SMAD4 Overexpression in Patients with Sleep Apnoea May Be Associated with Cardiometabolic Comorbidities
Abstract
1. Introduction
2. Experimental Section
2.1. Study Participants
2.2. PBMC Isolation and Culture
2.3. Intermittent Hypoxia In Vitro Model
2.4. HIF1α Inhibition
2.5. mRNA Isolation and Quantification
2.6. Determination of Plasma Levels of Soluble Proteins
2.7. Statistical Analyses
3. Results
3.1. Study Subjects
3.2. SMAD4 is Overexpressed in OSA Patients
3.3. Hypoxaemia Enhances SMAD4 Expression
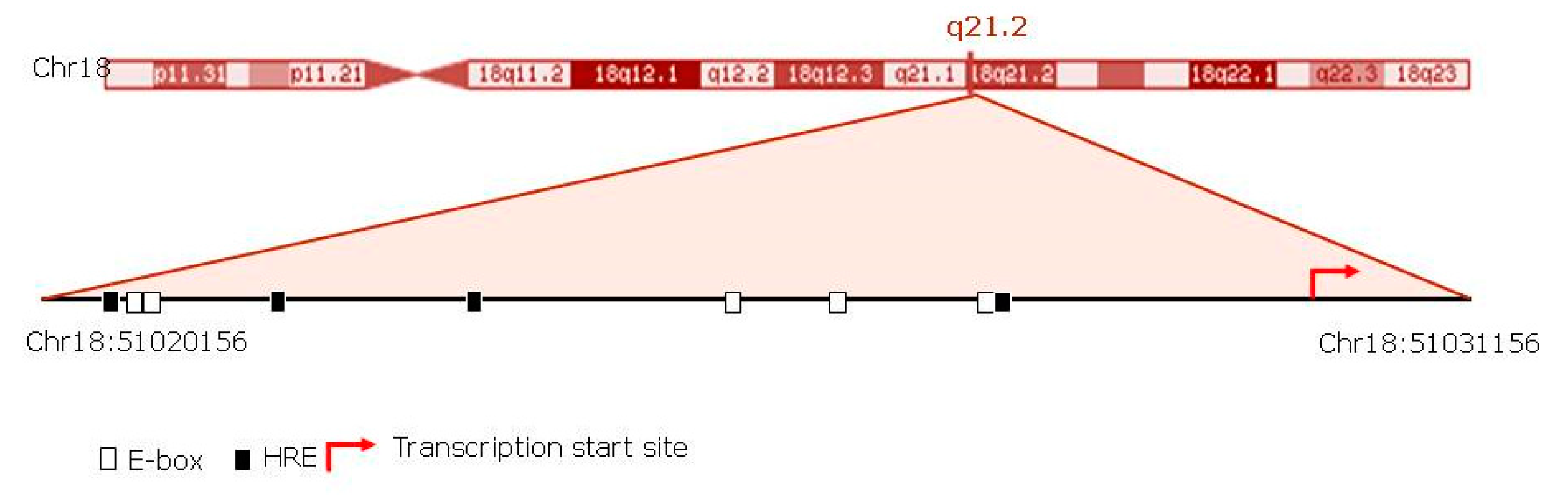
3.4. The Activity of Circadian Rhythm Genes is Associated with SMAD4 Expression
3.5. Association between Hypoxia and Circadian Genes
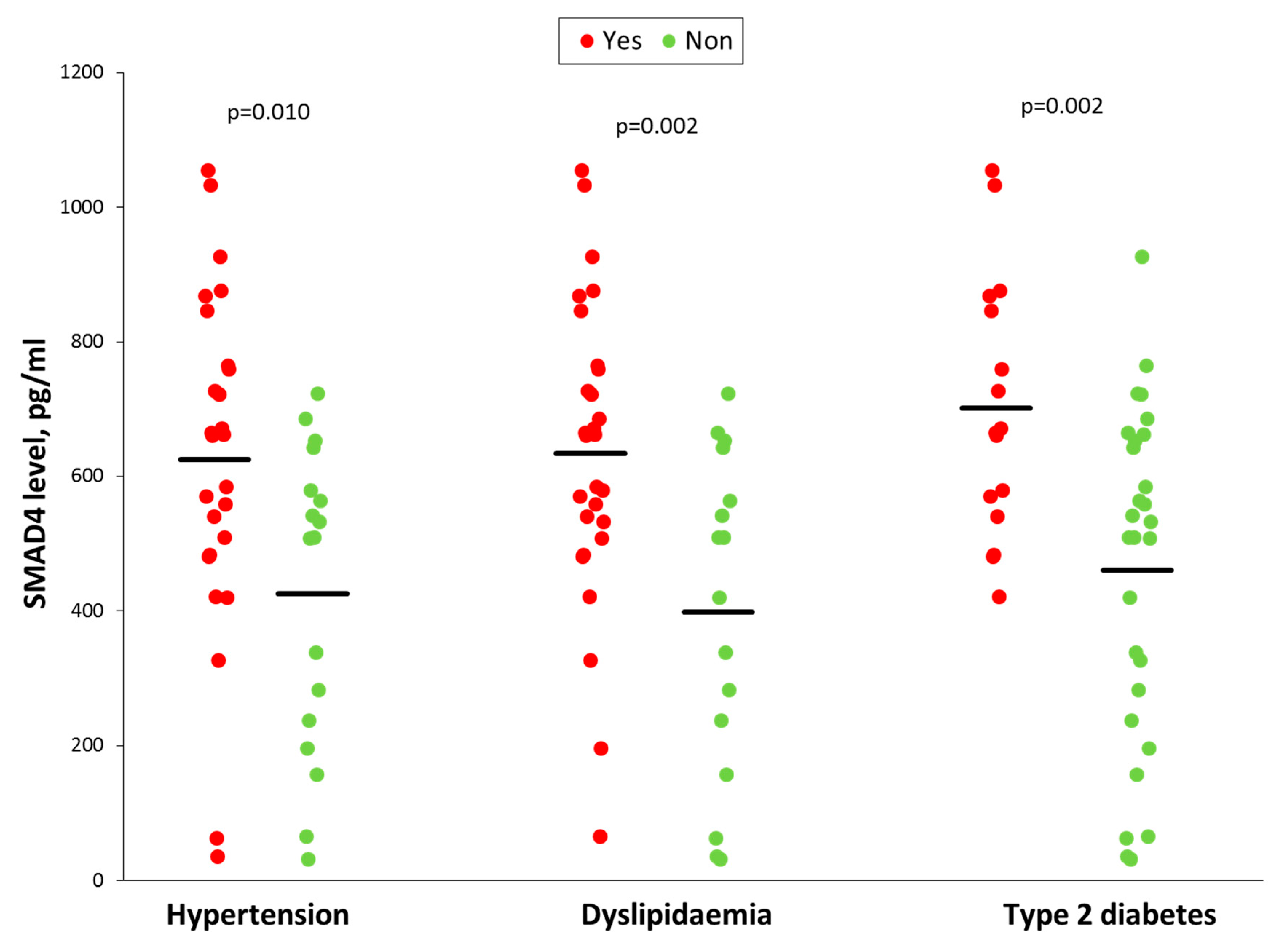
3.6. Smad4 Expression is Associated with a Higher Risk of OSA Cardiometabolic Morbidity
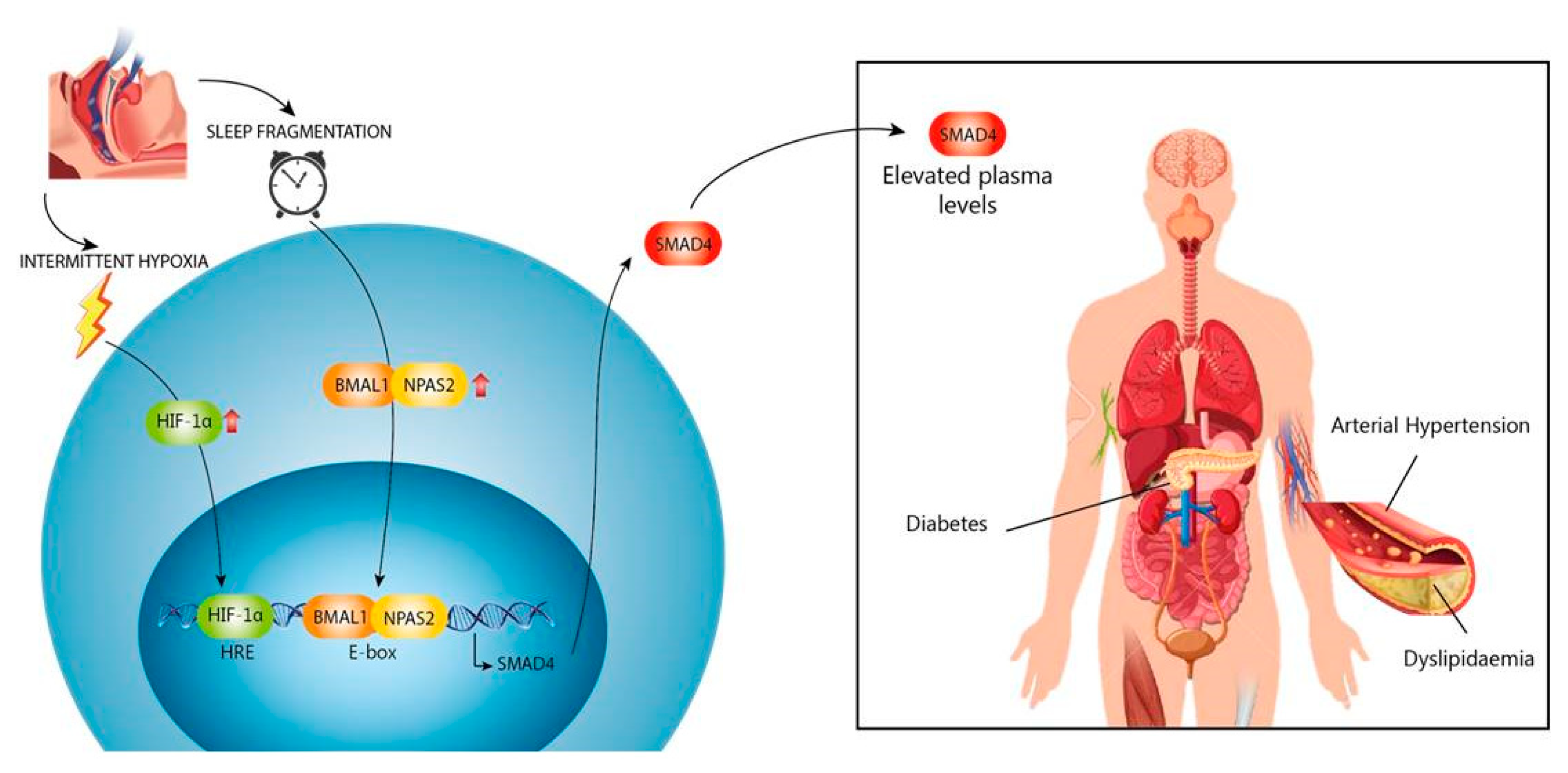
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Benjafield, A.V.; Ayas, N.T.; Eastwood, P.R.; Heinzer, R.; Ip, M.S.M.; Morrell, M.J.; Nunez, C.M.; Patel, S.R.; Penzel, T.; Pepin, J.L.; et al. Estimation of the global prevalence and burden of obstructive sleep apnoea: A literature-based analysis. Lancet Respir. Med. 2019, 7, 687–698. [Google Scholar] [CrossRef]
- Levy, P.; Kohler, M.; McNicholas, W.T.; Barbe, F.; McEvoy, R.D.; Somers, V.K.; Lavie, L.; Pepin, J.L. Obstructive sleep apnoea syndrome. Nat. Rev. Dis. Primers 2015, 1, 1–21. [Google Scholar] [CrossRef] [PubMed]
- Marin, J.M.; Carrizo, S.J.; Vicente, E.; Agusti, A.G. Long-term cardiovascular outcomes in men with obstructive sleep apnoea-hypopnoea with or without treatment with continuous positive airway pressure: An observational study. Lancet 2005, 365, 1046–1053. [Google Scholar] [CrossRef]
- Sanchez-de-la-Torre, M.; Campos-Rodriguez, F.; Barbe, F. Obstructive sleep apnoea and cardiovascular disease. Lancet Respir. Med. 2013, 1, 61–72. [Google Scholar] [CrossRef]
- Barros, D.; Garcia-Rio, F. Obstructive sleep apnea and dyslipidemia: From animal models to clinical evidence. Sleep 2019, 42, zsy236. [Google Scholar] [CrossRef]
- Hernandez-Jimenez, E.; Cubillos-Zapata, C.; Toledano, V.; Perez de Diego, R.; Fernandez-Navarro, I.; Casitas, R.; Carpio, C.; Casas-Martin, J.; Valentin, J.; Varela-Serrano, A.; et al. Monocytes inhibit NK activity via TGF-beta in patients with obstructive sleep apnoea. Eur. Respir. J. 2017, 49. [Google Scholar] [CrossRef]
- Tan, C.K.; Chong, H.C.; Tan, E.H.; Tan, N.S. Getting ‘Smad’ about obesity and diabetes. Nutr. Diabetes 2012, 2, e29. [Google Scholar] [CrossRef]
- Gordon, K.J.; Blobe, G.C. Role of transforming growth factor-beta superfamily signaling pathways in human disease. Biochim. Biophys. Acta 2008, 1782, 197–228. [Google Scholar] [CrossRef]
- Herder, C.; Zierer, A.; Koenig, W.; Roden, M.; Meisinger, C.; Thorand, B. Transforming growth factor-beta1 and incident type 2 diabetes: Results from the MONICA/KORA case-cohort study, 1984–2002. Diabetes Care 2009, 32, 1921–1923. [Google Scholar] [CrossRef]
- Shi, Y.; Massague, J. Mechanisms of TGF-beta signaling from cell membrane to the nucleus. Cell 2003, 113, 685–700. [Google Scholar] [CrossRef]
- Moren, A.; Imamura, T.; Miyazono, K.; Heldin, C.H.; Moustakas, A. Degradation of the tumor suppressor Smad4 by WW and HECT domain ubiquitin ligases. J. Biol. Chem. 2005, 280, 22115–22123. [Google Scholar] [CrossRef] [PubMed]
- Dupont, S.; Zacchigna, L.; Cordenonsi, M.; Soligo, S.; Adorno, M.; Rugge, M.; Piccolo, S. Germ-layer specification and control of cell growth by Ectodermin, a Smad4 ubiquitin ligase. Cell 2005, 121, 87–99. [Google Scholar] [CrossRef] [PubMed]
- Dupont, S.; Mamidi, A.; Cordenonsi, M.; Montagner, M.; Zacchigna, L.; Adorno, M.; Martello, G.; Stinchfield, M.J.; Soligo, S.; Morsut, L.; et al. FAM/USP9x, a deubiquitinating enzyme essential for TGFbeta signaling, controls Smad4 monoubiquitination. Cell 2009, 136, 123–135. [Google Scholar] [CrossRef] [PubMed]
- Massague, J.; Blain, S.W.; Lo, R.S. TGFbeta signaling in growth control, cancer, and heritable disorders. Cell 2000, 103, 295–309. [Google Scholar] [CrossRef]
- von Allmen, D.C.; Francey, L.J.; Rogers, G.M.; Ruben, M.D.; Cohen, A.P.; Wu, G.; Schmidt, R.E.; Ishman, S.L.; Amin, R.S.; Hogenesch, J.B.; et al. Circadian Dysregulation: The Next Frontier in Obstructive Sleep Apnea Research. Otolaryngol. Head Neck Surg. 2018, 159, 948–955. [Google Scholar] [CrossRef] [PubMed]
- Ko, C.H.; Takahashi, J.S. Molecular components of the mammalian circadian clock. Hum. Mol. Genet. 2006, 15 (Suppl. 2), R271–R277. [Google Scholar] [CrossRef]
- Haque, R.; Ali, F.G.; Biscoglia, R.; Abey, J.; Weller, J.; Klein, D.; Iuvone, P.M. CLOCK and NPAS2 have overlapping roles in the circadian oscillation of arylalkylamine N-acetyltransferase mRNA in chicken cone photoreceptors. J. Neurochem. 2010, 113, 1296–1306. [Google Scholar] [CrossRef]
- Takeda, N.; Maemura, K. The role of clock genes and circadian rhythm in the development of cardiovascular diseases. Cell Mol. Life Sci. 2015, 72, 3225–3234. [Google Scholar] [CrossRef]
- Grimaldi, B.; Bellet, M.M.; Katada, S.; Astarita, G.; Hirayama, J.; Amin, R.H.; Granneman, J.G.; Piomelli, D.; Leff, T.; Sassone-Corsi, P. PER2 controls lipid metabolism by direct regulation of PPARgamma. Cell Metab. 2010, 12, 509–520. [Google Scholar] [CrossRef]
- Young, M.E.; Razeghi, P.; Cedars, A.M.; Guthrie, P.H.; Taegtmeyer, H. Intrinsic diurnal variations in cardiac metabolism and contractile function. Circ. Res. 2001, 89, 1199–1208. [Google Scholar] [CrossRef]
- Cubillos-Zapata, C.; Almendros, I.; Diaz-Garcia, E.; Toledano, V.; Casitas, R.; Galera, R.; Lopez-Collazo, E.; Farre, R.; Gozal, D.; Garcia-Rio, F. Differential effect of intermittent hypoxia and sleep fragmentation on PD-1/PD-L1 upregulation. Sleep 2020, 43. [Google Scholar] [CrossRef] [PubMed]
- Cubillos-Zapata, C.; Avendano-Ortiz, J.; Hernandez-Jimenez, E.; Toledano, V.; Casas-Martin, J.; Varela-Serrano, A.; Torres, M.; Almendros, I.; Casitas, R.; Fernandez-Navarro, I.; et al. Hypoxia-induced PD-L1/PD-1 crosstalk impairs T-cell function in sleep apnoea. Eur. Respir. J. 2017, 50. [Google Scholar] [CrossRef] [PubMed]
- Cubillos-Zapata, C.; Balbas-Garcia, C.; Avendano-Ortiz, J.; Toledano, V.; Torres, M.; Almendros, I.; Casitas, R.; Zamarron, E.; Garcia-Sanchez, A.; Feliu, J.; et al. Age-dependent hypoxia-induced PD-L1 upregulation in patients with obstructive sleep apnoea. Respirology 2019, 24, 684–692. [Google Scholar] [CrossRef] [PubMed]
- Dumaine, J.E.; Ashley, N.T. Acute sleep fragmentation induces tissue-specific changes in cytokine gene expression and increases serum corticosterone concentration. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2015, 308, R1062–R1069. [Google Scholar] [CrossRef]
- Chen, W.D.; Yeh, J.K.; Peng, M.T.; Shie, S.S.; Lin, S.L.; Yang, C.H.; Chen, T.H.; Hung, K.C.; Wang, C.C.; Hsieh, I.C.; et al. Circadian CLOCK Mediates Activation of Transforming Growth Factor-beta Signaling and Renal Fibrosis through Cyclooxygenase 2. Am. J. Pathol. 2015, 185, 3152–3163. [Google Scholar] [CrossRef]
- Dong, C.; Gongora, R.; Sosulski, M.L.; Luo, F.; Sanchez, C.G. Regulation of transforming growth factor-beta1 (TGF-beta1)-induced pro-fibrotic activities by circadian clock gene BMAL1. Respir. Res. 2016, 17, 4. [Google Scholar] [CrossRef]
- Akagi, R.; Akatsu, Y.; Fisch, K.M.; Alvarez-Garcia, O.; Teramura, T.; Muramatsu, Y.; Saito, M.; Sasho, T.; Su, A.I.; Lotz, M.K. Dysregulated circadian rhythm pathway in human osteoarthritis: NR1D1 and BMAL1 suppression alters TGF-beta signaling in chondrocytes. Osteoarthr. Cartil. 2017, 25, 943–951. [Google Scholar] [CrossRef]
- Sato, F.; Sato, H.; Jin, D.; Bhawal, U.K.; Wu, Y.; Noshiro, M.; Kawamoto, T.; Fujimoto, K.; Seino, H.; Morohashi, S.; et al. Smad3 and Snail show circadian expression in human gingival fibroblasts, human mesenchymal stem cell, and in mouse liver. Biochem. Biophys. Res. Commun. 2012, 419, 441–446. [Google Scholar] [CrossRef]
- Bartels, N.K.; Borgel, J.; Wieczorek, S.; Buchner, N.; Hanefeld, C.; Bulut, D.; Mugge, A.; Rump, L.C.; Sanner, B.M.; Epplen, J.T. Risk factors and myocardial infarction in patients with obstructive sleep apnea: Impact of beta2-adrenergic receptor polymorphisms. BMC Med. 2007, 5, 1. [Google Scholar] [CrossRef]
- Spivak-Kroizman, T.R.; Hostetter, G.; Posner, R.; Aziz, M.; Hu, C.; Demeure, M.J.; Von Hoff, D.; Hingorani, S.R.; Palculict, T.B.; Izzo, J.; et al. Hypoxia triggers hedgehog-mediated tumor-stromal interactions in pancreatic cancer. Cancer Res. 2013, 73, 3235–3247. [Google Scholar] [CrossRef] [PubMed]
- Schwartz, D.L.; Powis, G.; Thitai-Kumar, A.; He, Y.; Bankson, J.; Williams, R.; Lemos, R.; Oh, J.; Volgin, A.; Soghomonyan, S.; et al. The selective hypoxia inducible factor-1 inhibitor PX-478 provides in vivo radiosensitization through tumor stromal effects. Mol. Cancer Ther. 2009, 8, 947–958. [Google Scholar] [CrossRef] [PubMed]
- Koh, M.Y.; Spivak-Kroizman, T.; Venturini, S.; Welsh, S.; Williams, R.R.; Kirkpatrick, D.L.; Powis, G. Molecular mechanisms for the activity of PX-478, an antitumor inhibitor of the hypoxia-inducible factor-1alpha. Mol. Cancer Ther. 2008, 7, 90–100. [Google Scholar] [CrossRef] [PubMed]
- Welsh, S.; Williams, R.; Kirkpatrick, L.; Paine-Murrieta, G.; Powis, G. Antitumor activity and pharmacodynamic properties of PX-478, an inhibitor of hypoxia-inducible factor-1alpha. Mol. Cancer Ther. 2004, 3, 233–244. [Google Scholar] [PubMed]
- Zhao, T.; Ren, H.; Jia, L.; Chen, J.; Xin, W.; Yan, F.; Li, J.; Wang, X.; Gao, S.; Qian, D.; et al. Inhibition of HIF-1alpha by PX-478 enhances the anti-tumor effect of gemcitabine by inducing immunogenic cell death in pancreatic ductal adenocarcinoma. Oncotarget 2015, 6, 2250–2262. [Google Scholar] [CrossRef]
- Chilov, D.; Hofer, T.; Bauer, C.; Wenger, R.H.; Gassmann, M. Hypoxia affects expression of circadian genes PER1 and CLOCK in mouse brain. FASEB J. 2001, 15, 2613–2622. [Google Scholar] [CrossRef]
- Gabryelska, A.; Sochal, M.; Turkiewicz, S.; Bialasiewicz, P. Relationship between HIF-1 and Circadian Clock Proteins in Obstructive Sleep Apnea Patients-Preliminary Study. J. Clin. Med. 2020, 9. [Google Scholar] [CrossRef]
- Peek, C.B.; Levine, D.C.; Cedernaes, J.; Taguchi, A.; Kobayashi, Y.; Tsai, S.J.; Bonar, N.A.; McNulty, M.R.; Ramsey, K.M.; Bass, J. Circadian Clock Interaction with HIF1alpha Mediates Oxygenic Metabolism and Anaerobic Glycolysis in Skeletal Muscle. Cell Metab. 2017, 25, 86–92. [Google Scholar] [CrossRef]
- Button, E.L.; Bersten, D.C.; Whitelaw, M.L. HIF has Biff—Crosstalk between HIF1a and the family of bHLH/PAS proteins. Exp. Cell Res. 2017, 356, 141–145. [Google Scholar] [CrossRef]
- Bartman, C.M.; Eckle, T. Circadian-Hypoxia Link and its Potential for Treatment of Cardiovascular Disease. Curr. Pharm. Des. 2019, 25, 1075–1090. [Google Scholar] [CrossRef]
- Hogenesch, J.B.; Gu, Y.Z.; Jain, S.; Bradfield, C.A. The basic-helix-loop-helix-PAS orphan MOP3 forms transcriptionally active complexes with circadian and hypoxia factors. Proc. Natl. Acad. Sci. USA 1998, 95, 5474–5479. [Google Scholar] [CrossRef]
- Wu, Y.; Tang, D.; Liu, N.; Xiong, W.; Huang, H.; Li, Y.; Ma, Z.; Zhao, H.; Chen, P.; Qi, X.; et al. Reciprocal Regulation between the Circadian Clock and Hypoxia Signaling at the Genome Level in Mammals. Cell Metab. 2017, 25, 73–85. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.; Lahens, N.F.; Ballance, H.I.; Hughes, M.E.; Hogenesch, J.B. A circadian gene expression atlas in mammals: Implications for biology and medicine. Proc. Natl. Acad. Sci. USA 2014, 111, 16219–16224. [Google Scholar] [CrossRef] [PubMed]
- Crnko, S.; Du Pre, B.C.; Sluijter, J.P.G.; Van Laake, L.W. Circadian rhythms and the molecular clock in cardiovascular biology and disease. Nat. Rev. Cardiol. 2019, 16, 437–447. [Google Scholar] [CrossRef] [PubMed]
- Xie, Z.; Su, W.; Liu, S.; Zhao, G.; Esser, K.; Schroder, E.A.; Lefta, M.; Stauss, H.M.; Guo, Z.; Gong, M.C. Smooth-muscle BMAL1 participates in blood pressure circadian rhythm regulation. J. Clin. Investig. 2015, 125, 324–336. [Google Scholar] [CrossRef]
- Lefta, M.; Campbell, K.S.; Feng, H.Z.; Jin, J.P.; Esser, K.A. Development of dilated cardiomyopathy in Bmal1-deficient mice. Am. J. Physiol. Heart Circ. Physiol. 2012, 303, H475–H485. [Google Scholar] [CrossRef]
- Englund, A.; Kovanen, L.; Saarikoski, S.T.; Haukka, J.; Reunanen, A.; Aromaa, A.; Lonnqvist, J.; Partonen, T. NPAS2 and PER2 are linked to risk factors of the metabolic syndrome. J. Circadian Rhythm. 2009, 7, 5. [Google Scholar] [CrossRef]
- Marques, F.Z.; Campain, A.E.; Tomaszewski, M.; Zukowska-Szczechowska, E.; Yang, Y.H.; Charchar, F.J.; Morris, B.J. Gene expression profiling reveals renin mRNA overexpression in human hypertensive kidneys and a role for microRNAs. Hypertension 2011, 58, 1093–1098. [Google Scholar] [CrossRef]
- Woon, P.Y.; Kaisaki, P.J.; Braganca, J.; Bihoreau, M.T.; Levy, J.C.; Farrall, M.; Gauguier, D. Aryl hydrocarbon receptor nuclear translocator-like (BMAL1) is associated with susceptibility to hypertension and type 2 diabetes. Proc. Natl. Acad. Sci. USA 2007, 104, 14412–14417. [Google Scholar] [CrossRef]
- Wang, Y.; Jiang, L.; Mo, X.; Lan, Y.; Yang, X.; Liu, X.; Zhang, J.; Zhu, L.; Liu, J.; Wu, X. Megakaryocytic Smad4 Regulates Platelet Function through Syk and ROCK2 Expression. Mol. Pharmacol. 2017, 92, 285–296. [Google Scholar] [CrossRef]
- Kaneko, K.; Yamada, T.; Tsukita, S.; Takahashi, K.; Ishigaki, Y.; Oka, Y.; Katagiri, H. Obesity alters circadian expressions of molecular clock genes in the brainstem. Brain Res. 2009, 1263, 58–68. [Google Scholar] [CrossRef]
- Al-Sarraf, I.A.K.; Kasabri, V.; Akour, A.; Naffa, R. Melatonin and cryptochrome 2 in metabolic syndrome patients with or without diabetes: A cross-sectional study. Horm. Mol. Biol. Clin. Investig. 2018, 35. [Google Scholar] [CrossRef] [PubMed]
- Ota, H.; Fujita, Y.; Yamauchi, M.; Muro, S.; Kimura, H.; Takasawa, S. Relationship Between Intermittent Hypoxia and Type 2 Diabetes in Sleep Apnea Syndrome. Int. J. Mol. Sci. 2019, 20, 4756. [Google Scholar] [CrossRef] [PubMed]
- Ota, H.; Itaya-Hironaka, A.; Yamauchi, A.; Sakuramoto-Tsuchida, S.; Miyaoka, T.; Fujimura, T.; Tsujinaka, H.; Yoshimoto, K.; Nakagawara, K.; Tamaki, S.; et al. Pancreatic beta cell proliferation by intermittent hypoxia via up-regulation of Reg family genes and HGF gene. Life Sci. 2013, 93, 664–672. [Google Scholar] [CrossRef] [PubMed]
- Uchiyama, T.; Ota, H.; Itaya-Hironaka, A.; Shobatake, R.; Yamauchi, A.; Sakuramoto-Tsuchida, S.; Makino, M.; Kimura, H.; Takeda, M.; Ohbayashi, C.; et al. Up-regulation of selenoprotein P and HIP/PAP mRNAs in hepatocytes by intermittent hypoxia via down-regulation of miR-203. Biochem. Biophys. Rep. 2017, 11, 130–137. [Google Scholar] [CrossRef]
- Drager, L.F.; Li, J.; Reinke, C.; Bevans-Fonti, S.; Jun, J.C.; Polotsky, V.Y. Intermittent hypoxia exacerbates metabolic effects of diet-induced obesity. Obesity 2011, 19, 2167–2174. [Google Scholar] [CrossRef]
- Uchiyama, T.; Itaya-Hironaka, A.; Yamauchi, A.; Makino, M.; Sakuramoto-Tsuchida, S.; Shobatake, R.; Ota, H.; Takeda, M.; Ohbayashi, C.; Takasawa, S. Intermittent Hypoxia Up-Regulates CCL2, RETN, and TNFalpha mRNAs in Adipocytes via Down-regulation of miR-452. Int. J. Mol. Sci. 2019, 20, 1960. [Google Scholar] [CrossRef]
- Takahashi, K.; Ueda, S.; Kobayashi, T.; Nishiyama, A.; Fujisawa, Y.; Sugaya, T.; Shiota, S.; Gohda, T.; Horikoshi, S.; Suzuki, Y. Chronic intermittent hypoxia-mediated renal sympathetic nerve activation in hypertension and cardiovascular disease. Sci. Rep. 2018, 8, 1–7. [Google Scholar] [CrossRef]
- Clementini, M.; Rossetti, P.H.; Penarrocha, D.; Micarelli, C.; Bonachela, W.C.; Canullo, L. Systemic risk factors for peri-implant bone loss: A systematic review and meta-analysis. Int. J. Oral Maxillofac. Surg. 2014, 43, 323–334. [Google Scholar] [CrossRef]
- Yadav, H.; Quijano, C.; Kamaraju, A.K.; Gavrilova, O.; Malek, R.; Chen, W.; Zerfas, P.; Zhigang, D.; Wright, E.C.; Stuelten, C.; et al. Protection from obesity and diabetes by blockade of TGF-beta/Smad3 signaling. Cell Metab. 2011, 14, 67–79. [Google Scholar] [CrossRef]
- Tan, C.K.; Leuenberger, N.; Tan, M.J.; Yan, Y.W.; Chen, Y.; Kambadur, R.; Wahli, W.; Tan, N.S. Smad3 deficiency in mice protects against insulin resistance and obesity induced by a high-fat diet. Diabetes 2011, 60, 464–476. [Google Scholar] [CrossRef]
- Wu, L.; Derynck, R. Essential role of TGF-beta signaling in glucose-induced cell hypertrophy. Dev. Cell 2009, 17, 35–48. [Google Scholar] [CrossRef] [PubMed]
- Zhao, M.; Mishra, L.; Deng, C.X. The role of TGF-beta/SMAD4 signaling in cancer. Int. J. Biol. Sci. 2018, 14, 111–123. [Google Scholar] [CrossRef] [PubMed]
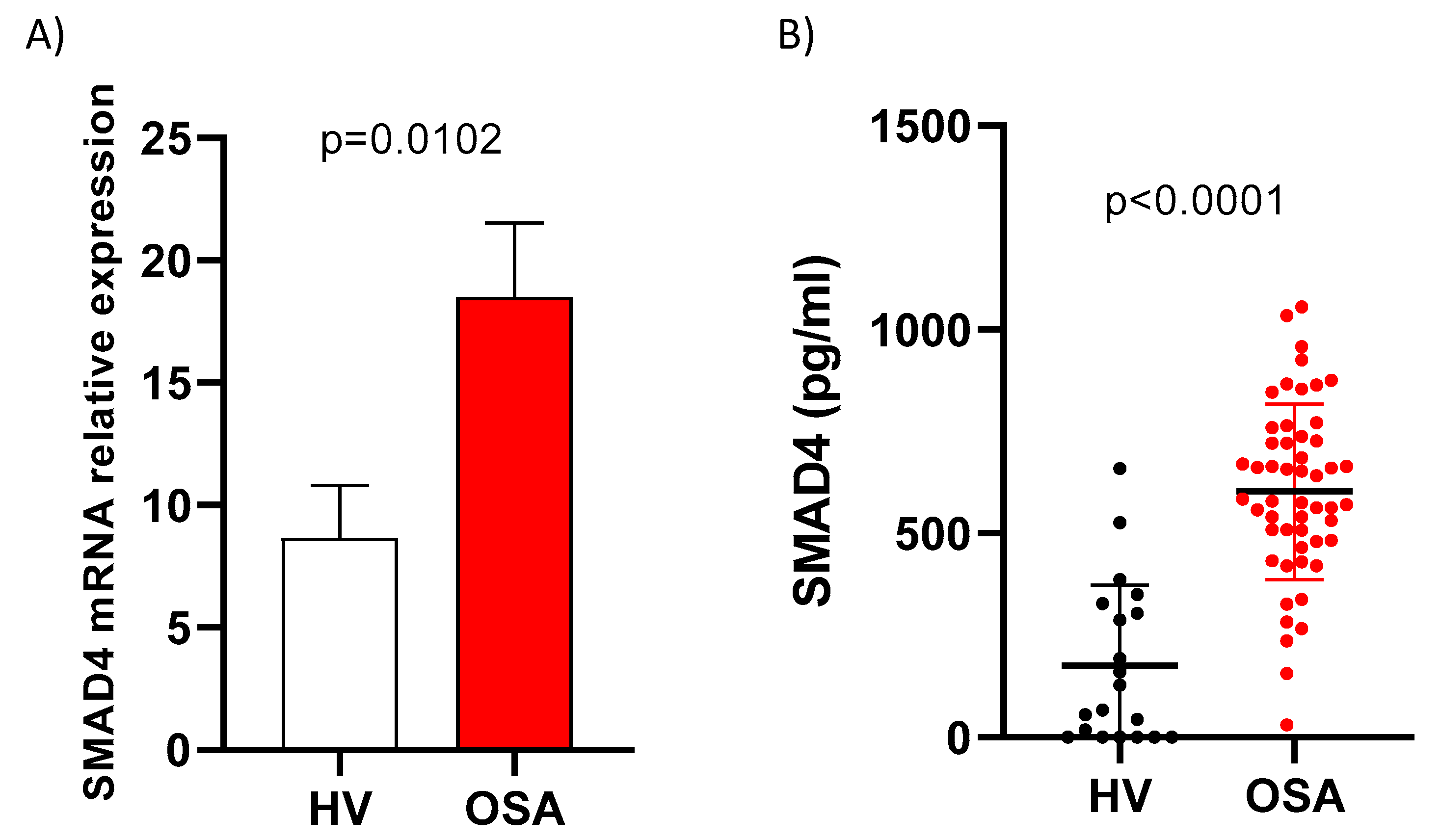
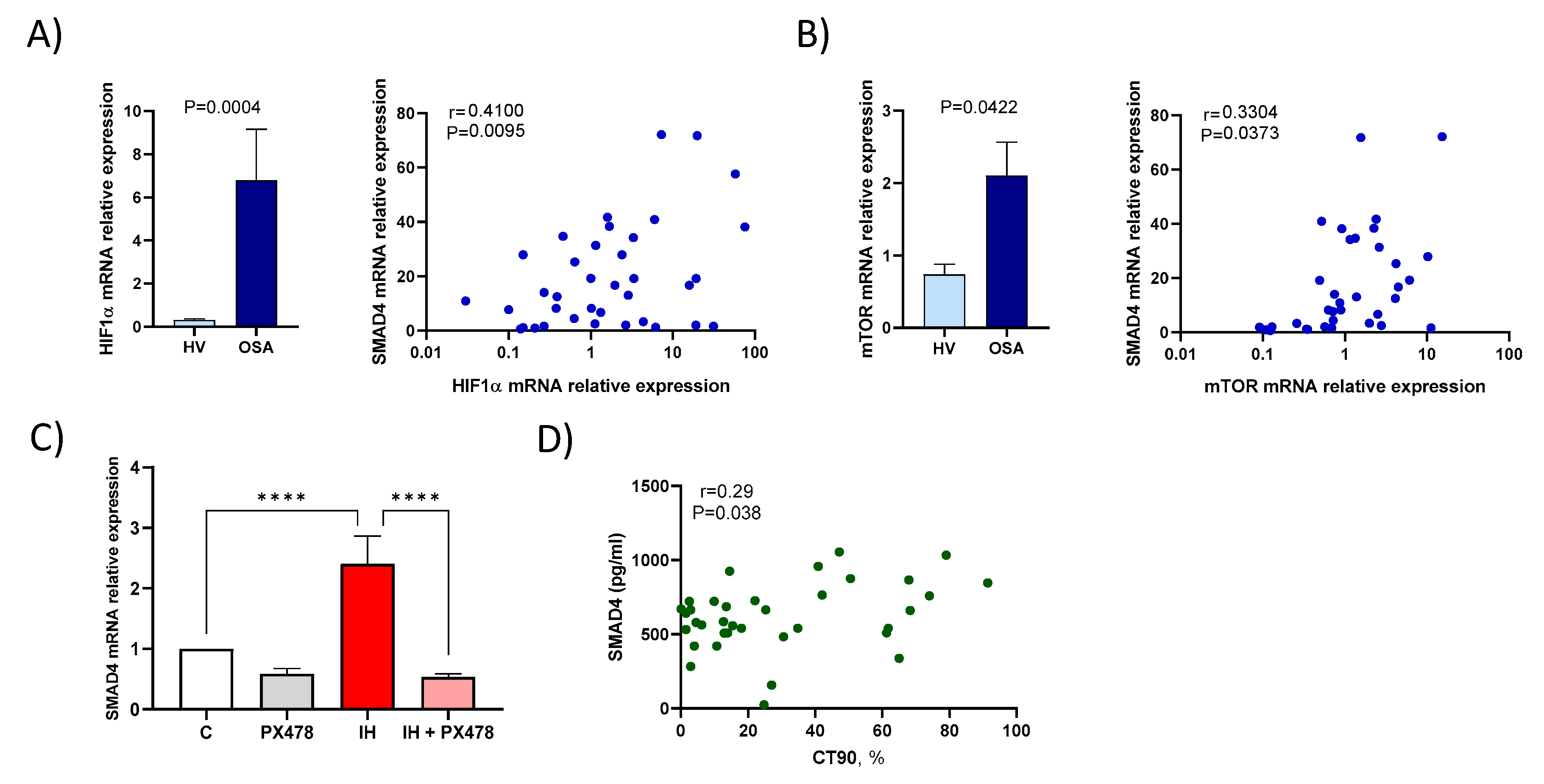
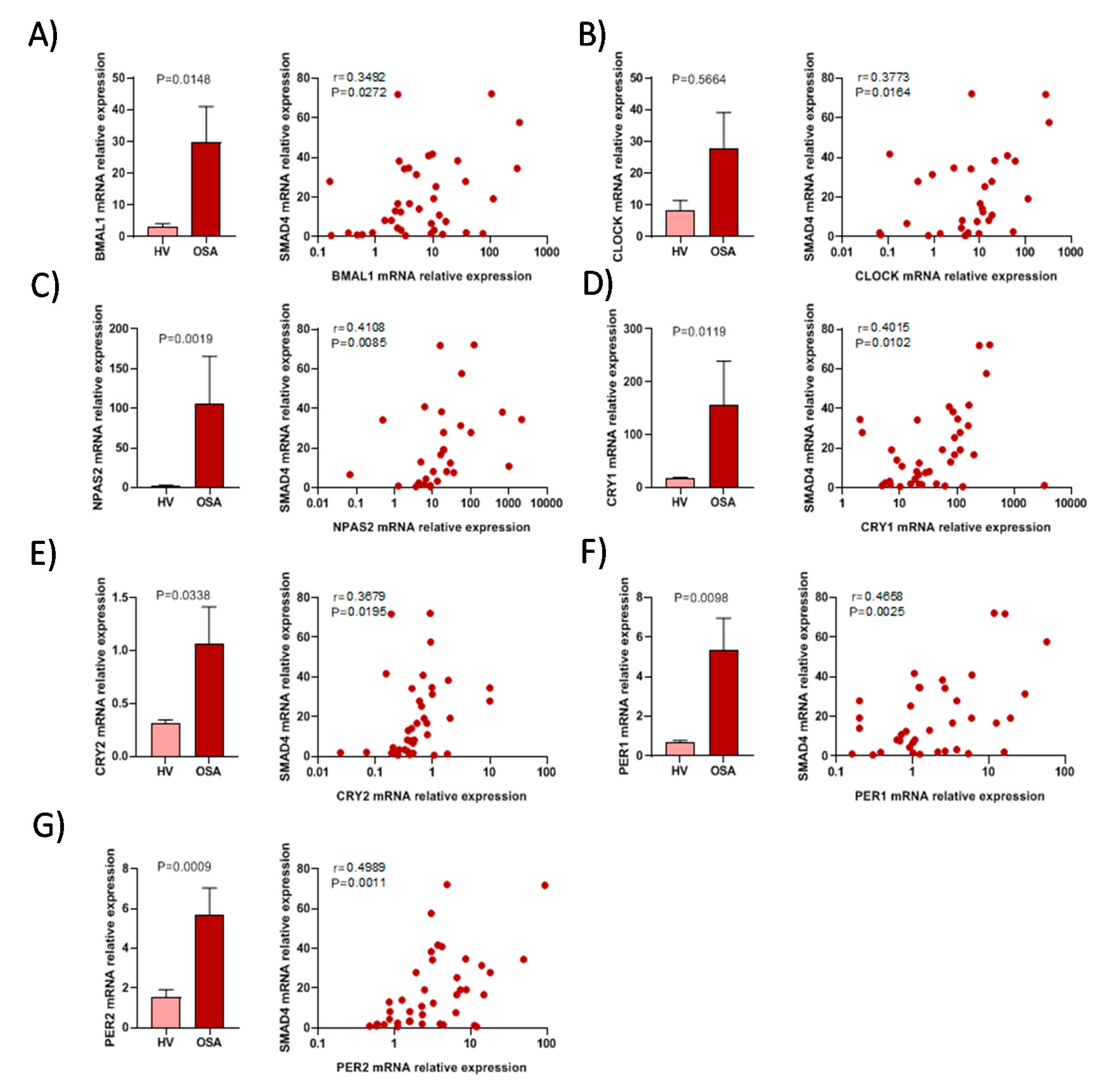
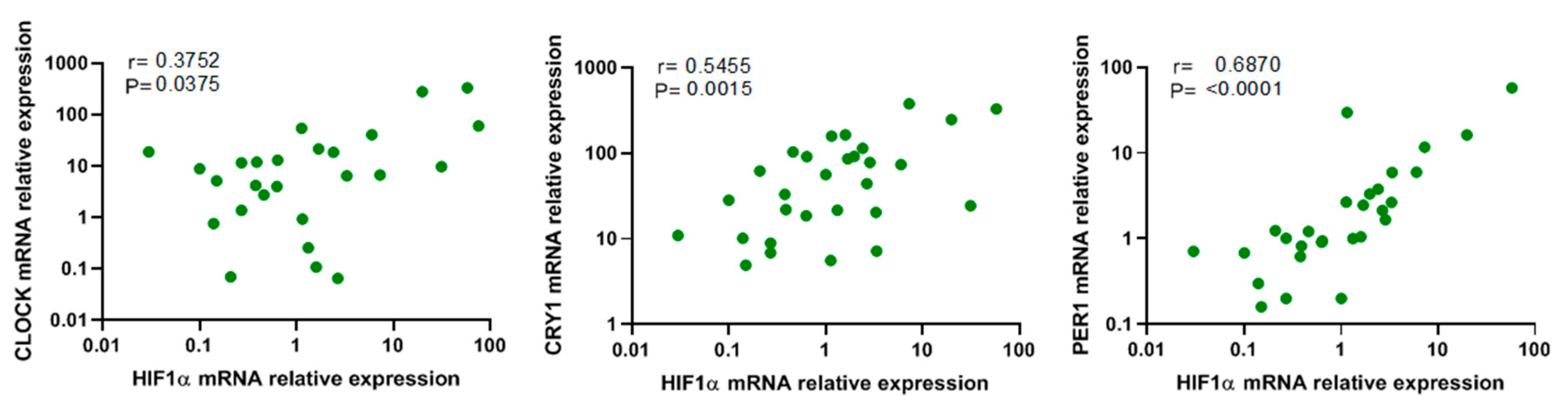
| Variable | Patients with Severe OSA (n = 52) | Healthy Volunteers (n = 26) | p-Value |
|---|---|---|---|
| Male Sex, n (%) | 34 (65) | 15 (58) | 0.462 |
| Age, Years | 61 ± 12 | 59 ± 11 | 0.631 |
| Body Mass Index, kg/m2 | 32.4 ± 5.4 | 29.3 ± 3.2 | 0.105 |
| Smoking Habit | |||
| Current Smoker | 14 (28) | 7 (27) | 0.100 |
| Former Smoker | 18 (35) | 8 (31) | |
| Never Smoker | 20 (37) | 11 (42) | |
| Epworth Sleepiness Scale | 8.5 ± 4.2 | 6.0 ± 2.8 | <0.001 |
| AHI, Events/h | 51.5 ± 16.8 | 2.8 ± 1.2 | <0.001 |
| Oxygen Desaturation Index, Events/h | 45.9 ± 20.4 | 4.2 ± 4.5 | <0.001 |
| Time Recorded with SaO2 <90%, % | 33.9 ± 29.4 | 4.4 ± 3.1 | <0.001 |
| Mean Nocturnal SaO2, % | 91 ± 3 | 93 ± 2 | 0.037 |
| Low Nocturnal SaO2, % | 75 ± 8 | 85 ± 5 | 0.001 |
| Comorbidities | |||
| Hypertension, n (%) | 28 (54) | 0 | <0.001 |
| Dyslipidaemia, n (%) | 29 (56) | 0 | <0.001 |
| Type 2 diabetes, n (%) | 16 (31) | 0 | <0.001 |
| Systolic BP, mmHg | 131 ± 19 | 125 ± 9 | 0.009 |
| Diastolic BP, mmHg | 80 ± 10 | 76 ± 8 | 0.043 |
| Cholesterol, mg/dL | 217 ± 51 | 186 ± 45 | <0.001 |
| HDL-cholesterol, mg/dL | 46 ± 15 | 49 ± 14 | 0.028 |
| LDL-cholesterol, mg/dL | 148 ± 51 | 118 ± 40 | <0.001 |
| Triglycerides, mg/dL | 153 ± 77 | 137 ± 61 | 0.037 |
| Fasting glycaemia, mg/dL | 119 ± 38 | 108 ± 26 | 0.089 |
| Haemoglobin A1c | 6.1 ± 1.1 | 5.4 ± 0.8 | 0.011 |
| Motif | Position | Matrix Score | Core Score | Sequence |
|---|---|---|---|---|
| EBOX | 51020730 (−) | 0.930 | 0.932 | ACACATGG |
| EBOX | 51020732 (−) | 1.000 | 0.987 | ACACATGG |
| EBOX | 51025411 (+) | 1.000 | 0.992 | ACATGTGT |
| EBOX | 51026204 (+) | 1.000 | 0.983 | GCATGTGT |
| EBOX | 51027428 (−) | 0.932 | 0.920 | CCACCTGC |
| HRE | 51027681 (−) | 1.000 | 0.989 | CACGTCC |
| HRE | 51021767 (+) | 1.000 | 0.956 | AACGTGG |
| HRE | 51020718 (+) | 1.000 | 1.000 | CACGTAC |
| HRE | 51023404 (−) | 1.000 | 1.000 | TACGTGA |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Díaz-García, E.; Jaureguizar, A.; Casitas, R.; García-Tovar, S.; Sánchez-Sánchez, B.; Zamarrón, E.; López-Collazo, E.; García-Río, F.; Cubillos-Zapata, C. SMAD4 Overexpression in Patients with Sleep Apnoea May Be Associated with Cardiometabolic Comorbidities. J. Clin. Med. 2020, 9, 2378. https://doi.org/10.3390/jcm9082378
Díaz-García E, Jaureguizar A, Casitas R, García-Tovar S, Sánchez-Sánchez B, Zamarrón E, López-Collazo E, García-Río F, Cubillos-Zapata C. SMAD4 Overexpression in Patients with Sleep Apnoea May Be Associated with Cardiometabolic Comorbidities. Journal of Clinical Medicine. 2020; 9(8):2378. https://doi.org/10.3390/jcm9082378
Chicago/Turabian StyleDíaz-García, Elena, Ana Jaureguizar, Raquel Casitas, Sara García-Tovar, Begoña Sánchez-Sánchez, Ester Zamarrón, Eduardo López-Collazo, Francisco García-Río, and Carolina Cubillos-Zapata. 2020. "SMAD4 Overexpression in Patients with Sleep Apnoea May Be Associated with Cardiometabolic Comorbidities" Journal of Clinical Medicine 9, no. 8: 2378. https://doi.org/10.3390/jcm9082378
APA StyleDíaz-García, E., Jaureguizar, A., Casitas, R., García-Tovar, S., Sánchez-Sánchez, B., Zamarrón, E., López-Collazo, E., García-Río, F., & Cubillos-Zapata, C. (2020). SMAD4 Overexpression in Patients with Sleep Apnoea May Be Associated with Cardiometabolic Comorbidities. Journal of Clinical Medicine, 9(8), 2378. https://doi.org/10.3390/jcm9082378





