Genetic Variants as Sudden-Death Risk Markers in Inherited Arrhythmogenic Syndromes: Personalized Genetic Interpretation
Abstract
1. Introduction
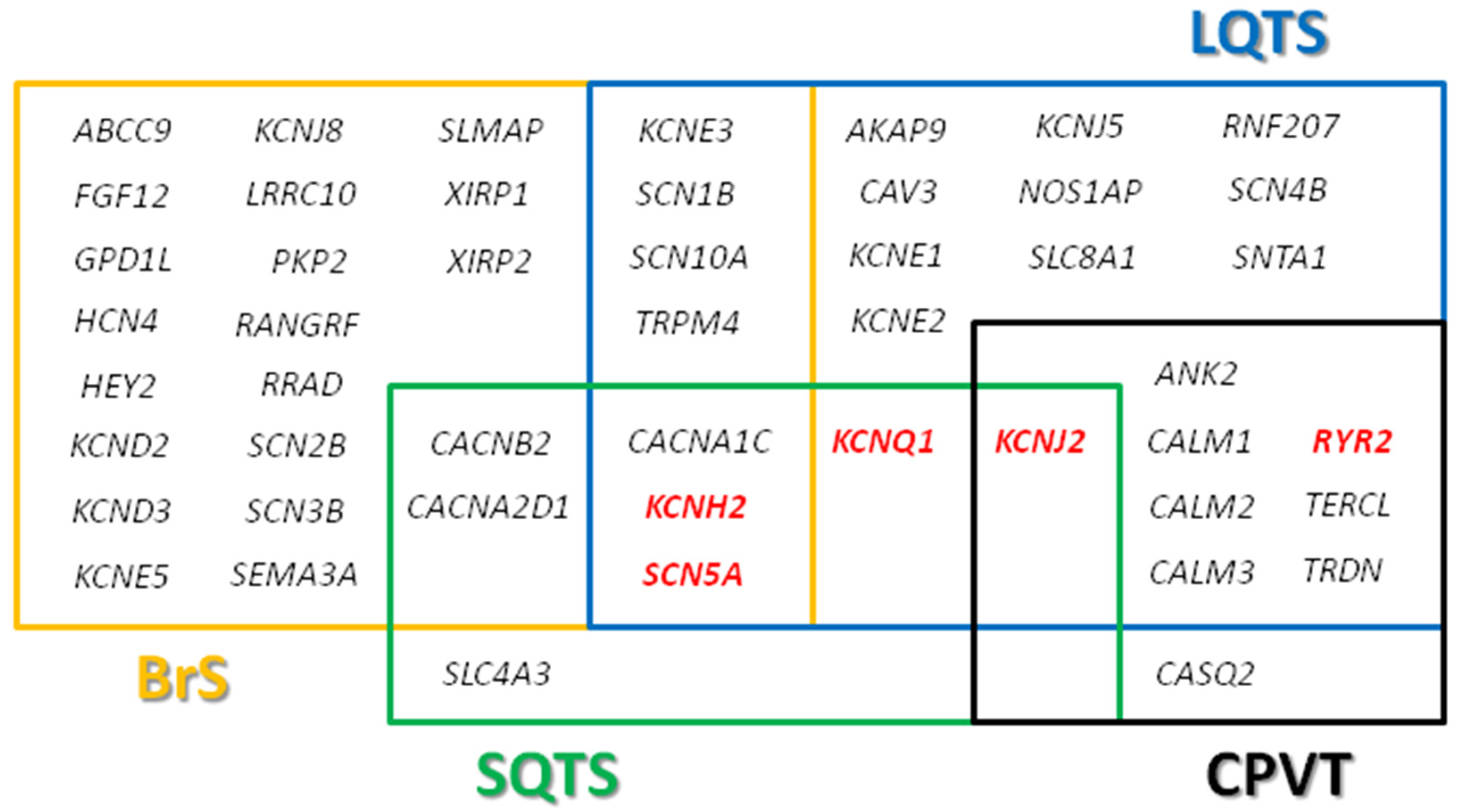
2. Long QT Syndrome
3. Brugada Syndrome
4. Catecholaminergic Polymorphic Ventricular Tachycardia
5. Short QT Syndrome
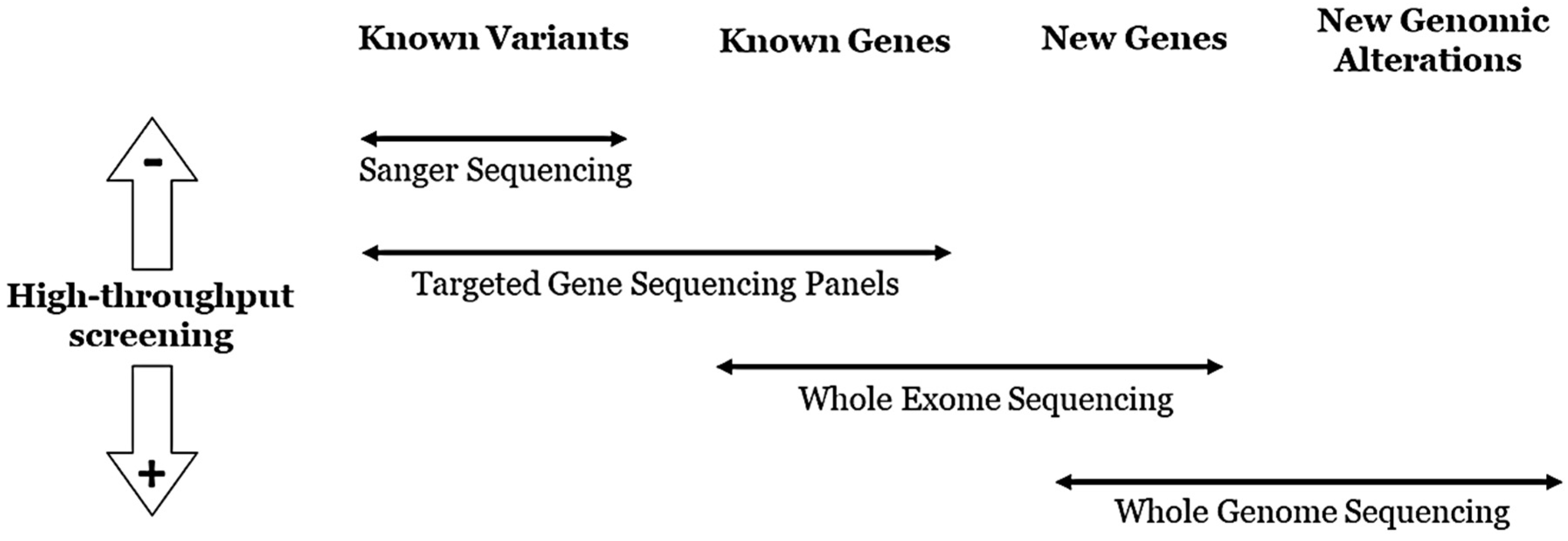
6. Genetic Diagnosis
7. Genetic Translation
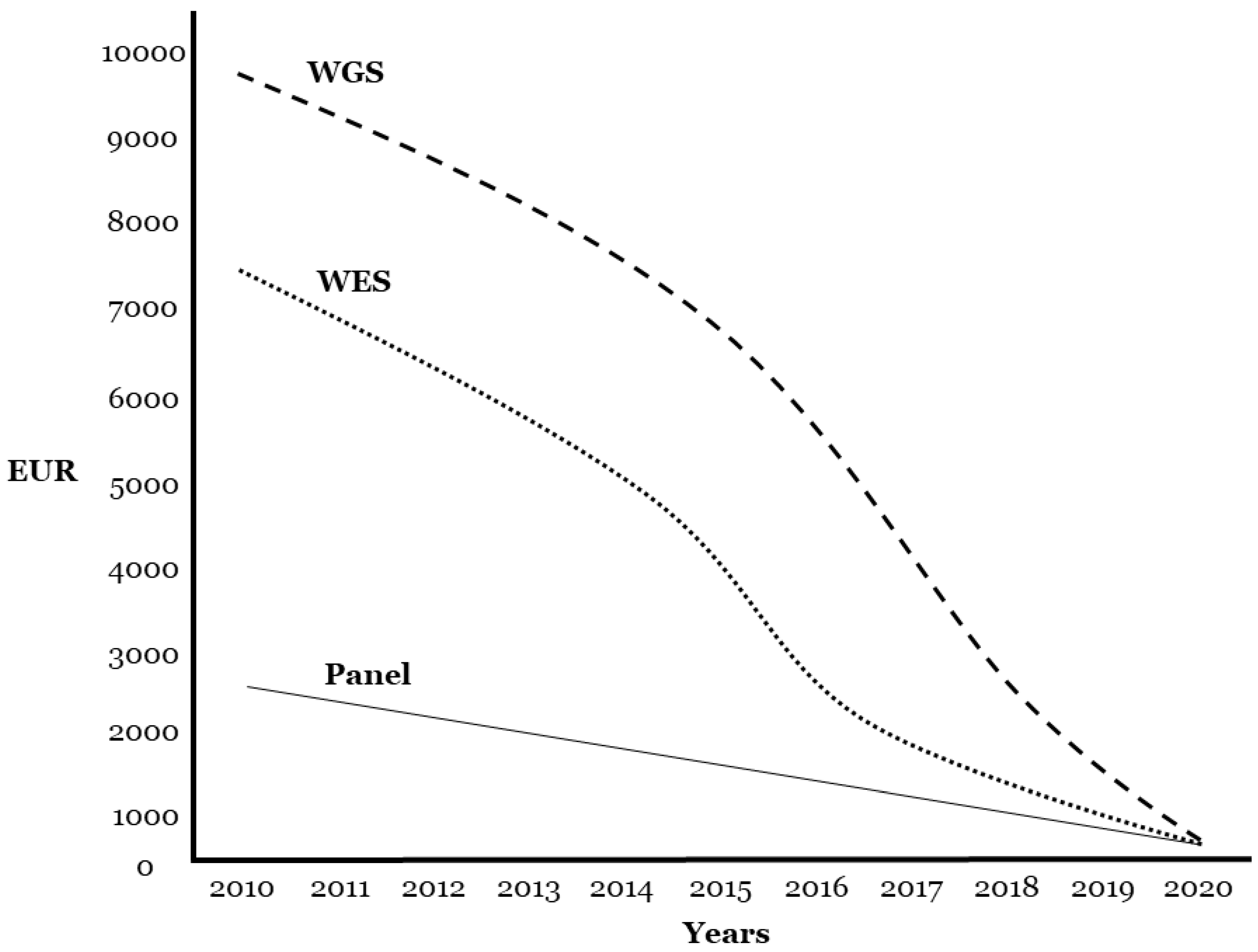
8. Technical Data
9. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Zipes, D.P.; Wellens, H.J. Sudden cardiac death. Circulation 1998, 98, 2334–2351. [Google Scholar] [CrossRef] [PubMed]
- Bagnall, R.D.; Weintraub, R.G.; Ingles, J.; Duflou, J.; Yeates, L.; Lam, L.; Davis, A.M.; Thompson, T.; Connell, V.; Wallace, J.; et al. A Prospective Study of Sudden Cardiac Death among Children and Young Adults. N. Engl. J. Med. 2016, 374, 2441–2452. [Google Scholar] [CrossRef] [PubMed]
- Bezzina, C.R.; Lahrouchi, N.; Silvia, P.G. Genetics of Sudden Cardiac Death. Circ. Res. 2015, 116, 1919–1936. [Google Scholar] [CrossRef] [PubMed]
- Singh, M.; Morin, D.P.; Link, M.S. Sudden cardiac death in Long QT syndrome (LQTS), Brugada syndrome, and catecholaminergic polymorphic ventricular tachycardia (CPVT). Prog. Cardiovasc. Dis. 2019, 62, 227–234. [Google Scholar] [CrossRef]
- Coll, M.; Perez-Serra, A.; Mates, J.; Del Olmo, B.; Puigmulé, M.; Fernandez-Falgueras, A.; Iglesias, A.; Pico, F.; Lopez, L.; Brugada, R.; et al. Incomplete Penetrance and Variable Expressivity: Hallmarks in Channelopathies Associated with Sudden Cardiac Death. Biology 2017, 7, 3. [Google Scholar] [CrossRef]
- Christensen, N.L.; Carter-Storch, R.; Bakkestrøm, R.; Dahl, J.S. Sudden cardiac death in asymptomatic aortic stenosis: Is the valve to blame? BMJ Case Rep. 2016, 2016. [Google Scholar] [CrossRef]
- Neubauer, J.; Lecca, M.R.; Russo, G.; Bartsch, C.; Medeiros-Domingo, A.; Berger, W.; Haas, C. Post-mortem whole-exome analysis in a large sudden infant death syndrome cohort with a focus on cardiovascular and metabolic genetic diseases. Eur. J. Hum. Genet. 2017, 25, 404–409. [Google Scholar] [CrossRef]
- Hershberger, R.E.; Givertz, M.M.; Ho, C.Y.; Judge, D.P.; Kantor, P.; McBride, K.L.; Morales, A.; Taylor, M.R.; Vatta, M.; Ware, S.M. Genetic Evaluation of Cardiomyopathy—A Heart Failure Society of America Practice Guideline. J. Card. Fail. 2018, 24, 281–302. [Google Scholar] [CrossRef]
- Hershberger, R.E.; Givertz, M.M.; Ho, C.Y.; Judge, D.P.; Kantor, P.F.; McBride, K.L.; Morales, A.; Taylor, M.R.G.; Vatta, M.; Ware, S.M.; et al. Genetic evaluation of cardiomyopathy: A clinical practice resource of the American College of Medical Genetics and Genomics (ACMG). Genet. Med. 2018, 20, 899–909. [Google Scholar] [CrossRef]
- Priori, S.G.; Blomström-Lundqvist, C. CardioPulse Articles2015 European Society of Cardiology Guidelines for the management of patients with ventricular arrhythmias and the prevention of sudden cardiac death summarized by co-chairs ‘Ten Commandments’ of 2015 European Society of Cardiology Guidelines for the management of patients with ventricular arrhythmias and the prevention of sudden cardiac deathAdolfo J. de Bold PhD OC FRSC: A pioneer in cardiovascular medicineNatriuretic peptides in 2015Teachable moment or missed opportunity? Eur. Hear. J. 2015, 36, 2757–2762. [Google Scholar] [CrossRef]
- Adler, A.; Novelli, V.; Amin, A.S.; Abiusi, E.; Care, M.; Nannenberg, E.A.; Feilotter, H.; Amenta, S.; Mazza, D.; Bikker, H.; et al. An International, Multicentered, Evidence-Based Reappraisal of Genes Reported to Cause Congenital Long QT Syndrome. Circulation 2020, 141, 418–428. [Google Scholar] [CrossRef] [PubMed]
- Giudicessi, J.R.; Roden, D.M.; Wilde, A.A.; Ackerman, M.J. Classification and Reporting of Potentially Proarrhythmic Common Genetic Variation in Long QT Syndrome Genetic Testing. Circulation 2018, 137, 619–630. [Google Scholar] [CrossRef]
- Janin, A.; Bessière, F.; Georgescu, T.; Chanavat, V.; Chevalier, P.; Millat, G. TRPM4 mutations to cause autosomal recessive and not autosomal dominant Brugada type 1 syndrome. Eur. J. Med. Genet. 2019, 62, 103527. [Google Scholar] [CrossRef] [PubMed]
- David, J.P.; Lisewski, U.; Crump, S.M.; Jepps, T.A.; Bocksteins, E.; Wilck, N.; Abbott, G.W. Deletion in mice of X-linked, Brugada syndrome- and atrial fibrillation-associated Kcne5 augments ventricular KV currents and predisposes to ventricular arrhythmia. FASEB J. 2019, 33, 2537–2552. [Google Scholar] [CrossRef] [PubMed]
- Hosseini, S.M.; Kim, R.; Udupa, S.; Costain, G.; Jobling, R.; Liston, E.; Jamal, S.M.; Szybowska, M.; Morel, C.F.; Bowdin, S.; et al. Reappraisal of Reported Genes for Sudden Arrhythmic Death. Circulation 2018, 138, 1195–1205. [Google Scholar] [CrossRef] [PubMed]
- Denham, N.; Pearman, C.M.; Ding, W.Y.; Waktare, J.; Gupta, D.; Snowdon, R.; Hall, M.; Cooper, R.; Modi, S.; Todd, D.; et al. Systematic re-evaluation of SCN5A variants associated with Brugada syndrome. J. Cardiovasc. Electrophysiol. 2018, 30, 118–127. [Google Scholar] [CrossRef]
- Campuzano, O.; Sarquella-Brugada, G.; Fernandez-Falgueras, A.; Cesar, S.; Coll, M.; Mates, J.; Arbelo, E.; Perez-Serra, A.; Del Olmo, B.; Jordá, P.; et al. Genetic interpretation and clinical translation of minor genes related to Brugada syndrome. Hum. Mutat. 2019, 40, 749–764. [Google Scholar] [CrossRef]
- Kim, C.W.; Aronow, W.S.; Dutta, T.; Frenkel, D.; Frishman, W.H. Catecholaminergic Polymorphic Ventricular Tachycardia. Cardiol. Rev. 2020. [Google Scholar] [CrossRef]
- Refaat, M.M.; Hotait, M.; Tseng, Z.H. Utility of the Exercise Electrocardiogram Testing in Sudden Cardiac Death Risk Stratification. Ann. Noninvasive Electrocardiol. 2014, 19, 311–318. [Google Scholar] [CrossRef]
- Wleklinski, M.J.; Kannankeril, P.J.; Knollmann, B.C. Molecular and tissue mechanisms of catecholaminergic polymorphic ventricular tachycardia. J. Physiol. 2020. [Google Scholar] [CrossRef]
- Crotti, L.; Spazzolini, C.; Tester, D.J.; Ghidoni, A.; Baruteau, A.-E.; Beckmann, B.-M.; Behr, E.R.; Bennett, J.; Bezzina, C.R.; A Bhuiyan, Z.; et al. Calmodulin mutations and life-threatening cardiac arrhythmias: Insights from the International Calmodulinopathy Registry. Eur. Hear. J. 2019, 40, 2964–2975. [Google Scholar] [CrossRef] [PubMed]
- Campuzano, O.; Sarquella-Brugada, G.; Cesar, S.; Arbelo, E.; Brugada, J.; Brugada, R. Recent Advances in Short QT Syndrome. Front. Cardiovasc. Med. 2018, 5, 149. [Google Scholar] [CrossRef] [PubMed]
- Campuzano, O.; Fernandez-Falgueras, A.; Maulen, X.L.; Sarquella-Brugada, G.; César, S.; Coll, M.; Mates, J.; Arbelo, E.; Jordà, P.; Perez-Serra, A.; et al. Short QT Syndrome: A Comprehensive Genetic Interpretation and Clinical Translation of Rare Variants. J. Clin. Med. 2019, 8, 1035. [Google Scholar] [CrossRef] [PubMed]
- Liu, R.; Zhang, P. Towards early detection of adverse drug reactions: Combining pre-clinical drug structures and post-market safety reports. BMC Med. Inform. Decis. Mak. 2019, 19, 1–9. [Google Scholar] [CrossRef]
- Schwarze, K.; Buchanan, J.; Taylor, J.C.; Wordsworth, S. Are whole-exome and whole-genome sequencing approaches cost-effective? A systematic review of the literature. Genet. Med. 2018, 20, 1122–1130. [Google Scholar] [CrossRef] [PubMed]
- Kalia, S.S.; on behalf of the ACMG Secondary Findings Maintenance Working Group; Adelman, K.; Bale, S.J.; Chung, W.K.; Eng, C.; Evans, J.P.; Herman, G.E.; Hufnagel, S.B.; Klein, T.E.; et al. Recommendations for reporting of secondary findings in clinical exome and genome sequencing, 2016 update (ACMG SF v2.0): A policy statement of the American College of Medical Genetics and Genomics. Genet. Med. 2016, 19, 249–255. [Google Scholar] [CrossRef]
- Dillon, O.J.; Melbourne Genomics Health Alliance; Lunke, S.; Stark, Z.; Yeung, A.; Thorne, N.; Gaff, C.; White, S.M.; Tan, T.Y. Exome sequencing has higher diagnostic yield compared to simulated disease-specific panels in children with suspected monogenic disorders. Eur. J. Hum. Genet. 2018, 26, 644–651. [Google Scholar] [CrossRef]
- Marschall, C.; Moscu-Gregor, A.; Klein, H.-G. Variant panorama in 1385 index patients and sensitivity of expanded next-generation sequencing panels in arrhythmogenic disorders. Cardiovasc. Diagn. Ther. 2019, 9, S292–S298. [Google Scholar] [CrossRef]
- Richards, S.; Aziz, N.; Bale, S.; Bick, D.; Das, S.; Gastier-Foster, J.; Voelkerding, K. Faculty Opinions recommendation of Standards and guidelines for the interpretation of sequence variants: A joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Fac. Opin. 2018, 17, 405–424. [Google Scholar]
- Bennett, J.S.; Bernhardt, M.; McBride, K.L.; Reshmi, S.C.; Zmuda, E.; Kertesz, N.J.; Garg, V.; Fitzgerald-Butt, S.; Kamp, A.N. Reclassification of Variants of Uncertain Significance in Children with Inherited Arrhythmia Syndromes is Predicted by Clinical Factors. Pediatr. Cardiol. 2019, 40, 1679–1687. [Google Scholar] [CrossRef]
- Campuzano, O.; Sarquella-Brugada, G.; Fernandez-Falgueras, A.; Coll, M.; Iglesias, A.; Ferrer-Costa, C.; Cesar, S.; Arbelo, E.; García-Álvarez, A.; Jordà, P.; et al. Reanalysis and reclassification of rare genetic variants associated with inherited arrhythmogenic syndromes. EBioMedicine 2020, 54, 102732. [Google Scholar] [CrossRef] [PubMed]
- Gando, I.; Yang, H.-Q.; Coetzee, W.A. Functional significance of channelopathy gene variants in unexplained death. Forensic Sci. Med. Pathol. 2018, 15, 437–444. [Google Scholar] [CrossRef] [PubMed]
- Riuró, H.; Campuzano, O.; Berne, P.; Arbelo, E.; Iglesias, A.; Pérez-Serra, A.; Coll-Vidal, M.; Partemi, S.; Mademont-Soler, I.; Picó, F.; et al. Genetic analysis, in silico prediction, and family segregation in long QT syndrome. Eur. J. Hum. Genet. 2014, 23, 79–85. [Google Scholar] [CrossRef] [PubMed]
- Olubando, D.; Hopton, C.; Eden, J.; Caswell, R.; Thomas, N.L.; Roberts, S.A.; Morris-Rosendahl, D.; Venetucci, L.; Newman, W. Classification and correlation of RYR2 missense variants in individuals with catecholaminergic polymorphic ventricular tachycardia reveals phenotypic relationships. J. Hum. Genet. 2020, 65, 531–539. [Google Scholar] [CrossRef]
- Jenewein, T.; Neumann, T.; Erkapic, D.; Kuniss, M.; Verhoff, M.A.; Thiel, G.; Kauferstein, S. Influence of genetic modifiers on sudden cardiac death cases. Int. J. Leg. Med. 2017, 132, 379–385. [Google Scholar] [CrossRef] [PubMed]
- Cerrone, M.; Remme, C.A.; Tadros, R.; Bezzina, C.R.; Delmar, M. Beyond the One Gene-One Disease Paradigm: Complex Genetics and Pleiotropy in Inheritable Cardiac Disorders. Circulation 2019, 140, 595–610. [Google Scholar] [CrossRef] [PubMed]
- Behr, E.R.; Savio-Galimberti, E.; Barc, J.; Holst, A.G.; Petropoulou, E.; Prins, B.; Jabbari, J.; Torchio, M.; Berthet, M.; Mizusawa, Y.; et al. Role of common and rare variants in SCN10A: Results from the Brugada syndrome QRS locus gene discovery collaborative study. Cardiovasc. Res. 2015, 106, 520–529. [Google Scholar] [CrossRef]
- Walsh, I.M.; Bowman, M.A.; Santarriaga, I.F.S.; Rodriguez, A.; Clark, P.L. Synonymous codon substitutions perturb cotranslational protein folding in vivo and impair cell fitness. Proc. Natl. Acad. Sci. USA 2020, 117, 3528–3534. [Google Scholar] [CrossRef]
- Malik, M. Drug-Induced QT/QTc Interval Shortening: Lessons from Drug-Induced QT/QTc Prolongation. Drug Saf. 2016, 39, 647–659. [Google Scholar] [CrossRef]
- Daimi, H.; Khelil, A.H.; Neji, A.; Ben Hamda, K.; Maaoui, S.; Aranega, A.; Chibani, J.B.; Franco, D. Role of SCN5A coding and non-coding sequences in Brugada syndrome onset: What’s behind the scenes? Biomed. J. 2019, 42, 252–260. [Google Scholar] [CrossRef]
- Hartman, P.; Beckman, K.; Silverstein, K.; Yohe, S.; Schomaker, M.; Henzler, C.; Onsongo, G.; Lam, H.C.; Munro, S.; Daniel, J.; et al. Next generation sequencing for clinical diagnostics: Five year experience of an academic laboratory. Mol. Genet. Metab. Rep. 2019, 19, 100464. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.; Gryazeva, T.; Wang, D.; Zhou, B.; Um, S.Y.; Eng, L.S.; Ruiter, K.; Rojas, L.; Williams, N.; Sampson, B.A.; et al. Using postmortem formalin fixed paraffin-embedded tissues for molecular testing of sudden cardiac death: A cautionary tale of utility and limitations. Forensic Sci. Int. 2020, 308, 110177. [Google Scholar] [CrossRef] [PubMed]
- Mates, J.; Mademont-Soler, I.; Fernandez-Falgueras, A.; Sarquella-Brugada, G.; Cesar, S.; Arbelo, E.; Fiol, V. Sudden Cardiac Death and Copy Number Variants: What Do We Know after 10 Years of Genetic Analysis? Forensic Sci. Int. Genet. 2020, 47, 102281. [Google Scholar] [CrossRef] [PubMed]
- Mademont-Soler, I.; Pinsach-Abuin, M.; Riuró, H.; Matés, J.; Pérez-Serra, A.; Coll, M.; Porres, J.M.; Del Olmo, B.; Iglesias, A.; Selga, E.; et al. Large Genomic Imbalances in Brugada Syndrome. PLoS ONE 2016, 11, e0163514. [Google Scholar] [CrossRef] [PubMed]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Campuzano, O.; Sarquella-Brugada, G.; Arbelo, E.; Cesar, S.; Jordà, P.; Pérez-Serra, A.; Toro, R.; Brugada, J.; Brugada, R. Genetic Variants as Sudden-Death Risk Markers in Inherited Arrhythmogenic Syndromes: Personalized Genetic Interpretation. J. Clin. Med. 2020, 9, 1866. https://doi.org/10.3390/jcm9061866
Campuzano O, Sarquella-Brugada G, Arbelo E, Cesar S, Jordà P, Pérez-Serra A, Toro R, Brugada J, Brugada R. Genetic Variants as Sudden-Death Risk Markers in Inherited Arrhythmogenic Syndromes: Personalized Genetic Interpretation. Journal of Clinical Medicine. 2020; 9(6):1866. https://doi.org/10.3390/jcm9061866
Chicago/Turabian StyleCampuzano, Oscar, Georgia Sarquella-Brugada, Elena Arbelo, Sergi Cesar, Paloma Jordà, Alexandra Pérez-Serra, Rocío Toro, Josep Brugada, and Ramon Brugada. 2020. "Genetic Variants as Sudden-Death Risk Markers in Inherited Arrhythmogenic Syndromes: Personalized Genetic Interpretation" Journal of Clinical Medicine 9, no. 6: 1866. https://doi.org/10.3390/jcm9061866
APA StyleCampuzano, O., Sarquella-Brugada, G., Arbelo, E., Cesar, S., Jordà, P., Pérez-Serra, A., Toro, R., Brugada, J., & Brugada, R. (2020). Genetic Variants as Sudden-Death Risk Markers in Inherited Arrhythmogenic Syndromes: Personalized Genetic Interpretation. Journal of Clinical Medicine, 9(6), 1866. https://doi.org/10.3390/jcm9061866






