Prediction of Adverse Events in Stable Non-Variceal Gastrointestinal Bleeding Using Machine Learning
Abstract
1. Introduction
2. Experimental Section
2.1. Data
2.2. Methods (Machine Learning Algorithms)
2.3. Analysis
3. Results
3.1. Baseline Statistics
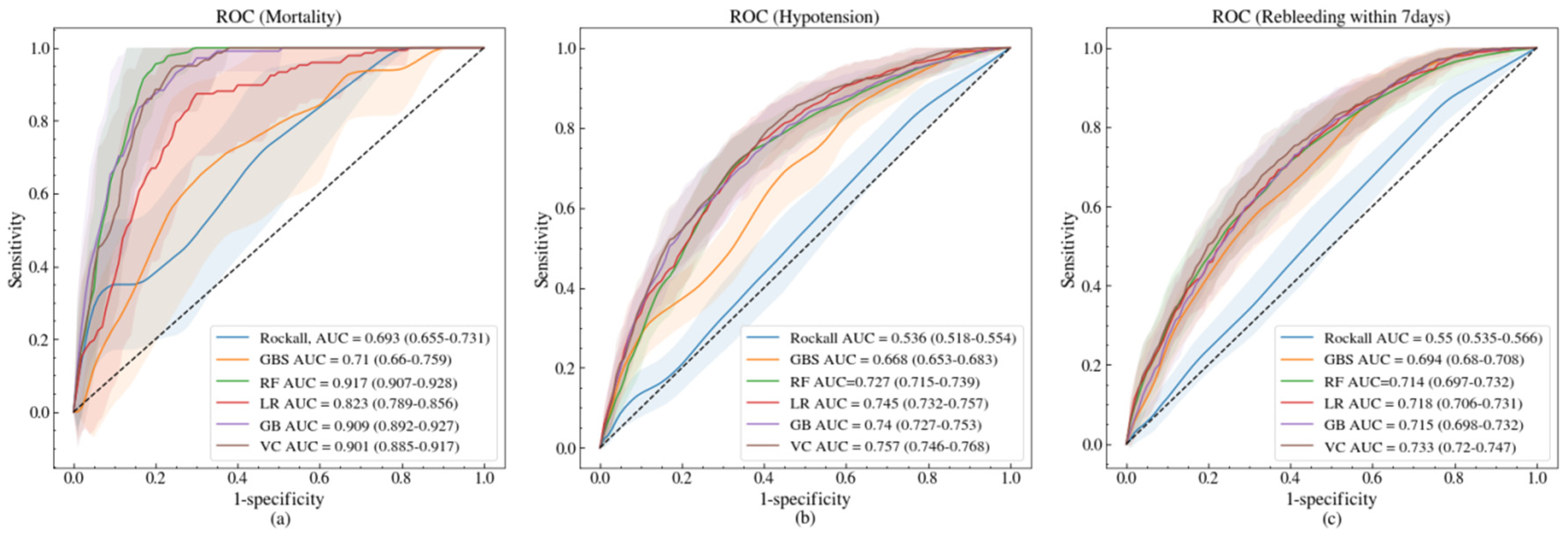
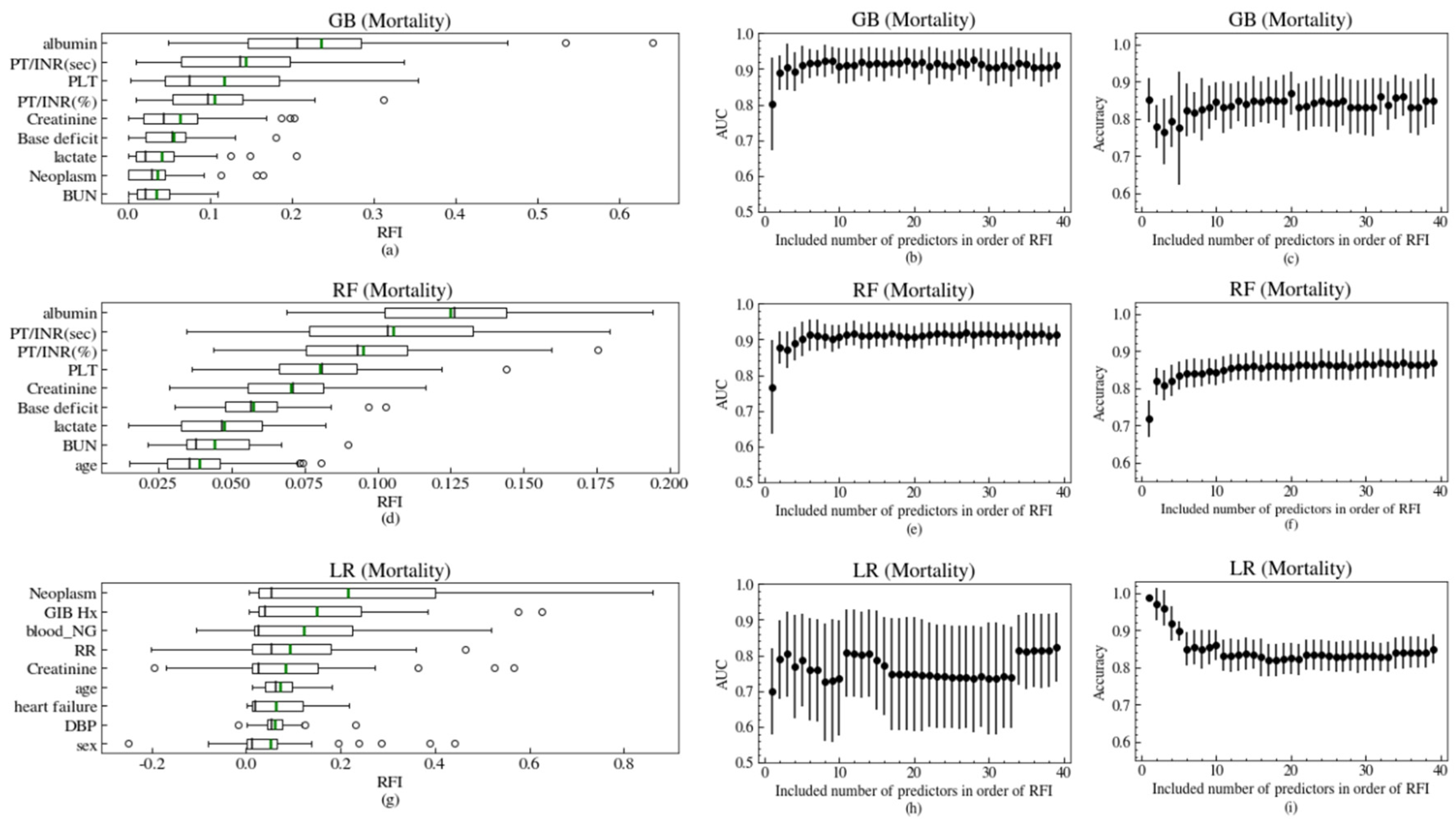
3.2. Model Performances
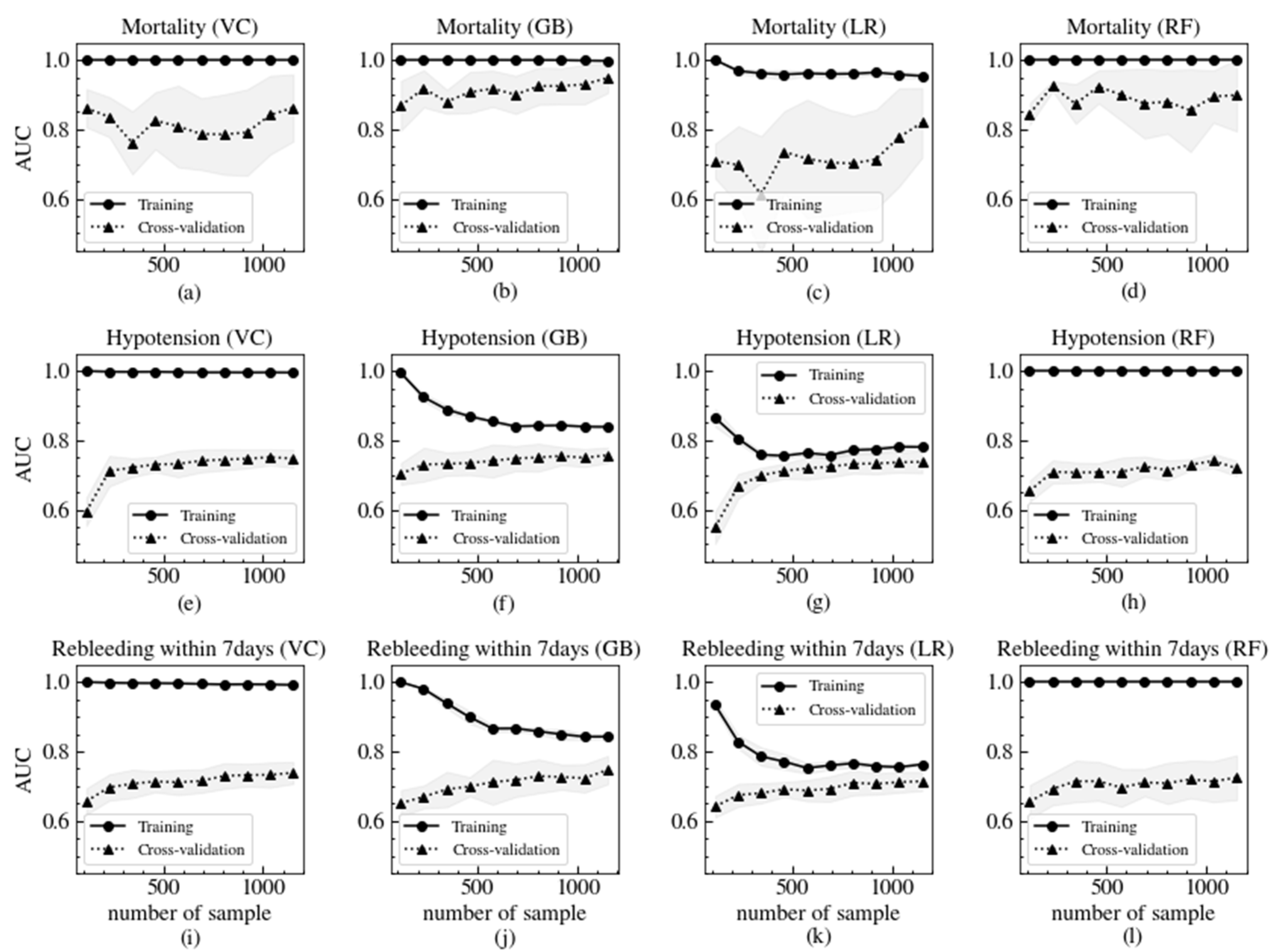
3.3. Model Validation
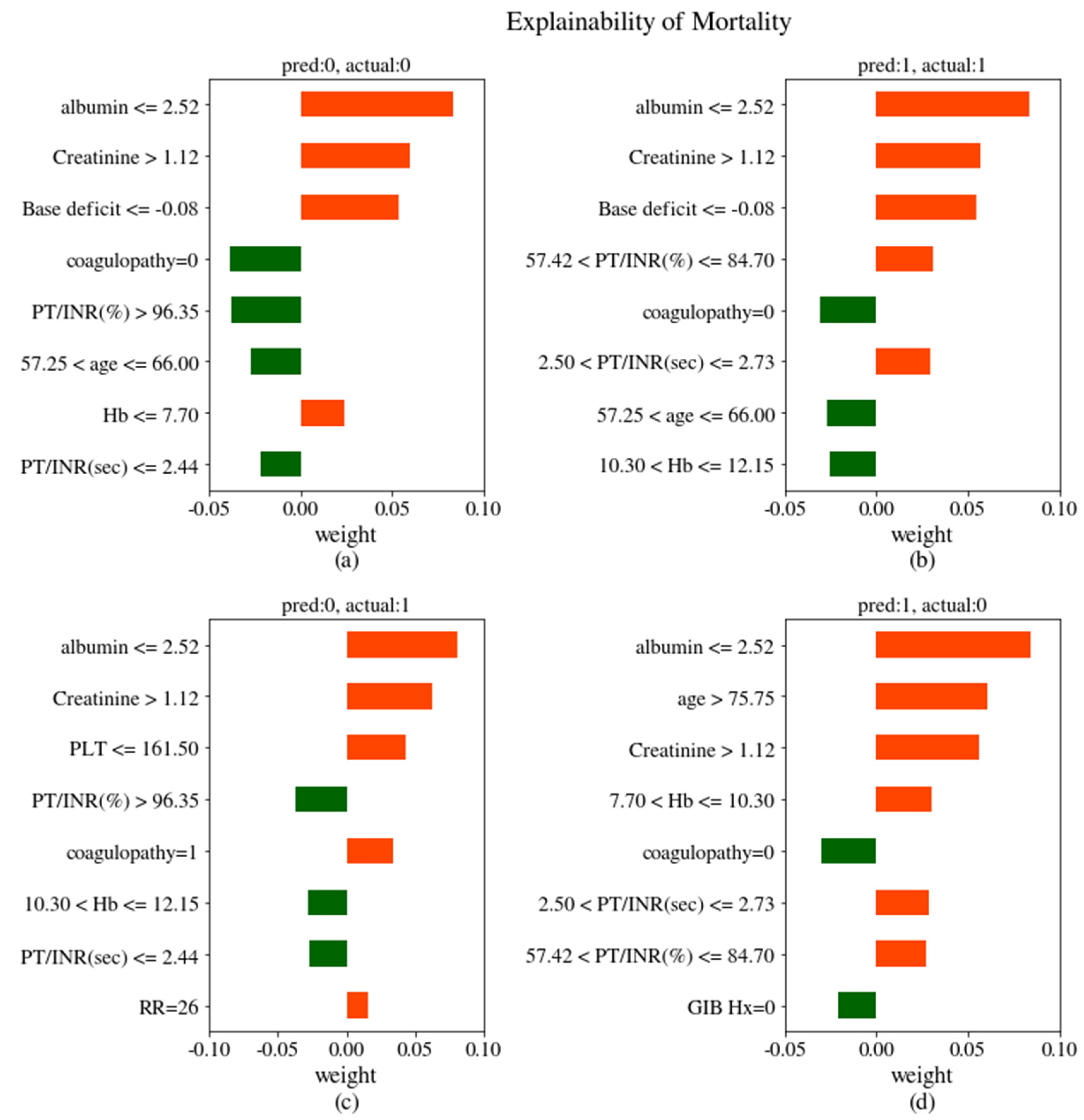
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Appendix A
| Official Variable Name | Convenient Variable Name |
|---|---|
| Demographics | |
| Age | age |
| Male:1 | sex |
| Comorbidities | |
| Diabetes Mellitus | DM |
| Hypertension | HTN |
| Cardiac disease | cardiac disease |
| Liver disease | liver disease |
| Coagulopathy | coagulopathy |
| Ischemic heart disease | IHD |
| Heart failure | heart failure |
| Neoplasm | Neoplasm |
| Chronic kidney disease | CKD |
| Previous GIB history | GIB Hx |
| COPD | COPD |
| Stroke | stroke |
| Associated symptom and signs | |
| Syncope | syncope |
| Hematemesis | Hematemesis |
| Melena, chief complaint | Melena |
| Melena on rectal examination | melena |
| Fresh blood on nasogastric tube | blood NG |
| Mental change | mental binary |
| Drug history Antiplatelet agent | offend antiplatelet |
| NSAIDs | offend NSAID |
| Anticoagulation | offend anticoagulation |
| Vital signs | |
| SBP (mmHg) | SBP |
| DBP (mmHg) | DBP |
| Heart rate (/min) | HR |
| Respiratory rate (/min) | RR |
| Body temperature (°C) | BT |
| Saturation of peripheral oxygen (%) | SpO2 |
| Laboratory findings | |
| Hemoglobin (g/dL) | Hb |
| Platelet count (×103/mm3) | PLT |
| PT/INR (%) | PT/INR (%) |
| PT/INR (s) | PT/INR (sec) |
| BUN (mg/dL) | BUN |
| Creatinine (mg/dL) | creatinine |
| Albumin (g/dL) | albumin |
| Lactate (mmol/L) | lactate |
| Base deficit (mmol/L) | Base deficit |
References
- Ko, B.; Kim, Y.J.; Jung, D.; Sohn, C.; Seo, D.; Lee, Y.S.; Lim, K.; Jung, H.-Y.; Kim, W. Early Risk Score for Predicting Hypotension in Normotensive Patients with Non-Variceal Upper Gastrointestinal Bleeding. J. Clin. Med. 2019, 8, 37. [Google Scholar] [CrossRef] [PubMed]
- Imperiale, T.F.; Dominitz, J.A.; Provenzale, D.T.; Boes, L.P.; Rose, C.M.; Bowers, J.C.; Musick, B.S.; Azzouz, F.; Perkins, S.M. Predicting poor outcome from acute upper gastrointestinal hemorrhage. Arch. Intern. Med. 2007, 167, 1291–1296. [Google Scholar] [CrossRef] [PubMed]
- Barkun, A.N.; Bardou, M.; Kuipers, E.J.; Sung, J.; Hunt, R.H.; Martel, M.; Sinclair, P. International Consensus upper Gastrointestinal Bleeding Conference Group. International consensus recommendations on the management of patients with nonvariceal upper gastrointestinal bleeding. Ann. Intern. Med. 2010, 152, 101–113. [Google Scholar] [CrossRef] [PubMed]
- Rockall, T.A.; Logan, R.F.; Devlin, H.B.; Northfield, T.C. Risk assessment after acute upper gastrointestinal haemorrhage. Gut 1996, 38, 316–321. [Google Scholar] [CrossRef]
- Blatchford, O.; Murray, W.R.; Blatchford, M. A risk score to predict need for treatment for uppergastrointestinal haemorrhage. Lancet 2000, 356, 1318–1321. [Google Scholar] [CrossRef]
- Cheng, D.W.; Lu, Y.W.; Teller, T.; Sekhon, H.K.; Wu, B.U. A modified Glasgow Blatchford Score improves risk stratification in upper gastrointestinal bleed: A prospective comparison of scoring systems. Aliment. Pharm. 2012, 36, 782–789. [Google Scholar] [CrossRef]
- Hyett, B.H.; Abougergi, M.S.; Charpentier, J.P.; Kumar, N.L.; Brozovic, S.; Claggett, B.L.; Travis, A.C.; Saltzman, J.R. The AIMS65 score compared with the Glasgow-Blatchford score in predicting outcomes in upper GI bleeding. YMGE 2013, 77, 551–557. [Google Scholar] [CrossRef]
- Stanley, A.J. Update on risk scoring systems for patients with upper gastrointestinal haemorrhage. WJG 2012, 18, 2739. [Google Scholar] [CrossRef]
- Ko, B.S.; Kim, W.Y.; Ryoo, S.M.; Ahn, S.; Sohn, C.H.; Seo, D.-W.; Lee, Y.S.; Lim, K.S.; Jung, H.-Y. Predicting the Occurrence of Hypotension in Stable Patients with Nonvariceal Upper Gastrointestinal Bleeding. Crit. Care Med. 2015, 43, 2409–2415. [Google Scholar] [CrossRef]
- Jung, D.H.; Ko, B.S.; Kim, Y.J.; Kim, W.Y. Comparison of risk scores and shock index in hemodynamically stable patients presenting to the emergency department with nonvariceal upper gastrointestinal bleeding. Eur. J. Gastroenterol. Hepatol. 2019, 31, 781–785. [Google Scholar] [CrossRef]
- Nanayakkara, S.; Fogarty, S.; Tremeer, M.; Ross, K.; Richards, B.; Bergmeir, C.; Xu, S.; Stub, D.; Smith, K.; Tacey, M.; et al. Characterising risk of in-hospital mortality following cardiac arrest using machine learning: A retrospective international registry study. PLoS Med. 2018, 15, e1002709. [Google Scholar] [CrossRef] [PubMed]
- Esteva, A.; Kuprel, B.; Novoa, R.A.; Ko, J.; Swetter, S.M.; Blau, H.M.; Thrun, S. Dermatologist-level classification of skin cancer with deep neural networks. Nature 2017, 542, 115–118. [Google Scholar] [CrossRef] [PubMed]
- Narula, S.; Shameer, K.; Omar, A.M.S.; Dudley, J.T.; Sengupta, P.P. Machine-Learning Algorithms to Automate Morphological and Functional Assessments in 2D Echocardiography. J. Am. Coll. Cardiol. 2016, 68, 2287–2295. [Google Scholar] [CrossRef] [PubMed]
- Fauw, J.; Ledsam, J.R.; Romera-Paredes, B.; Nikolov, S.; Tomasev, N.; Blackwell, S.; Askham, H.; Glorot, X.; O’Donoghue, B.; Visentin, D.; et al. Clinically applicable deep learning for diagnosis and referral in retinal disease. Nat. Med. 2018, 24, 1–15. [Google Scholar] [CrossRef]
- Shah, S.J.; Katz, D.H.; Selvaraj, S.; Burke, M.A.; Yancy, C.W.; Gheorghiade, M.; Bonow, R.O.; Huang, C.-C.; Deo, R.C. Phenomapping for novel classification of heart failure with preserved ejection fraction. Circulation 2015, 131, 269–279. [Google Scholar] [CrossRef] [PubMed]
- Shung, D.L.; Au, B.; Taylor, R.A.; Tay, J.K.; Laursen, S.B.; Stanley, A.J.; Dalton, H.R.; Ngu, J.; Schultz, M.; Laine, L. Validation of a Machine Learning Model That Outperforms Clinical Risk Scoring Systems for Upper Gastrointestinal Bleeding. Gastroenterology 2020, 158, 160–167. [Google Scholar] [CrossRef]
- Shung, D.; Simonov, M.; Gentry, M.; Au, B.; Laine, L. Machine Learning to Predict Outcomes in Patients with Acute Gastrointestinal Bleeding: A Systematic Review. Dig. Dis. Sci. 2019, 64, 2078–2087. [Google Scholar] [CrossRef]
- Stanley, A.J.; Laine, L.; Dalton, H.R.; Ngu, J.H.; Schultz, M.; Abazi, R.; Zakko, L.; Thornton, S.; Wilkinson, K.; Khor, C.J.L.; et al. Comparison of risk scoring systems for patients presenting with upper gastrointestinal bleeding: International multicentre prospective study. BMJ 2017, 356, i6432–i6438. [Google Scholar] [CrossRef]
- Cox, D.R. The Regression Analysis of Binary Sequences. J. R. Stat. Soc. Ser. B (Methodol.) 1959, 21, 238. [Google Scholar] [CrossRef]
- Breiman, L. Random forests, machine learning. Mach. Learn. 2001, 45, 5–32. [Google Scholar]
- Friedman, J. Greedy function approximation: A gradient boosting machine. Ann. Stat. 2001, 29, 1189–1232. [Google Scholar] [CrossRef]
- Opitz, D.W.; Maclin, R. Popular ensemble methods: An empirical study. J. Artif. Intell. Res. 1999, 11, 169–198. [Google Scholar]
- Ribeiro, M.T.; Singh, S. ‘Why should i trust you?’ Explaining the predictions of any classifier. In Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, New York, NY, USA, 24–27 August 2016; Volume 13, pp. 1135–1144. [Google Scholar] [CrossRef]
- Laine, L.; Jensen, D.M. Management of Patients with Ulcer Bleeding. Am. J. Gastroenterol. 2012, 107, 345–360. [Google Scholar] [CrossRef] [PubMed]
- Wong, A.; Young, A.T.; Liang, A.S.; Gonzales, R.; Douglas, V.C.; Hadley, D. Development and Validation of an Electronic Health Record-Based Machine Learning Model to Estimate Delirium Risk in Newly Hospitalized Patients Without Known Cognitive Impairment. JAMA Netw. Open 2018, 1, e181018. [Google Scholar] [CrossRef] [PubMed]
- Wilson, F.P.; Shashaty, M.; Testani, J.; Aqeel, I.; Borovskiy, Y.; Ellenberg, S.S.; Feldman, H.I.; Fernandez, H.; Gitelman, Y.; Lin, J.; et al. Automated, electronic alerts for acute kidney injury: A single-blind, parallel-group, randomised controlled trial. Lancet 2015, 385, 1966–1974. [Google Scholar] [CrossRef]
- Bzdok, D.; Altman, N.; Krzywinski, M. Statistics versus machine learning. Nat. Methods 2018, 15, 233–234. [Google Scholar] [CrossRef]
- Rotondano, G.; Cipolletta, L.; Grossi, E.; Koch, M.; Intraligi, M.; Buscema, M.; Marmo, R. Progetto Nazionale Emorragie Digestive IROUGB. Artificial neural networks accurately predict mortality in patients with nonvariceal upper GI bleeding. Gastrointest. Endosc. 2011, 73, 218–226.e2. [Google Scholar] [CrossRef]
- Lyles, T.; Elliott, A.; Rockey, D.C. A risk scoring system to predict in-hospital mortality in patients with cirrhosis presenting with upper gastrointestinal bleeding. J. Clin. Gastroenterol. 2014, 48, 712–720. [Google Scholar] [CrossRef]
- Lee, H.H.; Park, J.M.; Han, S.; Park, S.M.; Kim, H.Y.; Oh, J.H.; Kim, C.W.; Yoon, S.K.; Choi, M.-G. A simplified prognostic model to predict mortality in patients with acute variceal bleeding. Dig. Liver Dis. 2018, 50, 247–253. [Google Scholar] [CrossRef]
- Horibe, M.; Iwasaki, E.; Bazerbachi, F.; Kaneko, T.; Matsuzaki, J.; Minami, K.; Masaoka, T.; Hosoe, N.; Ogura, Y.; Namiki, S.; et al. Horibe GI bleeding prediction score: A simple score for triage decision-making in patients with suspected upper GI bleeding. Gastrointest. Endosc. 2020. [Google Scholar] [CrossRef]
- Krawczyk, B. Learning from imbalanced data: Open challenges and future directions. Prog. Artif. Intell. 2016, 5, 221–232. [Google Scholar] [CrossRef]
- Mani, I.; Zhang, J. kNN approach to unbalanced data distributions: A case study involving information extraction. In Proceedings of the Workshop on Learning from Imbalanced Datasets, Washington, DC, USA, 21 August 2003. [Google Scholar]
- Tomek, I. Two modifications of CNN. Syst. ManCybern. IEEE Trans. 2010, 6, 769–772. [Google Scholar]
| Characteristic | Mortality | Hypotension | Rebleeding within 7 days | ||||||
|---|---|---|---|---|---|---|---|---|---|
| No (n = 1422) | Yes (n = 17) | p-Value | No (n = 1297) | Yes (n = 142) | p-Value | No (n = 1305) | Yes (n = 134) | p-Value | |
| Demographics | |||||||||
| Age | 63 (52–74) | 75 (59–80) | 0.011 | 64 (52–74) | 63 (54–73) | 0.900 | 64 (52–74) | 64.5 (56–76) | 0.062 |
| Male | 426 (30.0) | 7 (41.2) | 0.461 | 405 (31.2) | 28 (19.7) | 0.006 | 401 (30.7) | 32 (23.9) | 0.122 |
| Comorbidities | |||||||||
| Diabetes mellitus | 304 (21.4) | 5 (29.4) | 0.614 | 269 (20.7) | 40 (28.2) | 0.052 | 276 (21.1) | 33 (24.6) | 0.410 |
| Hypertension | 538 (37.8) | 4 (23.5) | 0.338 | 490 (37.8) | 52 (36.6) | 0.857 | 488 (37.4) | 54 (40.3) | 0.571 |
| Cardiac disease | 313 (22.0) | 5 (29.4) | 0.662 | 286 (22.1) | 32 (22.5) | 0.980 | 296 (22.7) | 22 (16.4) | 0.120 |
| Liver disease | 64 (4.5) | 1 (5.9) | 0.753 | 60 (4.6) | 5 (3.5) | 0.697 | 55 (4.2) | 10 (7.5) | 0.132 |
| Coagulopathy | 267(18.8) | 9 (52.9) | 0.001 | 239 (18.4) | 37 (26.1) | 0.038 | 235 (18.0) | 41 (30.6) | <0.001 |
| Ischemic heart disease | 188 (13.2) | 3 (17.6) | 0.861 | 174 (13.4) | 17 (13.2) | 0.725 | 179 (13.7) | 10 (9.0) | 0.158 |
| Heart failure | 52 (3.7) | 3 (17.6) | 0.019 | 46 (3.5) | 9 (6.3) | 0.157 | 50 (3.8) | 5 (3.7) | 0.858 |
| Neoplasm | 187 (13.2) | 9 (52.9) | <0.001 | 168 (13.0) | 28 (19.7) | 0.036 | 165 (12.6) | 31 (23.1) | 0.001 |
| Chronic kidney disease | 123 (8.6) | 4 (23.5) | 0.086 | 118 (9.0) | 9 (6.3) | 0.345 | 114 (8.7) | 13 (9.7) | 0.829 |
| Previous GIB history | 229 (16.1) | 8 (47.1) | 0.002 | 209 (16.1) | 28 (19.7) | 0.327 | 210 (16.1) | 27 (20.1) | 0.279 |
| COPD | 29 (2.0) | 1 (5.9) | 0.804 | 27 (2.1) | 3 (2.1) | 0.776 | 27 (2.1) | 3 (2.2) | 0.852 |
| Stroke | 126 (8.9) | 1 (5.9) | 0.999 | 117 (9.0) | 10 (7.0) | 0.527 | 118 (9.0) | 9 (6.7) | 0.457 |
| Associated symptom and signs | |||||||||
| Syncope | 52 (3.7) | 1 (5.9) | 0.870 | 43 (3.3) | 10 (7.0) | 0.045 | 43 (3.3) | 10 (7.5) | 0.028 |
| Hematemesis | 500 (35.2) | 8 (47.1) | 0.444 | 449 (34.6) | 59 (41.5) | 0.122 | 449 (34.4) | 59 (44.0) | 0.034 |
| Melena, chief complaint | 922 (64.8) | 9 (52.9) | 0.444 | 848 (65.4) | 83 (58.5) | 0.122 | 856 (63.6) | 75 (56.0) | 0.034 |
| Melena on rectal examination | 650 (45.7) | 8 (47.1) | 0.893 | 571 (44.0) | 87 (61.3) | <0.001 | 576 (44.1) | 82 (61.2) | <0.001 |
| Fresh blood on nasogastric tube | 155 (10.9) | 5 (29.4) | 0.043 | 125 (9.6) | 35 (24.6) | <0.001 | 127 (9.8) | 33 (24.6) | <0.001 |
| Mental change | 29 (2.0) | 4 (23.5) | <0.001 | 28 (2.2) | 5 (3.5) | 0.463 | 29 (3.2) | 4 (3.0) | 0.796 |
| Drug history Antiplatelet agent | 275 (19.3) | 0 (0.0) | 0.088 | 255 (19.7) | 20 (14.1) | 0.136 | 259 (19.8) | 16 (11.9) | 0.036 |
| NSAIDs | 47 (3.3) | 0 (0.0) | 0.940 | 44 (3.3) | 3 (2.1) | 0.571 | 45 (3.4) | 2 (1.5) | 0.338 |
| Anticoagulation | 103 (7.2) | 2 (11.8) | 0.808 | 91 (7.0) | 14 (9.9) | 0.286 | 92 (7.0) | 13 (9.7) | 0.342 |
| Vital signs | |||||||||
| SBP (mmHg) | 123 (111–141) | 118 (110–130) | 0.534 | 125 (112–142) | 109 (101–118) | <0.001 | 124 (111–141) | 116 (107–132) | <0.001 |
| DBP (mmHg) | 75 (67–86) | 72 (62–86) | 0.703 | 76 (68–86) | 68 (62–77) | <0.001 | 76 (68–86) | 73 (65–81) | 0.009 |
| Heart rate (/min) | 89 (76–103) | 100 (86–118) | 0.048 | 89 (76–103) | 90 (77–107) | 0.229 | 89 (76–103) | 92 (79–105) | 0.149 |
| Respiratory rate (/min) | 20 (18–20) | 20 (20–20) | 0.045 | 20 (18–20) | 20 (18.5–20) | 0.229 | 20 (18–20) | 20 (20–20) | 0.331 |
| Body temperature (°C) | 36.5 (36.2–36.8) | 36.5 (36–36.9) | 0.835 | 36.5 (36.2–36.9) | 36.4 (36–36.6) | <0.001 | 36.5 (36.2–36.9) | 36.5 (36–36.8) | 0.187 |
| Saturation of peripheral oxygen, room air (%) | 99 (97–100) | 99 (96–99) | 0.325 | 99 (97–100) | 99 (97–100) | 0.880 | 99 (97–100) | 99 (98–100) | 0.851 |
| Laboratory findings | |||||||||
| Hemoglobin (g/dL) | 10.3 (8.3–12.5) | 8.5 (7.2–10.3) | 0.018 | 10.4 (8.4–12.6) | 9.4 (7.4–11.5) | <0.001 | 10.5 (8.4–12.6) | 8.7 (7.2–10.6) | <0.001 |
| Platelet count (×103/mm3) | 223 (169–276) | 104 (69–239) | 0.011 | 223 (169–277) | 211 (160–260) | 0.209 | 225 (170–277) | 191 (153–260) | 0.002 |
| PT/INR (%) | 90.9 (79.2–103) | 62.9 (51.6–78.6) | <0.001 | 91.0 (79.3–103.2) | 88.1 (70–97.4) | 0.003 | 91.2 (79.5–103) | 85.8 (70.3–97.4) | <0.001 |
| PT/INR (s) | 2.47 (2.42–2.54) | 2.7 (2.6–2.8) | <0.001 | 2.47 (2.42–2.54) | 2.5 (2.4–2.6) | 0.004 | 2.47 (2.42–2.54) | 2.5 (2.4–2.6) | 0.001 |
| BUN (mg/dL) | 24.0 (15.0–38.0) | 35.0 (24.0–62.0) | 0.006 | 23.0 (15.0–37.0) | 31.0 (22.0–43.0) | <0.001 | 23.0 (15.0–37.0) | 31.0 (22.0–45.8) | <0.001 |
| Creatinine (mg/dL) | 0.8 (0.7–1.1) | 1.2 (1.0–2.9) | 0.015 | 0.8 (0.7–1.1) | 0.9 (0.7–1.2) | 0.156 | 0.0 (0.0–0.8 (0.7–1.1)1.0) | 0.9 (0.7–1.2) | 0.134 |
| Albumin (g/dL) | 3.3 (2.8–3.7) | 2.3 (1.9–2.5) | <0.001 | 3.3 (2.9–3.8) | 3.0 (2.5–3.4) | <0.001 | 3.4 (2.9–3.8) | 2.8 (2.3–3.3) | <0.001 |
| Lactate (mmol/L) | 1.3 (0.9–2.1) | 2.2 (1.1–5.7) | 0.051 | 1.3 (0.9–2.0) | 1.6 (1.2–2.6) | <0.001 | 1.3 (0.9–2.0) | 1.6 (1.1–2.4) | 0.013 |
| Base deficit (mmol/L) | 1.5 (−0.5–3.5) | −0.9 (−6.8–6.0) | 0.433 | 1.7 (−0.5–3.7) | 0.8 (−1.7–2.8) | <0.001 | 1.7 (−0.5–3.6) | 0.5 (−0.8–2.8) | 0.002 |
| Risk scores | |||||||||
| Glasgow-Blatchford Bleeding Score | 10 (6–12) | 13 (11–14) | 0.003 | 10 (6–12) | 12 (9–14) | <0.001 | 10 (6–12) | 12 (10–14) | <0.001 |
| Pre-endoscopy Rockall | 1 (1–3) | 2 (1–6) | 0.005 | 1 (1–3) | 1 (1–3) | 0.147 | 1 (1–3) | 2 (1–3) | 0.049 |
| Outcomes | Models Scorings | Rockall Score | GBS | Logistic Regression Classifier | Random Forest Classifier | Gradient Boosting Classifier | Voting Classifier | Actual Positive Ratio (%) |
|---|---|---|---|---|---|---|---|---|
| Mortality | Predicted Positive ratio (%, 95% CI) | 29.3 (28.4–30.2) | 25.2 (24.4–26.0) | 14.7 (13.4–15.9) | 14.6 (13.5–15.6) | 15.4 (13.8–17.0) | 16.5 (15.8–17.1) | 1.18 |
| AUC (95% CI) | 0.694 (0.649–0.738) | 0.715 (0.676–0.754) | 0.826 (0.792–0.861) | 0.909 (0.896–0.921) | 0.911 (0.893–0.930) | 0.908 (0.893–0.922) | ||
| Brier score (95% CI) | 0.294 (0.285–0.303) | 0.250 (0.242–0.258) | 0.129 (0.120–0.139) | 0.034 (0.032–0.036) | 0.035 (0.032–0.039) | 0.050 (0.047–0.054) | ||
| Log loss (95% CI) | 27.0 (26.7–27.3) | 34.1 (34.0–34.1) | 0.510 (0.435–0.563) | 0.142 (0.137–0.147) | 0.126 (0.116–0.137) | 0.180 (0.171–0.189) | ||
| Hypotension | Predicted Positive ratio (%, 95% CI) | 45.0 (44.1–45.9) | 35.0 (34.1–35.8) | 26.1 (25.1–27.2) | 22.56 (21.52–23.59) | 16.8 (15.4–18.2) | 19.7 (18.7–20.8) | 9.87 |
| AUC (95% CI) | 0.536 (0.516–0.555) | 0.668 (0.656–0.680) | 0.747 (0.737–0.758) | 0.739 (0.727–0.751) | 0.756 (0.743–0.768) | 0.766 (0.755–0.777) | ||
| Brier score (95% CI) | 0.233 (0.226 0.240) | 0.273 (0.267–0.279) | 0.194 (0.191–0.198) | 0.0834 (0.0826–0.08416) | 0.082 (0.081–0.084) | 0.092 (0.091–0.093) | ||
| Log loss (95% CI) | 25.0 (24.8–25.3) | 31.1 (31.0–31.1) | 0.569 (0.559–0.579) | 0.329 (0.310–0.348) | 0.289 (0.283–0.295) | 0.319 (0.316–0.322) | ||
| Rebleeding within 7 days | Predicted Positive ratio (%, 95% CI) | 45.0 (44.3 45.8) | 35.0 (33.8 36.1) | 31.6 (30.6 32.7) | 27.4 (26.2 28.5) | 20.7 (19.0 22.3) | 21.3 (20.2 22.4) | 9.31 |
| AUC (95% CI) | 0.550 (0.534–0.566) | 0.694 (0.680–0.708) | 0.712 (0.698–0.726) | 0.707 (0.689–0.725) | 0.717 (0.699–0.734) | 0.729 (0.714–0.745) | ||
| Brier score (95% CI) | 0.223 (0.215–0.231) | 0.258 (0.249–0.266) | 0.201 (0.197–0.206) | 0.081 (0.080–0.082) | 0.081 (0.080–0.082) | 0.092 (0.090–0.093) | ||
| Log loss (95% CI) | 25.0 (24.8–25.2) | 31.3 (31.2–31.3) | 0.589 (0.578–0.599) | 0.319 (0.295–0.344) | 0.287 (0.281–0.293) | 0.323 (0.319–0.327) |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Seo, D.-W.; Yi, H.; Park, B.; Kim, Y.-J.; Jung, D.H.; Woo, I.; Sohn, C.H.; Ko, B.S.; Kim, N.; Kim, W.Y. Prediction of Adverse Events in Stable Non-Variceal Gastrointestinal Bleeding Using Machine Learning. J. Clin. Med. 2020, 9, 2603. https://doi.org/10.3390/jcm9082603
Seo D-W, Yi H, Park B, Kim Y-J, Jung DH, Woo I, Sohn CH, Ko BS, Kim N, Kim WY. Prediction of Adverse Events in Stable Non-Variceal Gastrointestinal Bleeding Using Machine Learning. Journal of Clinical Medicine. 2020; 9(8):2603. https://doi.org/10.3390/jcm9082603
Chicago/Turabian StyleSeo, Dong-Woo, Hahn Yi, Beomhee Park, Youn-Jung Kim, Dae Ho Jung, Ilsang Woo, Chang Hwan Sohn, Byuk Sung Ko, Namkug Kim, and Won Young Kim. 2020. "Prediction of Adverse Events in Stable Non-Variceal Gastrointestinal Bleeding Using Machine Learning" Journal of Clinical Medicine 9, no. 8: 2603. https://doi.org/10.3390/jcm9082603
APA StyleSeo, D.-W., Yi, H., Park, B., Kim, Y.-J., Jung, D. H., Woo, I., Sohn, C. H., Ko, B. S., Kim, N., & Kim, W. Y. (2020). Prediction of Adverse Events in Stable Non-Variceal Gastrointestinal Bleeding Using Machine Learning. Journal of Clinical Medicine, 9(8), 2603. https://doi.org/10.3390/jcm9082603






