Identification of a miRNA Based-Signature Associated with Acute Coronary Syndrome: Evidence from the FLORINF Study
Abstract
:1. Introduction
2. Experimental Section
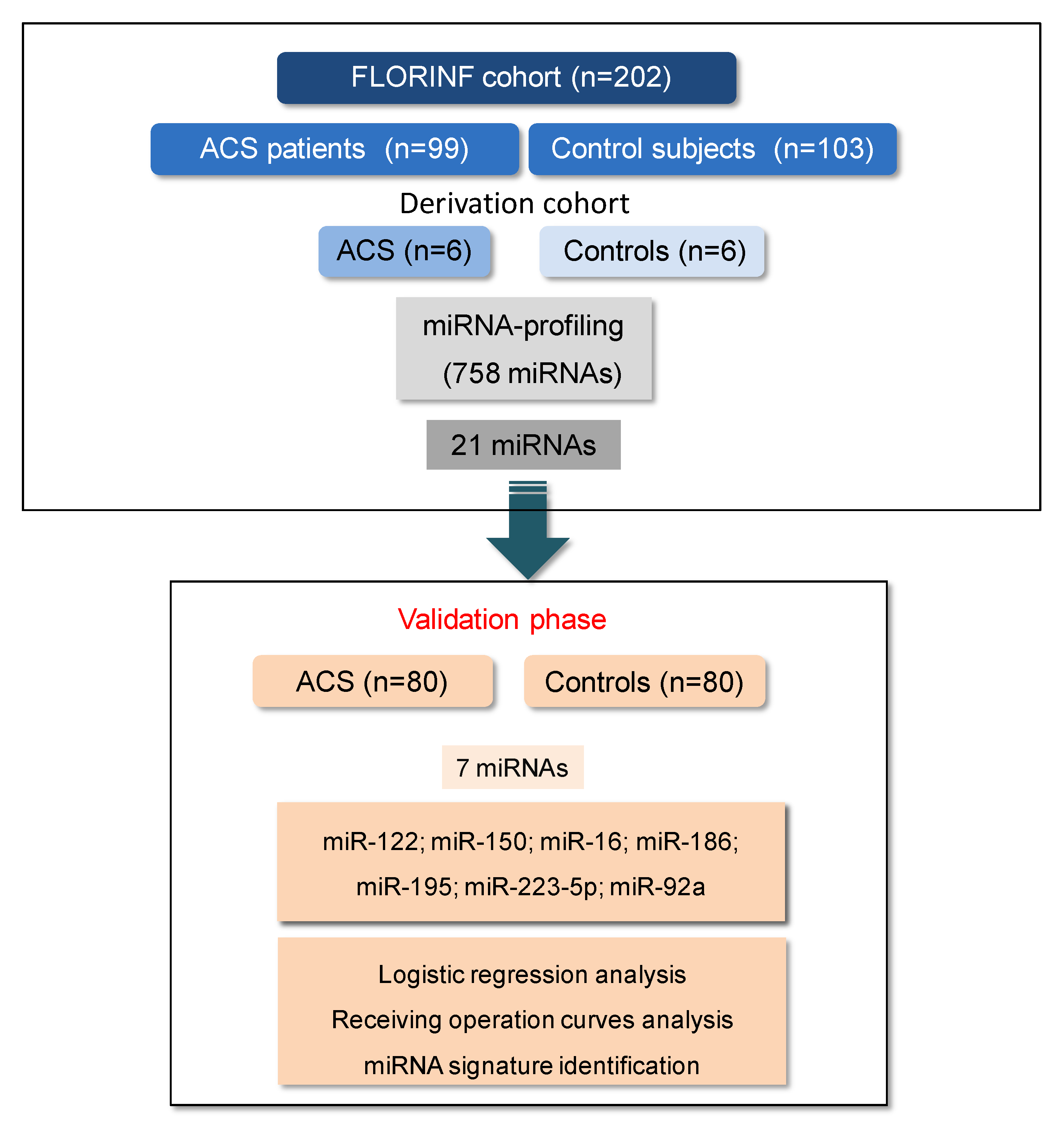
2.1. Study Population
2.2. Total RNA Isolation and Quality Control
2.3. MiRNA Expression Profiling
2.4. MiRNA Validation Phase
2.5. Statistical Analysis
3. Results
3.1. Screening of MiRNAs in the Derivation Cohort and Validation of Candidate miRNAs
3.2. Multivariate Analysis and Improved Predictive Value of the Clinical Model by Addition of the Selected miRNAs
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| ACS | Acute coronary syndrome |
| AMI | Acute myocardial infarction |
| AUC | Area under the curve |
| CAD | Coronary artery disease |
| CVR | Cardiovascular risk |
| MiRNA | Micro RNA |
| RT-qPCR | Reverse transcription-quantitative polymerase chain reaction |
| ROC | Receiver operating characteristic |
| SCORE | Systematic Coronary Risk Evaluation |
References
- Timmis, A.; Townsend, N.; Gale, C.; Grobbee, R.; Maniadakis, N.; Flather, M.; Wilkins, E.; Wright, L.; Vos, R.; Bax, J.; et al. European Society of Cardiology: Cardiovascular Disease Statistics 2017. Eur. Heart J. 2018, 39, 508–579. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bartel, D.P. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 2004, 116, 281–297. [Google Scholar] [CrossRef] [Green Version]
- Welten, S.M.; Goossens, E.A.; Quax, P.H.; Nossent, A.Y. The multifactorial nature of microRNAs in vascular remodelling. Cardiovasc. Res. 2016, 110, 6–22. [Google Scholar] [CrossRef]
- Thum, T.; Condorelli, G. Long noncoding RNAs and microRNAs in cardiovascular pathophysiology. Circ. Res. 2015, 116, 751–762. [Google Scholar] [CrossRef]
- Creemers, E.E.; Tijsen, A.J.; Pinto, Y.M. Circulating microRNAs: Novel biomarkers and extracellular communicators in cardiovascular disease? Circ. Res. 2012, 110, 483–495. [Google Scholar] [CrossRef]
- Boon, R.A.; Vickers, K.C. Intercellular transport of microRNAs. Arterioscler Thromb. Vasc. Biol. 2013, 33, 186–192. [Google Scholar] [CrossRef] [Green Version]
- Gupta, S.K.; Bang, C.; Thum, T. Circulating microRNAs as biomarkers and potential paracrine mediators of cardiovascular disease. Circ. Cardiovasc. Genet. 2010, 3, 484–488. [Google Scholar] [CrossRef] [Green Version]
- Faccini, J.; Ruidavets, J.B.; Cordelier, P.; Martins, F.; Maoret, J.J.; Bongard, V.; Ferrieres, J.; Roncalli, J.; Elbaz, M.; Vindis, C. Circulating miR-155, miR-145 and let-7c as diagnostic biomarkers of the coronary artery disease. Sci. Rep. 2017, 7, 42916. [Google Scholar] [CrossRef] [Green Version]
- Knuuti, J.; Wijns, W.; Saraste, A.; Capodanno, D.; Barbato, E.; Funck-Brentano, C.; Prescott, E.; Storey, R.F.; Deaton, C.; Cuisset, T.; et al. 2019 ESC Guidelines for the diagnosis and management of chronic coronary syndromes. Eur. Heart J. 2020, 41, 407–477. [Google Scholar] [CrossRef]
- Amar, J.; Lelouvier, B.; Servant, F.; Lluch, J.; Burcelin, R.; Bongard, V.; Elbaz, M. Blood Microbiota Modification After Myocardial Infarction Depends Upon Low-Density Lipoprotein Cholesterol Levels. J. Am. Heart Assoc. 2019, 8, e011797. [Google Scholar] [CrossRef]
- Buscail, L.; Bournet, B.; Vernejoul, F.; Cambois, G.; Lulka, H.; Hanoun, N.; Dufresne, M.; Meulle, A.; Vignolle-Vidoni, A.; Ligat, L.; et al. First-in-man phase 1 clinical trial of gene therapy for advanced pancreatic cancer: Safety, biodistribution, and preliminary clinical findings. Mol. Ther.: J. Am. Soc. Gene Ther. 2015, 23, 779–789. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fichtlscherer, S.; De Rosa, S.; Fox, H.; Schwietz, T.; Fischer, A.; Liebetrau, C.; Weber, M.; Hamm, C.W.; Roxe, T.; Muller-Ardogan, M.; et al. Circulating microRNAs in patients with coronary artery disease. Circ. Res. 2010, 107, 677–684. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hellemans, J.; Mortier, G.; De Paepe, A.; Speleman, F.; Vandesompele, J. qBase relative quantification framework and software for management and automated analysis of real-time quantitative PCR data. Genome Biol. 2007, 8, R19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Navickas, R.; Gal, D.; Laucevicius, A.; Taparauskaite, A.; Zdanyte, M.; Holvoet, P. Identifying circulating microRNAs as biomarkers of cardiovascular disease: A systematic review. Cardiovasc. Res. 2016, 111, 322–337. [Google Scholar] [CrossRef]
- Wang, S.S.; Wu, L.J.; Li, J.J.; Xiao, H.B.; He, Y.; Yan, Y.X. A meta-analysis of dysregulated miRNAs in coronary heart disease. Life Sci. 2018, 215, 170–181. [Google Scholar] [CrossRef]
- Colpaert, R.M.W.; Calore, M. MicroRNAs in Cardiac Diseases. Cells 2019, 8, 737. [Google Scholar] [CrossRef] [Green Version]
- Gao, W.; He, H.W.; Wang, Z.M.; Zhao, H.; Lian, X.Q.; Wang, Y.S.; Zhu, J.; Yan, J.J.; Zhang, D.G.; Yang, Z.J.; et al. Plasma levels of lipometabolism-related miR-122 and miR-370 are increased in patients with hyperlipidemia and associated with coronary artery disease. Lipids Health Dis. 2012, 11, 55. [Google Scholar] [CrossRef] [Green Version]
- Bostjancic, E.; Zidar, N.; Glavac, D. MicroRNAs and cardiac sarcoplasmic reticulum calcium ATPase-2 in human myocardial infarction: Expression and bioinformatic analysis. BMC Genom. 2012, 13, 552. [Google Scholar] [CrossRef] [Green Version]
- Cortez-Dias, N.; Costa, M.C.; Carrilho-Ferreira, P.; Silva, D.; Jorge, C.; Calisto, C.; Pessoa, T.; Robalo Martins, S.; de Sousa, J.C.; da Silva, P.C.; et al. Circulating miR-122-5p/miR-133b Ratio Is a Specific Early Prognostic Biomarker in Acute Myocardial Infarction. Circ. J.: Off. J. Jpn. Circ. Soc. 2016, 80, 2183–2191. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Chang, W.; Zhang, Y.; Zhang, L.; Ding, H.; Qi, H.; Xue, S.; Yu, H.; Hu, L.; Liu, D.; et al. Circulating miR-22-5p and miR-122-5p are promising novel biomarkers for diagnosis of acute myocardial infarction. J. Cell. Physiol. 2019, 234, 4778–4786. [Google Scholar] [CrossRef]
- Huang, X.; Huang, F.; Yang, D.; Dong, F.; Shi, X.; Wang, H.; Zhou, X.; Wang, S.; Dai, S. Expression of microRNA-122 contributes to apoptosis in H9C2 myocytes. J. Cell. Mol. Med. 2012, 16, 2637–2646. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Li, H.; Chen, S.; Li, Y.; Cui, Z.; Ma, J. Knockdown of MicroRNA-122 Protects H9c2 Cardiomyocytes from Hypoxia-Induced Apoptosis and Promotes Autophagy. Med. Sci. Monit. Int. Medical J. Exp. Clin. Res. 2017, 23, 4284–4290. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Devaux, Y.; Vausort, M.; McCann, G.P.; Zangrando, J.; Kelly, D.; Razvi, N.; Zhang, L.; Ng, L.L.; Wagner, D.R.; Squire, I.B. MicroRNA-150: A novel marker of left ventricular remodeling after acute myocardial infarction. Circ. Cardiovasc. Genet. 2013, 6, 290–298. [Google Scholar] [CrossRef] [Green Version]
- Li, H.; Zhang, P.; Li, F.; Yuan, G.; Wang, X.; Zhang, A.; Li, F. Plasma miR-22-5p, miR-132-5p, and miR-150-3p Are Associated with Acute Myocardial Infarction. BioMed. Res. Int. 2019, 2019, 5012648. [Google Scholar] [CrossRef] [Green Version]
- Liu, W.; Liu, Y.; Zhang, Y.; Zhu, X.; Zhang, R.; Guan, L.; Tang, Q.; Jiang, H.; Huang, C.; Huang, H. MicroRNA-150 Protects Against Pressure Overload-Induced Cardiac Hypertrophy. J. Cell. Biochem. 2015, 116, 2166–2176. [Google Scholar] [CrossRef]
- Ma, J.L.; Guo, W.L.; Chen, X.M. Overexpressing microRNA-150 attenuates hypoxia-induced human cardiomyocyte cell apoptosis by targeting glucose-regulated protein-94. Mol. Med. Rep. 2018, 17, 4181–4186. [Google Scholar] [CrossRef]
- Tang, Y.; Wang, Y.; Park, K.M.; Hu, Q.; Teoh, J.P.; Broskova, Z.; Ranganathan, P.; Jayakumar, C.; Li, J.; Su, H.; et al. MicroRNA-150 protects the mouse heart from ischaemic injury by regulating cell death. Cardiovasc. Res. 2015, 106, 387–397. [Google Scholar] [CrossRef] [Green Version]
- Linsley, P.S.; Schelter, J.; Burchard, J.; Kibukawa, M.; Martin, M.M.; Bartz, S.R.; Johnson, J.M.; Cummins, J.M.; Raymond, C.K.; Dai, H.; et al. Transcripts targeted by the microRNA-16 family cooperatively regulate cell cycle progression. Mol. Cell. Biol. 2007, 27, 2240–2252. [Google Scholar] [CrossRef] [Green Version]
- Duisters, R.F.; Tijsen, A.J.; Schroen, B.; Leenders, J.J.; Lentink, V.; van der Made, I.; Herias, V.; van Leeuwen, R.E.; Schellings, M.W.; Barenbrug, P.; et al. miR-133 and miR-30 regulate connective tissue growth factor: Implications for a role of microRNAs in myocardial matrix remodeling. Circ. Res. 2009, 104, 170–178, 176p following 178. [Google Scholar] [CrossRef] [Green Version]
- Liu, J.; Sun, F.; Wang, Y.; Yang, W.; Xiao, H.; Zhang, Y.; Lu, R.; Zhu, H.; Zhuang, Y.; Pan, Z.; et al. Suppression of microRNA-16 protects against acute myocardial infarction by reversing beta2-adrenergic receptor down-regulation in rats. Oncotarget 2017, 8, 20122–20132. [Google Scholar] [CrossRef] [Green Version]
- Jaguszewski, M.; Osipova, J.; Ghadri, J.R.; Napp, L.C.; Widera, C.; Franke, J.; Fijalkowski, M.; Nowak, R.; Fijalkowska, M.; Volkmann, I.; et al. A signature of circulating microRNAs differentiates takotsubo cardiomyopathy from acute myocardial infarction. Eur. Heart J. 2014, 35, 999–1006. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Devaux, Y.; Vausort, M.; McCann, G.P.; Kelly, D.; Collignon, O.; Ng, L.L.; Wagner, D.R.; Squire, I.B. A panel of 4 microRNAs facilitates the prediction of left ventricular contractility after acute myocardial infarction. PLoS ONE 2013, 8, e70644. [Google Scholar] [CrossRef]
- van Rooij, E.; Sutherland, L.B.; Liu, N.; Williams, A.H.; McAnally, J.; Gerard, R.D.; Richardson, J.A.; Olson, E.N. A signature pattern of stress-responsive microRNAs that can evoke cardiac hypertrophy and heart failure. Proc. Natl. Acad. Sci. USA 2006, 103, 18255–18260. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zampetaki, A.; Attia, R.; Mayr, U.; Gomes, R.S.; Phinikaridou, A.; Yin, X.; Langley, S.R.; Willeit, P.; Lu, R.; Fanshawe, B.; et al. Role of miR-195 in aortic aneurysmal disease. Circ. Res. 2014, 115, 857–866. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Long, G.; Wang, F.; Duan, Q.; Yang, S.; Chen, F.; Gong, W.; Yang, X.; Wang, Y.; Chen, C.; Wang, D.W. Circulating miR-30a, miR-195 and let-7b associated with acute myocardial infarction. PLoS ONE 2012, 7, e50926. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gao, C.K.; Liu, H.; Cui, C.J.; Liang, Z.G.; Yao, H.; Tian, Y. Roles of MicroRNA-195 in cardiomyocyte apoptosis induced by myocardial ischemia-reperfusion injury. J. Genet. 2016, 95, 99–108. [Google Scholar] [CrossRef]
- Chen, C.; Jia, K.Y.; Zhang, H.L.; Fu, J. MiR-195 enhances cardiomyocyte apoptosis induced by hypoxia/reoxygenation injury via downregulating c-myb. Eur. Rev. Med. Pharmacol. Sci. 2016, 20, 3410–3416. [Google Scholar]
| ACS (n = 80) | Controls (n = 80) | p Value | ||
|---|---|---|---|---|
| Gender (%) | Male | 82 | 58 | 0.001 |
| Female | 18 | 42 | ||
| Age (years) | 56.9 ± 9.4 | 60.1 ± 7.9 | 0.023 | |
| Obesity (%) | 16 | 32 | 0.008 | |
| Dyslipideamia (%) | 58 | 77 | 0.002 | |
| Diabetes (%) | 19 | 24 | 0.86 | |
| Hypertension (%) | 42 | 85 | <0.0001 | |
| Current smoker (%) | 40 | 15 | <0.0001 | |
| Heredity (%) | 29 | 34 | 0.76 | |
| Blood glucose (mmol/L) | 116.8 ± 41.4 | 111.7 ± 58.4 | 0.03 | |
| Triglycerides (mg/dL) | 146.1 ± 86.4 | 137.1 ± 70.6 | 0.59 | |
| Total cholesterol (mg/dL) | 196.8 ± 44.5 | 199.2 ± 44.5 | 0.458 | |
| LDL-cholesterol (mg/dL) a | 120.1 ± 38.1 | 119.3 ± 40.3 | 0.96 | |
| HDL-cholesterol (mg/dL) b | 47.7 ± 11.7 | 52.8 ± 15.6 | 0.048 | |
| Medical treatment at admsission | ||||
| Beta blocker agents (%) | 17 | 27 | 0.12 | |
| ACE inhibitors (%) c | 16 | 25 | 0.11 | |
| Antiplatelet agents (%) | 17 | 26 | 0.16 | |
| Statins (%) | 25 | 48 | 0.001 | |
| ARA II (%) d | 10 | 41 | <0.0001 | |
| Antidiabetic treatment (%) | 10 | 20 | 0.07 | |
| ACS (n = 80) Mean −ΔCt | SD | Controls (n = 80) Mean −ΔCt | SD | Fold Change | p Value | |
|---|---|---|---|---|---|---|
| let 7c | −6.834 | 1.299 | −7.06 | 1.247 | 1.270 | 0.2598 |
| let 7g | −4.94 | 1.336 | −4.935 | 1.448 | 0.926 | 0.8921 |
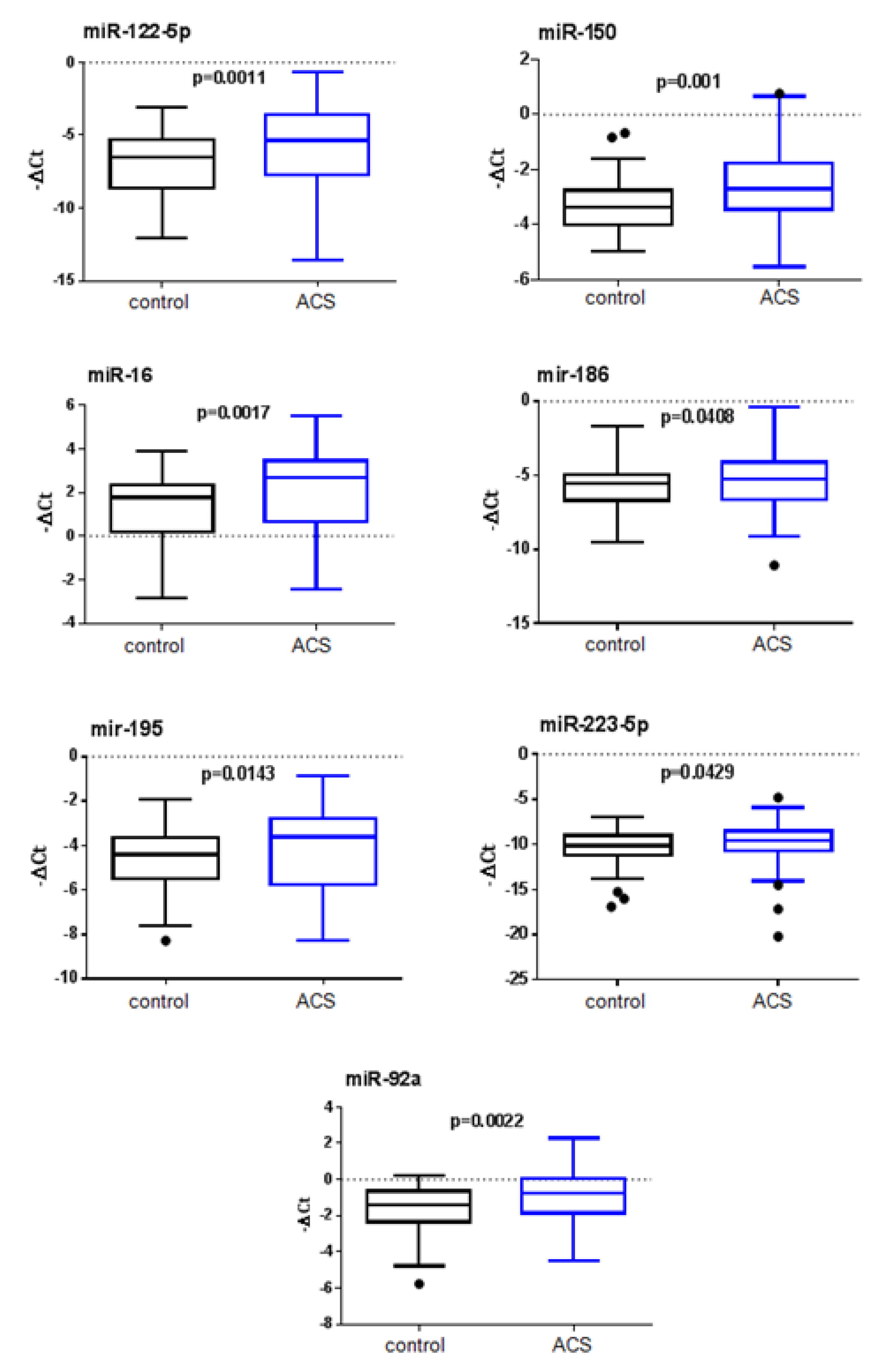
| miR-122 | −5.718 | 2.747 | −6.9 | 2.261 | 2.258 | 0.0011 |
| miR-126 | −1.261 | 1.692 | −1.521 | 1.401 | 1.010 | 0.395 |
| miR-133a | −7.481 | 2.398 | −7.551 | 2.07 | 1.010 | 0.5427 |
| miR-139-5p | −6.822 | 1.525 | −7.147 | 1.295 | 0.962 | 0.2484 |
| miR-145 | −4.728 | 1.552 | −4.536 | 1.74 | 0.861 | 0.4899 |
| miR-146a | −2.238 | 1.733 | −2.523 | 1.637 | 1.057 | 0.3253 |
| miR-146b | −4.513 | 1.54 | −4.58 | 1.5 | 1.000 | 0.828 |
| miR-150 | −2.494 | 1.46 | −3.311 | 0.847 | 1.575 | 0.001 * |
| miR-155 | −7.001 | 1.536 | −7.204 | 1.39 | 0.877 | 0.5137 |
| miR-16 | 1.948 | 2.023 | 1.393 | 1.448 | 1.858 | 0.0017 * |
| miR-186 | −5.35 | 1.879 | −5.815 | 1.661 | 1.238 | 0.0408 |
| miR-195 | −4.187 | 1.839 | −4.572 | 1.348 | 1.723 | 0.0143 * |
| miR-21 | −3.686 | 1.356 | −3.738 | 1.393 | 1.073 | 0.7035 |
| miR-223-5p | −9.845 | 2.331 | −10.34 | 1.864 | 1.510 | 0.0429 |
| miR-223-3p | −0.516 | 2.182 | −1.054 | 1.805 | 1.431 | 0.0825 |
| miR-30b | −1.898 | 1.401 | −1.762 | 1.386 | 0.831 | 0.5495 |
| miR-30c | −3.239 | 1.626 | −3.341 | 1.48 | 0.971 | 0.7111 |
| miR-574-3p | −4.622 | 1.768 | −5.1 | 1.346 | 1.283 | 0.0582 * |
| miR-92a | −1.009 | 1.554 | −1.59 | 1.229 | 1.551 | 0.0022 * |
| Unadjusted | Adjusted | |||||
|---|---|---|---|---|---|---|
| OR | 95%CI | p Value | OR | 95%CI | p Value | |
| miR-122 | ||||||
| t2 vs. t1 | 0.68 | 0.31–1.50 | 0.34 | 0.79 | 0.26–2.37 | 0.67 |
| t3 vs. t1 | 2.89 | 1.29–6.46 | 0.0096 | 3.94 | 1.28–12.1 | 0.017 |
| Tertiles −7.0 and −5.15 | ||||||
| miR-150 | ||||||
| t2 vs. t1 | 1.23 | 0.56–2.70 | 0.61 | 1.17 | 0.38–3.64 | 0.79 |
| t3 vs. t1 | 4.84 | 2.11–11.1 | 0.001 | 4.39 | 1.40–13.8 | 0.011 |
| Tertiles −3.57 and −2.69 | ||||||
| miR-16 | ||||||
| t2 vs. t1 | 0.58 | 0.26–1.28 | 0.17 | 0.70 | 0.22–2.18 | 0.54 |
| t3 vs. t1 | 3.48 | 1.53–7.91 | 0.0029 | 3.59 | 1.16–11.1 | 0.027 |
| Tertiles 1.29 and 2.67 | ||||||
| miR-195 | ||||||
| t2 vs. t1 | 0.61 | 0.28–1.34 | 0.22 | 1.01 | 0.33–3.04 | 0.99 |
| t3 vs. t1 | 3.39 | 1.49–7.72 | 0.0036 | 5.22 | 1.52–17.9 | 0.009 |
| Tertiles −4.86 and −3.54 | ||||||
| miR-92a | ||||||
| t2 vs. t1 | 1.15 | 0.53–2.52 | 0.73 | 1.72 | 0.53–5.61 | 0.37 |
| t3 vs. t1 | 3.08 | 1.39–6.85 | 0.0057 | 3.31 | 1.07–10.2 | 0.038 |
| Tertiles −1.73 and −0.62 | ||||||
| miR-186 | ||||||
| t2 vs. t1 | 0.64 | 0.30–1.41 | 0.27 | 0.61 | 0.20–1.86 | 0.39 |
| t3 vs. t1 | 2.37 | 1.08–5.22 | 0.032 | 2.47 | 0.85–7.17 | 0.09 |
| Tertiles −6.11 and −4.83 | ||||||
| miR-223-5p | ||||||
| t2 vs. t1 | 1.08 | 0.50–2.32 | 0.85 | 0.94 | 0.31–2.84 | 0.92 |
| t3 vs. t1 | 1.4 | 0.65–3.00 | 0.39 | 1.63 | 0.57–4.66 | 0.37 |
| Tertiles −1.57 and 0 | ||||||
| AUC | 95% CI | p Value | |
|---|---|---|---|
| Clinical model | 0.882 | (0.830–0.933) | - |
| + miR-122 | 0.903 | (0.856–0.950) | 0.13 |
| + miR-150 | 0.907 | (0.861–0.953) | 0.10 |
| + miR-195 | 0.887 | (0.842–0.946) | 0.30 |
| + miR-92a | 0.900 | (0.853–0.951) | 0.13 |
| + miR-16 | 0.896 | (0.845–0.946) | 0.27 |
| + miR-122+miR-150 | 0.911 | (0.866–0.956) | 0.07 |
| + miR-122+miR-195 | 0.915 | (0.873–0.956) | 0.07 |
| + miR-122+miR92a | 0.904 | (0.857–0.951) | 0.15 |
| + miR-122+miR16 | 0.909 | (0.684–0.954) | 0.09 |
| + miR-150+miR92a | 0.906 | (0.859–0.953) | 0.11 |
| + miR-150+miR-195 | 0.909 | (0.866–0.951) | 0.11 |
| + miR-150+miR16 | 0.909 | (0.866–0.953) | 0.07 |
| + miR-195+miR-92a | 0.906 | (0.862–0.951) | 0.15 |
| + miR-195+miR-16 | 0.908 | (0.865–0.951) | 0.10 |
| + miR-16+miR-92a | 0.901 | (0.855–0.947) | 0.30 |
| + miR-122+miR-150+miR-92a | 0.913 | (0.869–0.958) | 0.06 |
| + miR-122+miR-150+miR-195 | 0.925 | (0.886–0.964) | 0.02 |
| + miR-122+miR-150+miR-16 | 0.915 | (0.871–0.958) | 0.06 |
| + miR-150+miR-92a+miR-195 | 0.914 | (0.872–0.956) | 0.07 |
| + miR-150+miR-92a+miR-16 | 0.911 | (0.867–0.955) | 0.08 |
| + miR-16+miR-195+miR-92a | 0.909 | (0.865–0.952) | 0.15 |
| + miR-122+miR-195+miR-92a+miR-16 | 0.919 | (0.877–0.960) | 0.06 |
| + miR-122+miR-150+miR-195+miR-92a | 0.927 | (0.889–0.966) | 0.01 |
| + miR-122+miR-150+miR-195+miR-16 | 0.924 | (0.885–0.933) | 0.003 |
| + miR-150+miR-195+miR-92a+miR-16 | 0.917 | (0.876–0.958) | 0.07 |
| +miR-122+miR-150+-miR-195+miR-92a+miR- 16 | 0.928 | (0.890–0.966) | 0.02 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Elbaz, M.; Faccini, J.; Laperche, C.; Grousset, E.; Roncalli, J.; Ruidavets, J.-B.; Vindis, C. Identification of a miRNA Based-Signature Associated with Acute Coronary Syndrome: Evidence from the FLORINF Study. J. Clin. Med. 2020, 9, 1674. https://doi.org/10.3390/jcm9061674
Elbaz M, Faccini J, Laperche C, Grousset E, Roncalli J, Ruidavets J-B, Vindis C. Identification of a miRNA Based-Signature Associated with Acute Coronary Syndrome: Evidence from the FLORINF Study. Journal of Clinical Medicine. 2020; 9(6):1674. https://doi.org/10.3390/jcm9061674
Chicago/Turabian StyleElbaz, Meyer, Julien Faccini, Clémence Laperche, Elisa Grousset, Jérôme Roncalli, Jean-Bernard Ruidavets, and Cécile Vindis. 2020. "Identification of a miRNA Based-Signature Associated with Acute Coronary Syndrome: Evidence from the FLORINF Study" Journal of Clinical Medicine 9, no. 6: 1674. https://doi.org/10.3390/jcm9061674
APA StyleElbaz, M., Faccini, J., Laperche, C., Grousset, E., Roncalli, J., Ruidavets, J.-B., & Vindis, C. (2020). Identification of a miRNA Based-Signature Associated with Acute Coronary Syndrome: Evidence from the FLORINF Study. Journal of Clinical Medicine, 9(6), 1674. https://doi.org/10.3390/jcm9061674







