Abstract
Diabetic kidney disease (DKD) is the leading cause of chronic kidney disease (CKD). Elucidating the mechanisms underlying proteinuria in DKD is crucial because it is a common problem in DKD-related mortality and morbidity. MicroRNAs (miRs) associated with DKD have been detected in experimental diabetes models and in patients with both diabetes and CKD. Here, we aimed to investigate pathologic miRs in diabetic nephropathy (DN) by prospectively following six nephrotic, biopsy-proven isolated DN patients (enrolled between August 2015 and July 2017) for one year. The urinary exosomes were isolated at the time of the biopsy and the contained miRs were analyzed by next-generation sequencing. The results were compared to the control group, composed of age-, gender-, and CKD stage-matched patients with proteinuric CKD who did not present diabetes. Among the 72 identified miRs, we investigated eight (miR-188-5p, miR-150-3p, miR-760, miR-3677-3p, miR-548ah-3p, miR-548p, miR-320e, and miR-23c) exhibiting the strongest upregulation (13–15 fold) and two (miR-133a-3p and miR-153-3p) with the strongest downregulation (7–9 fold). The functional analysis of these miRs showed that they were involved in known and novel pathways of DN, supporting their pathologic roles. The bioinformatics-based prediction of the target genes of these miRs will inspire future research on the mechanisms underlying DN pathogenesis.
1. Introduction
Diabetic kidney disease (DKD) has emerged as a leading cause of chronic kidney disease (CKD) worldwide. It occurs in approximately 30% and 40% of patients with type 1 and 2 diabetes mellitus, respectively [1,2,3], and is more common in low- and lower-middle-income countries [4]. The clinical diagnosis of DKD is based on the measurement of the estimated glomerular filtration rate (eGFR) and the albuminuria/proteinuria, along with clinical observations, including diabetes duration and the presence of diabetic retinopathy [5,6]. The major threats presented by DKD are risks of mortality, cardiovascular disease, end-stage renal disease, and infections, and a common risk factor for these DKD-related complications is proteinuria [7,8,9]. A better understanding of the mechanisms underlying proteinuria in DKD may help to develop novel treatment strategies for this aspect of DKD.
Nephrotic-range proteinuria is the most severe form of proteinuria in diabetic patients. It may result from isolated diabetic nephropathy (DN), non-diabetic renal disease, or combined nephropathies [10,11,12]. Renal biopsy is currently the only diagnostic tool able to delineate the renal pathologies in diabetic patients [12]. However, the invasiveness and cost of this technique, as well as the risk of tissue-sampling errors limit its general applicability to the diabetes patient population [13,14]. Non-invasive urine analysis is an appropriate approach to assess functional changes because urine contains proteins from the filtered plasma. However, only a few of these proteins are derived from the kidney, and structural changes in this organ are poorly detected through urine [15]. Urinary exosomes are normally secreted by cells from all nephron segments and mostly carry protein, mRNA, and microRNA (miR) markers of renal dysfunction and structural injury [16]. Urinary-exosomal miR analysis has emerged as a promising tool for the discovery of DKD biomarkers [15,17]. To date, the reported urinary-exosomal miR profiles for DKD are likely to be analyses based on a mixture of isolated DN, non-diabetic renal disease and combined nephropathies. In the current study, we examined the urinary-exosomal miR signatures of the nephrotic, biopsy-proven, isolated DN patients.
2. Materials and Methods
2.1. Patients
Fifteen diabetic patients with nephrotic range proteinuria, who underwent a renal biopsy in our hospital between August 2015 and July 2017, were screened for this study. The diabetic patients with biopsy-proven non-diabetic renal disease or combined nephropathies were excluded. Other exclusion criteria included any febrile illness during the last three months, chronic inflammatory or rheumatic disease, hepatitis, HIV infection, and hereditary renal disease. The six patients fulfilling these criteria were prospectively followed at our nephrology outpatient clinic for one year. The control patients were non-diabetic CKD patients whose urinary protein-to-creatinine ratio (UPCR) ranged from 500 to 1000 mg/g. Blood samples and 20 mL of voided midstream urine were collected for routine laboratory testing. Due to the variability in the urinary protein excretion, two out of three specimens collected within a 3- to 6-month period should provide abnormal results before considering that a patient developed an increased UPCR. The urine samples were used immediately for exosomal miR separation. This study was approved by the Chang Gung Medical Foundation Institutional Review Board. All methods were performed in accordance with the relevant guidelines and regulations, and informed consent was obtained from all participants.
2.2. Isolation of Urinary-Exosomal RNA
Urinary-exosomal RNA was isolated using the exosome RNA isolation kit (Norgen-Biotek, Thorold, USA) according to the manufacturer’s protocol, as previously described [18]. Briefly, the RNA was extracted from 1 mL of urine. The RNA extractions were eluted in 100 μL and stored at −80 °C until use. The purified RNA was quantified at OD260nm by using a ND-1000 spectrophotometer (NanoDrop Technology, Wilmington, NC, USA) and was qualitated by using a Bioanalyzer 2100 (Agilent Technology, USA) with the RNA 6000 labchip kit (Agilent Technologies, San Jose, CA, USA).
2.3. Library Preparation and Sequencing
The small RNA library construction and deep sequencing was commercially carried out at a biotechnology company (Welgene, Taipei, Taiwan). The samples were prepared using the Illumina sample preparation kit according to the TruSeq Small RNA Sample Preparation Guide. The 3′ and 5′ adaptors were ligated to the total RNA and reverse transcription, and followed by a polymerase chain reaction (PCR) amplification. The enriched cDNA constructs were size-fractionated and purified on a 6% polyacrylamide gel electrophoresis and on the bands containing the 18–40 nucleotide-RNA fragments (140–155 nucleotides in length with both adapters). The libraries were sequenced on an Illumina instrument (75SE cycle single-read). The sequencing data were processed with the Illumina software.
2.4. Small RNA- Sequencing Analysis
Initially, the sequences generated went through a filtering process to obtain the qualified reads. Trimmomatic [19] was implemented to trim or remove the reads according to the quality score. The qualified reads, after filtering low-quality data, were analyzed using miRDeep2 [20] to clip the 3’ adapter sequence and discard the reads shorter than 18 nucleotides, before aligning the reads to the human genome from University of California, Santa Cruz (UCSC). Only the reads that mapped perfectly to the genome five or fewer times were used for miR-detection, since miRs usually map to few genomic locations. MiRDeep2 estimates the expression levels of known miRs. The miRs with two-fold changes in expression and p value < 0.05 were selected for the target-genes prediction by using the miRmap database.
The output files were further annotated by adding gene-functional descriptions and gene ontology (GO) classifications. The reference genomes and gene annotations were retrieved from the Ensembl database (http://asia.ensembl.org/index.html). The GO term and fold enrichment, or the depletion for the gene lists of significantly up- and down-regulated genes in the urine exosomes, were determined. The GO analysis for significant genes was performed using the Kyoto Encyclopedia of Genes and Genomes (KEGG, http://www.genome.jp/kegg/) and the National Institutes of Health (NIH) Database for Annotation, Visualization and Integrated Discovery (DAVID) Bioinformatics Resources 6.7.
3. Results
3.1. Patient Characteristics
According to the inclusion and exclusion criteria, six patients were prospectively followed at our nephrology outpatient clinic for a year. The clinical characteristics of the six study patients are summarized in Table 1. Their urine samples were collected for exosome isolation and analyzed by next-generation sequencing. At biopsy, Patient 4 was at Stage 1 CKD; meanwhile, Patients 1 and 2 were at Stage 4; and Patients 3, 5, and 6 were at Stage 3. All the patients had nephrotic range proteinuria. Their UPCR ranged from 5470.6 to 20364.4 mg/g. Four of the six patients had poor blood-sugar control. Their glycated- hemoglobin (HbA1C) levels were between 10.4% and 14.9%. All the study patients were on standard medications, including anti-diabetic agents, renin-angiotensin system (RAS) blockades, and statins. Patient 1 was on a low dose of methylprednisolone (2 mg/day).

Table 1.
Characteristics and clinical data of the study subjects.
3.2. Urinary-Exosomal miR Signatures
When comparing the next-generation sequencing data with age-, gender-, and CKD stage-matched controls, we identified 72 miRs showing significant differences in abundance (Figure 1). Among the 72 miRs identified, 22 were up-regulated and 50 were down-regulated. Eight miRs (miR-188-5p, miR-150-3p, miR-760, miR-3677-3p, miR-548ah-3p, miR-548p, miR-320e, and miR-23c) showed the strongest up-regulation (13–15 fold) and two (miR-133a-3p, miR-153-3p) showed the strongest down-regulation (7–9 folds). These deregulated miRs exhibited a correlated expression pattern between DN and control (Figure 2). The attributed functions of the deregulated miRs, based on published data, are summarized in Table 2.
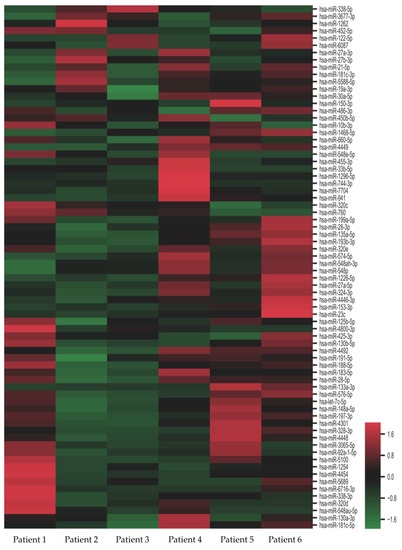
Figure 1.
Heat map of differentially-expressed microRNAs (miRs) in nephrotic diabetic nephropathy (DN) patients. Fold change of expression levels were normalized to the mean signal intensities of the controls. Red and green colors represent fold change up- and down-regulation, respectively, as indicated by the linear scale bar.
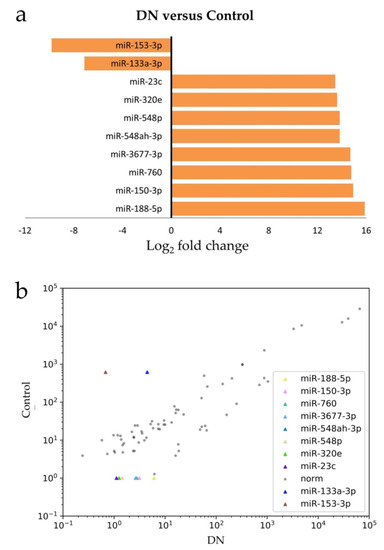
Figure 2.
The most deregulated miRs in DN: (a) the differentially-expressed miRs in DN (versus control) are shown in Log2 fold change, with a positive value referring to up-regulation and a negative value meaning down-regulation; (b) the scatter plot shows that these deregulated miRs exhibited correlated-expression pattern between DN and control.

Table 2.
MicroRNAs (miRs) with the highest fold change in expression levels in nephroticdiabetic nephropathy (DN) patients.
The strongest up-regulation that we identified in the urine exosomes of the nephrotic, isolated DN patients was with miR-188-5p. This miR has been reported to regulate the high glucose-induced epithelial-mesenchymal transition of HK-2 cells through the PTEN/PI3K/Akt pathway [21]. The second most strongly up-regulated miR was miR-150-3p. It has been reported to be up-regulated in the circulating exosomes of clinically-diagnosed DN patients [22]. In addition, we described for the first time that miR-760, miR-3677-3p, miR-548ah-3p, miR-548p, miR-320e, and miR-23c were up-regulated in the DN patients. Although miR-133a-3p and miR-153-3p have been reported to have implications in pig kidneys [24] and lupus nephritis [25], this is the first time they were reported in the nephrotic DN patients.
3.3. Functional Analysis of the Identified miRs
Using the KEGG and NIH DAVID Bioinformatics Resources, we obtained the predicted pathways targeted by the miRs that were identified in nephrotic DN patients. The P-values of significance-over-background are shown alongside the results of the analysis (Table 3). These pathways included PI3K-Akt-signaling, mitogen-activated protein kinase (MAPK)-signaling, human papillomavirus infection, RAS-signaling, endocytosis, and pathways regulating the pluripotency of stem cells. In these pathways, the target molecules of the miRs we examined are summarized in Table 4. Many of the listed target genes are novel and not yet characterized in experimental diabetes models. In the PI3K-Akt-signaling pathway, miR-548p was predicted to target EIF4E, FGFR1 and PRLR. Meanwhile, miR-548ah-3p was involved in the PI3K-Akt-signaling pathway by targeting CCND1, PRKAA2 and PPP2R5E. In the MAPK-signaling pathway, the predicted-target genes included MAP3K7, FGFR1, MKNK2, TGFBR1, RPS6KA3, NF1, AKT3, STK4 and FGF1. The miR-23c, miR-548p, miR-548ah-3p, and miR-320e played roles in the human papillomavirus infection pathway through their target genes. By targeting different genes, miR-548ah-3p played roles in all these functional pathways. Except for the endocytosis pathway, an interesting finding was that miR-320e regulated AKT3, which was the hub of the other five pathways.

Table 3.
Predicted pathways associated with the identified miRs in nephrotic DN patients.

Table 4.
Target genes, functional pathways and the miRs in nephrotic DN patients.
4. Discussion
Although various miRs have been reported to be related to diabetic nephropathy, these results need to be cautiously interpreted. Some studies have identified miRs in cultured kidney cells [26], whereas others have analyzed urinary-exosomal miRs from patients who have simultaneously suffered from diabetes and CKD [27,28]. In cultured podocytes, high glucose was shown to induce podocyte dedifferentiation through the up-regulation of miR-193a [29]. TGF-β1 was reported to up-regulate miR-192 in cultured glomerular mesangial cells and in glomeruli from streptozotocin-induced diabetic mice. The specific reduction of renal miR-192 by using a locked nucleic acid–modified inhibitor of miR-192 decreases renal fibrosis [30,31]. However, in cultured human proximal tubular cells (HK-2 cells), TGF-β1 led to a 51% reduction in miR-192 [26]. The miRs identified in these studies, including miR-192, miR-193a, miR-362-3p, miR-877-3p, miR-150-5p, and miR-15a-5p, could be informative but not necessarily have a role in the pathogenesis of human DKD. In our study, we examined the miR signature in nephrotic, biopsy-proven, isolated DN and found a completely different set of miRs. We demonstrated that miR-188-5p, miR-150-3p, miR-760, miR-3677-3p, miR-548ah-3p, miR-548p, miR-320e, and miR-23c were significantly up-regulated in nephrotic DN patients. A significant down-regulation of miR-133a-3p and miR-153-3p was also found in our study patients.
In a cell culture model, miR-188-5p is shown to regulate high glucose-induced epithelial-mesenchymal transition (EMT) in human proximal tubular cell lines through the PTEN/PI3K/Akt pathway [21]. By analyzing miRs from the patients’ urine exosomes, we demonstrated the pathologic role of miR-188-5p in nephrotic and biopsy-proven isolated DN. Notably, our analysis, based on KEGG and NIH DAVID Bioinformatics Resources, showed that the PI3K/Akt-signaling pathway appeared to play pathologic roles in isolated DN (Table 3). The circulating-exosomal miR-150-3p is reported to be related to albuminuria in patients with DN [22]. In line with this observation, our findings on up-regulated miR-150-3p revealed its pathologic role in isolated DN. Further experiments on the function of our newly-reported miRs are required to characterize their pathologic roles in isolated DN.
In addition to the PI3K-Akt-signaling pathway, we also showed that MAPK-signaling, human papillomavirus infection, Ras-signaling, endocytosis, and pathways regulating the pluripotency of stem cells could be the mechanisms underpinning DN. Some of these pathways have been reported in various experimental diabetes models. MAPK-signaling plays crucial roles in DN because it is related to inflammatory, oxidative, and apoptotic processes [32]; evidence of its activation has been demonstrated on the kidney biopsies of diabetic patients [33]. High glucose levels are known to induce Ras/Rac1-dependent superoxide formation in rat mesangial cells [34,35]. Although we were unable to specify the intra-renal location of our identified miRs, our functional analysis supported the findings derived from experimental diabetes and highlighted the pathologic roles of Ras-signaling in DN patients. Furthermore, the functional analysis of our identified miRs revealed a participation of the endocytosis pathway. This was of particular interest because it reflected the fact that all our study patients were nephrotic. In cell culture models, high glucose levels are known to reduce the megalin-mediated albumin endocytosis in renal proximal tubule cells [36]. Additionally, in renal proximal tubule cells, the inhibition of Akt is known to abrogate insulin-induced albumin endocytosis [37].
By using KEGG pathway prediction, we identified human papillomavirus infection and pathways regulating the pluripotency of stem cells as two novel pathways underpinning DN. In our results, AKT3, targeted by miR-320e (Table 4), served as the signaling hub linking the known and novel mechanisms of DN. Akt2 is known to protect podocytes in rodent diabetic models [38,39]. However, in humans, AKT3, not AKT2, is far more abundant in glomeruli than in tubules [40]. As we investigated pathologic miRs and pathways underpinning heavy proteinuria in DN, our findings on miR-320e and AKT3 were of particular importance.
Our study analyzed miRs from liquid (urine) biopsy specimens in clinical nephrotic, biopsy-proven DN patients. Compared to the previously-reported DN-related miRs, these newly-reported miRs could be the true regulators in the pathogenesis of DN. Functional analysis, based on our identified miRs, disclosed known and new pathways underpinning DN pathogenesis. These findings will inspire future research into the mechanisms underlying the pathogenesis of DN.
A major limitation of our study was the lack of real-time quantitative PCR-based validation of our identified miRs. However, unlike the known-reference miRs in cervical cancer and colorectal cancer [41,42], the reference miRs in DN remain unknown. An alternative approach to overcome this problem we would like to suggest is to test our reported deregulated miRs in various clinical and experimental models.
5. Conclusions
In conclusion, we reported a new urinary-exosomal miR signature in nephrotic, biopsy-proven DN patients. Their functional roles in DN pathogenesis were partly- supported by known pathways in previously-reported experimental diabetes models. Further experiments are required to investigate the functional roles of our newly-found target genes of each miR in the pathogenesis of DN.
Author Contributions
Conceptualization, W.-C.L. and C.-T.L.; Methodology, W.-C.L. and P.-T.L.; Software, P.-T.L.; Validation, W.-H.K., H.-Y.N. and C.-T.L.; Formal analysis, P.-T.L.; Investigation, T.T.-Y.C. and W.-H.K.; Resources, W.-C.L. and L.-C.L.; Data curation, W.-C.L.; Writing—original draft preparation, W.-C.L.; Writing—review and editing, W.-C.L. and C.-T.L.; Visualization, W.-C.L.; Supervision, C.-T.L.; Project administration, W.-C.L. and H.-Y.N.; Funding acquisition, C.-T.L. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by Kaohsiung Chang Gung Memorial Hospital, grant numbers CORPG8F1011, CORPG8F1012, and CMRPG8J0621.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
References
- De Boer, I.H.; Rue, T.C.; Hall, Y.N.; Heagerty, P.J.; Weiss, N.S.; Himmelfarb, J. Temporal trends in the prevalence of diabetic kidney disease in the United States. JAMA 2011, 305, 2532–2539. [Google Scholar] [CrossRef]
- Lee, W.C.; Lee, Y.T.; Li, L.C.; Ng, H.Y.; Kuo, W.H.; Lin, P.T.; Liao, Y.C.; Chiou, T.T.; Lee, C.T. The Number of Comorbidities Predicts Renal Outcomes in Patients with Stage 3(-)5 Chronic Kidney Disease. J. Clin. Med. 2018, 7. [Google Scholar] [CrossRef]
- Reutens, A.T. Epidemiology of diabetic kidney disease. Med. Clin. North Am. 2013, 97, 1–18. [Google Scholar] [CrossRef]
- Xie, Y.; Bowe, B.; Mokdad, A.H.; Xian, H.; Yan, Y.; Li, T.; Maddukuri, G.; Tsai, C.Y.; Floyd, T.; Al-Aly, Z. Analysis of the Global Burden of Disease study highlights the global, regional, and national trends of chronic kidney disease epidemiology from 1990 to 2016. Kidney Int. 2018, 94, 567–581. [Google Scholar] [CrossRef]
- American Diabetes, A. 11. Microvascular Complications and Foot Care: Standards of Medical Care in Diabetes-2019. Diabetes Care 2019, 42, S124–S138. [Google Scholar] [CrossRef]
- Umanath, K.; Lewis, J.B. Update on Diabetic Nephropathy: Core Curriculum 2018. Am. J. Kidney Dis. 2018, 71, 884–895. [Google Scholar] [CrossRef]
- Kitai, Y.; Doi, Y.; Osaki, K.; Sugioka, S.; Koshikawa, M.; Sugawara, A. Nephrotic range proteinuria as a strong risk factor for rapid renal function decline during pre-dialysis phase in type 2 diabetic patients with severely impaired renal function. Clin. Exp. Nephrol. 2015, 19, 1037–1043. [Google Scholar] [CrossRef]
- Agrawal, V.; Marinescu, V.; Agarwal, M.; McCullough, P.A. Cardiovascular implications of proteinuria: An indicator of chronic kidney disease. Nat. Rev. Cardiol. 2009, 6, 301–311. [Google Scholar] [CrossRef]
- Berni, E.; Pritchard, N.; Jenkins-Jones, S.; Ambery, P.; Jain, M.; Jermutus, L.; Scott, L.A.; Currie, C.J. Hospital admissions for severe infections in people with chronic kidney disease in relation to renal disease severity and diabetes status. Endocrinol. Diabetes Metab. 2018, 1, e00029. [Google Scholar] [CrossRef]
- Christensen, P.K.; Larsen, S.; Horn, T.; Olsen, S.; Parving, H.H. Causes of albuminuria in patients with type 2 diabetes without diabetic retinopathy. Kidney Int. 2000, 58, 1719–1731. [Google Scholar] [CrossRef]
- Kveder, R.; Kajtna-Koselj, M.; Rott, T.; Bren, A.F. Nephrotic syndrome in patients with diabetes mellitus is not always associated with diabetic nephropathy. Nephrol. Dial. Transplant. 2001, 16 (Suppl. 6), 86–87. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Grujicic, M.; Salapura, A.; Basta-Jovanovic, G.; Figurek, A.; Micic-Zrnic, D.; Grbic, A. Non-Diabetic Kidney Disease in Patients with Type 2 Diabetes Mellitus-11-Year Experience from a Single Center. Med. Arch. 2019, 73, 87–91. [Google Scholar] [CrossRef] [PubMed]
- Rush, D. Protocol transplant biopsies: An underutilized tool in kidney transplantation. Clin. J. Am. Soc. Nephrol. 2006, 1, 138–143. [Google Scholar] [CrossRef]
- Moledina, D.G.; Luciano, R.L.; Kukova, L.; Chan, L.; Saha, A.; Nadkarni, G.; Alfano, S.; Wilson, F.P.; Perazella, M.A.; Parikh, C.R. Kidney Biopsy-Related Complications in Hospitalized Patients with Acute Kidney Disease. Clin. J. Am. Soc. Nephrol. 2018, 13, 1633–1640. [Google Scholar] [CrossRef]
- Gudehithlu, K.P.; Garcia-Gomez, I.; Vernik, J.; Brecklin, C.; Kraus, M.; Cimbaluk, D.J.; Hart, P.; Dunea, G.; Arruda, J.A.; Singh, A.K. In Diabetic Kidney Disease Urinary Exosomes Better Represent Kidney Specific Protein Alterations Than Whole Urine. Am. J. Nephrol. 2015, 42, 418–424. [Google Scholar] [CrossRef] [PubMed]
- Ludwig, A.K.; Giebel, B. Exosomes: Small vesicles participating in intercellular communication. Int. J. Biochem. Cell Biol. 2012, 44, 11–15. [Google Scholar] [CrossRef] [PubMed]
- Eissa, S.; Matboli, M.; Aboushahba, R.; Bekhet, M.M.; Soliman, Y. Urinary exosomal microRNA panel unravels novel biomarkers for diagnosis of type 2 diabetic kidney disease. J. Diabetes Complicat. 2016, 30, 1585–1592. [Google Scholar] [CrossRef]
- Tain, Y.L.; Huang, L.T.; Chan, J.Y.; Lee, C.T. Transcriptome analysis in rat kidneys: Importance of genes involved in programmed hypertension. Int. J. Mol. Sci. 2015, 16, 4744–4758. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Friedlander, M.R.; Mackowiak, S.D.; Li, N.; Chen, W.; Rajewsky, N. miRDeep2 accurately identifies known and hundreds of novel microRNA genes in seven animal clades. Nucleic. Acids Res. 2012, 40, 37–52. [Google Scholar] [CrossRef]
- Xue, M.; Cheng, Y.; Han, F.; Chang, Y.; Yang, Y.; Li, X.; Chen, L.; Lu, Y.; Sun, B.; Chen, L. Triptolide Attenuates Renal Tubular Epithelial-mesenchymal Transition Via the MiR-188-5p-mediated PI3K/AKT Pathway in Diabetic Kidney Disease. Int. J. Biol. Sci. 2018, 14, 1545–1557. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.; Bae, Y.U.; Jeon, J.S.; Noh, H.; Park, H.K.; Byun, D.W.; Han, D.C.; Ryu, S.; Kwon, S.H. The circulating exosomal microRNAs related to albuminuria in patients with diabetic nephropathy. J. Transl. Med. 2019, 17, 236. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Zeng, L.; Cao, C.; Lu, C.; Lian, W.; Han, J.; Zhang, X.; Zhang, J.; Tang, T.; Li, M. Long noncoding RNA MALAT1 regulates renal tubular epithelial pyroptosis by modulated miR-23c targeting of ELAVL1 in diabetic nephropathy. Exp. Cell Res. 2017, 350, 327–335. [Google Scholar] [CrossRef] [PubMed]
- Marin, D.E.; Braicu, C.; Dumitrescu, G.; Pistol, G.C.; Cojocneanu, R.; Neagoe, I.B.; Taranu, I. MicroRNA profiling in kidney in pigs fed ochratoxin A contaminated diet. Ecotoxicol. Environ. Saf. 2019, 184, 109637. [Google Scholar] [CrossRef]
- Navarro-Quiroz, E.; Pacheco-Lugo, L.; Navarro-Quiroz, R.; Lorenzi, H.; Espana-Puccini, P.; Diaz-Olmos, Y.; Almendrales, L.; Olave, V.; Gonzalez-Torres, H.; Diaz-Perez, A.; et al. Profiling analysis of circulating microRNA in peripheral blood of patients with class IV lupus nephritis. PLoS ONE 2017, 12, e0187973. [Google Scholar] [CrossRef]
- Krupa, A.; Jenkins, R.; Luo, D.D.; Lewis, A.; Phillips, A.; Fraser, D. Loss of MicroRNA-192 promotes fibrogenesis in diabetic nephropathy. J. Am. Soc. Nephrol. 2010, 21, 438–447. [Google Scholar] [CrossRef]
- Delic, D.; Eisele, C.; Schmid, R.; Baum, P.; Wiech, F.; Gerl, M.; Zimdahl, H.; Pullen, S.S.; Urquhart, R. Urinary Exosomal miRNA Signature in Type II Diabetic Nephropathy Patients. PLoS ONE 2016, 11, e0150154. [Google Scholar] [CrossRef]
- Xie, Y.; Jia, Y.; Cuihua, X.; Hu, F.; Xue, M.; Xue, Y. Urinary Exosomal MicroRNA Profiling in Incipient Type 2 Diabetic Kidney Disease. J. Diabetes Res. 2017, 2017, 6978984. [Google Scholar] [CrossRef]
- Mishra, A.; Ayasolla, K.; Kumar, V.; Lan, X.; Vashistha, H.; Aslam, R.; Hussain, A.; Chowdhary, S.; Marashi Shoshtari, S.; Paliwal, N.; et al. Modulation of apolipoprotein L1-microRNA-193a axis prevents podocyte dedifferentiation in high-glucose milieu. Am. J. Physiol. Renal. Physiol. 2018, 314, F832–F843. [Google Scholar] [CrossRef]
- Kato, M.; Zhang, J.; Wang, M.; Lanting, L.; Yuan, H.; Rossi, J.J.; Natarajan, R. MicroRNA-192 in diabetic kidney glomeruli and its function in TGF-beta-induced collagen expression via inhibition of E-box repressors. Proc. Natl. Acad. Sci. USA 2007, 104, 3432–3437. [Google Scholar] [CrossRef]
- Putta, S.; Lanting, L.; Sun, G.; Lawson, G.; Kato, M.; Natarajan, R. Inhibiting microRNA-192 ameliorates renal fibrosis in diabetic nephropathy. J. Am. Soc. Nephrol. 2012, 23, 458–469. [Google Scholar] [CrossRef] [PubMed]
- Malik, S.; Suchal, K.; Khan, S.I.; Bhatia, J.; Kishore, K.; Dinda, A.K.; Arya, D.S. Apigenin ameliorates streptozotocin-induced diabetic nephropathy in rats via MAPK-NF-kappaB-TNF-alpha and TGF-beta1-MAPK-fibronectin pathways. Am. J. Physiol. Renal. Physiol. 2017, 313, F414–F422. [Google Scholar] [CrossRef] [PubMed]
- Adhikary, L.; Chow, F.; Nikolic-Paterson, D.J.; Stambe, C.; Dowling, J.; Atkins, R.C.; Tesch, G.H. Abnormal p38 mitogen-activated protein kinase signalling in human and experimental diabetic nephropathy. Diabetologia 2004, 47, 1210–1222. [Google Scholar] [CrossRef] [PubMed]
- Tung, C.W.; Hsu, Y.C.; Shih, Y.H.; Chang, P.J.; Lin, C.L. Glomerular mesangial cell and podocyte injuries in diabetic nephropathy. Nephrology 2018, 23 (Suppl. 4), 32–37. [Google Scholar] [CrossRef]
- Lin, C.L.; Wang, F.S.; Kuo, Y.R.; Huang, Y.T.; Huang, H.C.; Sun, Y.C.; Kuo, Y.H. Ras modulation of superoxide activates ERK-dependent fibronectin expression in diabetes-induced renal injuries. Kidney Int. 2006, 69, 1593–1600. [Google Scholar] [CrossRef]
- Peruchetti, D.B.; Silva-Aguiar, R.P.; Siqueira, G.M.; Dias, W.B.; Caruso-Neves, C. High glucose reduces megalin-mediated albumin endocytosis in renal proximal tubule cells through protein kinase B O-GlcNAcylation. J. Biol. Chem. 2018, 293, 11388–11400. [Google Scholar] [CrossRef]
- Coffey, S.; Costacou, T.; Orchard, T.; Erkan, E. Akt Links Insulin Signaling to Albumin Endocytosis in Proximal Tubule Epithelial Cells. PLoS ONE 2015, 10, e0140417. [Google Scholar] [CrossRef]
- Coward, R.; Fornoni, A. Insulin signaling: Implications for podocyte biology in diabetic kidney disease. Curr. Opin. Nephrol. Hypertens 2015, 24, 104–110. [Google Scholar] [CrossRef]
- Wang, H.; Misaki, T.; Taupin, V.; Eguchi, A.; Ghosh, P.; Farquhar, M.G. GIV/girdin links vascular endothelial growth factor signaling to Akt survival signaling in podocytes independent of nephrin. J. Am. Soc. Nephrol. 2015, 26, 314–327. [Google Scholar] [CrossRef]
- Uhlen, M.; Fagerberg, L.; Hallstrom, B.M.; Lindskog, C.; Oksvold, P.; Mardinoglu, A.; Sivertsson, A.; Kampf, C.; Sjostedt, E.; Asplund, A.; et al. Proteomics. Tissue-based map of the human proteome. Science 2015, 347, 1260419. [Google Scholar] [CrossRef]
- Leitao Mda, C.; Coimbra, E.C.; de Lima Rde, C.; Guimaraes Mde, L.; Heraclio Sde, A.; Silva Neto Jda, C.; de Freitas, A.C. Quantifying mRNA and microRNA with qPCR in cervical carcinogenesis: A validation of reference genes to ensure accurate data. PLoS ONE 2014, 9, e111021. [Google Scholar] [CrossRef] [PubMed]
- Eriksen, A.H.; Andersen, R.F.; Pallisgaard, N.; Sorensen, F.B.; Jakobsen, A.; Hansen, T.F. MicroRNA Expression Profiling to Identify and Validate Reference Genes for the Relative Quantification of microRNA in Rectal Cancer. PLoS ONE 2016, 11, e0150593. [Google Scholar] [CrossRef] [PubMed]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).