4.5 ± 2.9 |
| Primary tumor site | |
| Abdomen/Pelvis | 117 (90) |
| Thorax/Cervix | 12 (9) |
| Unknown | 1 (1) |
| Primary tumor volume b (cm3) | 345 (67–510) |
| MYCN status | |
| Nonamplified | 95 (73) |
| Amplified | 29 (22) |
| Unknown | 6 (5) |
| Cytogenetics | |
| 1p deletion (n = 74) | 16 (22) |
| 11q deletion (n = 79) | 23 (29) |
| 17q gain (n = 74) | 22 (29) |
| Pathology | |
| Undifferentiated | 15 (12) |
| Poorly differentiated | 65 (50) |
| Differentiating | 22 (17) |
| Ganglioneuroblastoma | 19 (15) |
| Unknown | 9 (7) |
| INPC | |
| Favorable | 28 (22) |
| Unfavorable | 93 (72) |
| Unknown | 9 (7) |
| Curie score at diagnosis b | 12.5 (3–25) |
| Metastatic site | |
| Bone | 116 (89) |
| Bone marrow | 92 (71) |
| Distant lymph node | 66 (49) |
| Lung | 14 (11) |
| CNS | 3 (2) |
| Liver | 17 (13) |
| Skin | 5 (4) |
| Other c | 47 (36) |
| Variables | Low (n = 104) | High (n = 26) | p-Value |
|---|---|---|---|
| Sex, No. (%) | 0.964 | ||
| Male | 61 (59) | 16 (62) | |
| Female | 43 (41) | 10 (38) | |
| Age at diagnosis, mean ± Std, years | 4.6 ± 3.1 | 3.9 ± 2.0 | 0.134 |
| Primary tumor volume (cm3) b | 245 (1–2305) | 109 (9–1421) | 0.030 |
| Residual tumor volume (%) a, b | 28 (2–98) | 31 (10–98) | 0.833 |
| Reduction in Curie score, mean ± Std (%) c | 44 ± 37 | 48 ± 36 | 0.620 |
| Response to induction treatment, No. (%) | |||
| Complete response | 42 (40) | 9 (35) | |
| Very good partial response | 23 (22) | 6 (23) | |
| Partial response | 33 (32) | 8 (31) | |
| Mixed response | 1 (1) | 1 (4) | |
| Stable disease | 3 (3) | 1 (4) | |
| Progressive disease | 2 (2) | 1 (4) | |
| MYCN status, No. (%) | 0.762 | ||
| Nonamplified | 74 (71) | 21 (81) | |
| Amplified | 24 (23) | 5 (19) | |
| Unknown | 6 (6) | 0 | |
| Cytogenetics, No. (%) | |||
| 1p deletion (n = 74) | 14 (18) | 2 (15) | 0.920 |
| 11q deletion (n = 79) | 18 (27) | 5 (38) | 0.633 |
| 17q gain (n = 79) | 18 (27) | 4 (33) | 0.912 |
| INPC (n = 121) | 1.000 | ||
| Favorable | 22 (23) | 6 (25) | |
| Unfavorable | 75 (77) | 18 (75) | |
| Serum LDH (IU/L) b | 1302 (261–10000) | 1479 (597–14435) | 0.221 |
| Serum ferritin (ng/mL) b | 224 (20–3771) | 400 (74–1245) | 0.025 |
| Serum NSE (ng/mL) b | 97 (11–1081) | 99 (25–1815) | 0.426 |
| 24 h urine VMA (mg/day) b | 14.8 (0.4–588.4) | 40.0 (1.7–100) | 0.001 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Seo, E.S.; Lee, E.-j.; Lee, B.; Shin, M.; Cho, Y.-S.; Hyun, J.K.; Cho, H.W.; Ju, H.Y.; Yoo, K.H.; Koo, H.H.; et al. Metastatic Burden Defines Clinically and Biologically Distinct Subgroups of Stage 4 High-Risk Neuroblastoma. J. Clin. Med. 2020, 9, 2730. https://doi.org/10.3390/jcm9092730
Seo ES, Lee E-j, Lee B, Shin M, Cho Y-S, Hyun JK, Cho HW, Ju HY, Yoo KH, Koo HH, et al. Metastatic Burden Defines Clinically and Biologically Distinct Subgroups of Stage 4 High-Risk Neuroblastoma. Journal of Clinical Medicine. 2020; 9(9):2730. https://doi.org/10.3390/jcm9092730
Chicago/Turabian StyleSeo, Eun Seop, Eun-jin Lee, Boram Lee, Muheon Shin, Young-Seok Cho, Ju Kyung Hyun, Hee Won Cho, Hee Young Ju, Keon Hee Yoo, Hong Hoe Koo, and et al. 2020. "Metastatic Burden Defines Clinically and Biologically Distinct Subgroups of Stage 4 High-Risk Neuroblastoma" Journal of Clinical Medicine 9, no. 9: 2730. https://doi.org/10.3390/jcm9092730
APA StyleSeo, E. S., Lee, E.-j., Lee, B., Shin, M., Cho, Y.-S., Hyun, J. K., Cho, H. W., Ju, H. Y., Yoo, K. H., Koo, H. H., Lee, J. W., & Sung, K. W. (2020). Metastatic Burden Defines Clinically and Biologically Distinct Subgroups of Stage 4 High-Risk Neuroblastoma. Journal of Clinical Medicine, 9(9), 2730. https://doi.org/10.3390/jcm9092730





