Concomitant TP53 Mutation Confers Worse Prognosis in EGFR-Mutated Non-Small Cell Lung Cancer Patients Treated with TKIs
Abstract
1. Introduction
2. Materials and Methods
2.1. EGFR and TP53 Mutation Analysis
2.2. Response Evaluation
2.3. Statistical Analyses
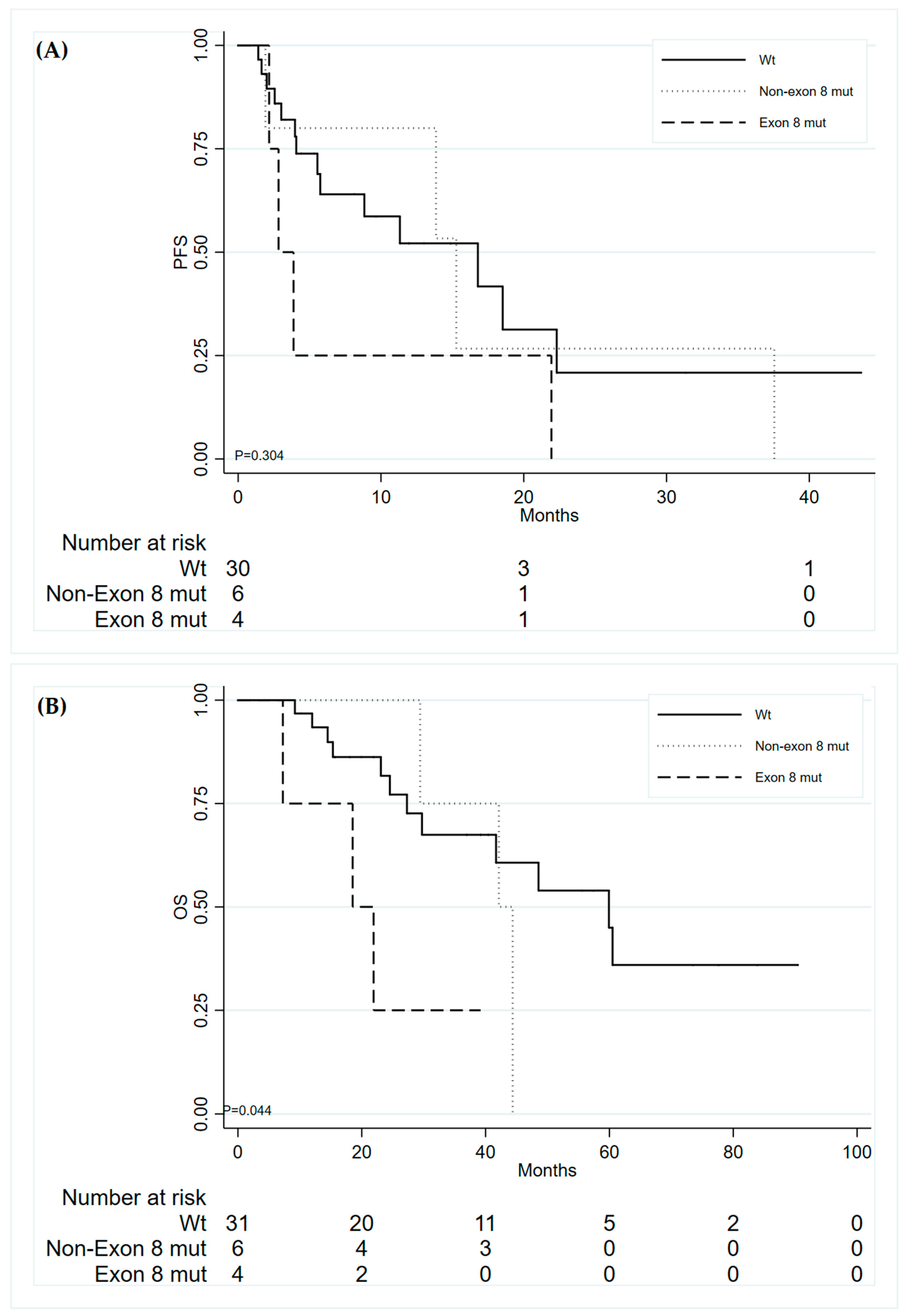
3. Results
3.1. Clinico-Pathologic and Molecular Features of Patients
3.2. Patients Outcome in Relation to EGFR Mutations
3.3. Patients Outcome in Relation to TP53 Mutations
3.4. Multivariate Analysis of the Role of TP53 Mutation: Combined Cohorts of Patients
3.5. TP53 Mutations in Relation to Responsiveness to Third Generation TKIs: Combined Cohorts of Patients
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Rosell, R.; Carcereny, E.; Gervais, R.; Vergnenègre, A.; Massuti, B.; Felip, E.; Palmero, R.; Garcia-Gomez, R.; Pallares, C.; Sanchez, J.M.; et al. Erlotinib versus standard chemotherapy as first-line treatment for European patients with advanced EGFR mutation-positive non-small-cell lung cancer (EURTAC): a multicentre, open-label, randomised phase 3 trial. Lancet Oncol. 2012, 13, 239–246. [Google Scholar] [CrossRef]
- Mok, T.S.; Wu, Y.-L.; Thongprasert, S.; Yang, J.C.-H.; Chu, D.-T.; Saijo, N.; Sunpaweravong, P.; Han, B.; Margono, B.; Ichinose, Y.; et al. Gefitinib or Carboplatin–Paclitaxel in Pulmonary Adenocarcinoma. New Engl. J. Med. 2009, 361, 947–957. [Google Scholar] [CrossRef]
- Yang, J.C.-H.; Wu, Y.-L.; Schuler, M.; Sebastian, M.; Popat, S.; Yamamoto, N.; Zhou, C.; Hu, C.-P.; O’Byrne, K.; Feng, J.; et al. Afatinib versus cisplatin-based chemotherapy for EGFR mutation-positive lung adenocarcinoma (LUX-Lung 3 and LUX-Lung 6): analysis of overall survival data from two randomised, phase 3 trials. Lancet Oncol. 2015, 16, 141–151. [Google Scholar] [CrossRef]
- Mok, T.S.; Wu, Y.-L.; Ahn, M.-J.; Garassino, M.C.; Kim, H.R.; Ramalingam, S.R.; Shepherd, F.A.; He, Y.; Akamatsu, H.; Theelen, W.S.; et al. Osimertinib or Platinum-Pemetrexed in EGFR T790M-Positive Lung Cancer. New Engl. J. Med. 2016, 376, 629–640. [Google Scholar] [CrossRef]
- Soria, J.-C.; Ohe, Y.; Vansteenkiste, J.; Reungwetwattana, T.; Chewaskulyong, B.; Lee, K.H.; Dechaphunkul, A.; Imamura, F.; Nogami, N.; Kurata, T.; et al. Osimertinib in UntreatedEGFR-Mutated Advanced Non–Small-Cell Lung Cancer. New Engl. J. Med. 2018, 378, 113–125. [Google Scholar] [CrossRef]
- Canale, M.; Petracci, E.; Delmonte, A.; Chiadini, E.; Dazzi, C.; Papi, M.; Capelli, L.; Casanova, C.; De Luigi, N.; Mariotti, M.; et al. Impact of TP53 Mutations on Outcome in EGFR-Mutated Patients Treated with First-Line Tyrosine Kinase Inhibitors. Clin. Cancer Res. 2016, 23, 2195–2202. [Google Scholar] [CrossRef]
- Kim, Y.; Lee, B.; Shim, J.H.; Lee, S.-H.; Park, W.-Y.; Choi, Y.-L.; Sun, J.-M.; Ahn, J.S.; Ahn, M.-J.; Park, K. Concurrent Genetic Alterations Predict the Progression to Target Therapy in EGFR-Mutated Advanced NSCLC. J. Thorac. Oncol. 2019, 14, 193–202. [Google Scholar] [CrossRef]
- VanderLaan, P.A.; Rangachari, D.; Mockus, S.M.; Spotlow, V.; Reddi, H.V.; Malcolm, J.; Costa, D.B. Mutations in TP53, PIK3CA, PTEN and other genes in EGFR mutated lung cancers: Correlation with clinical outcomes. Lung Cancer 2018, 106, 17–21. [Google Scholar] [CrossRef]
- Molina-Vila, M.A.; Bertran-Alamillo, J.; Gascó, A.; Mayo-de-las-Casas, C.; Sánchez-Ronco, M.; Pujantell-Pastor, L.; Majem, M. Nondisruptive p53 Mutations Are Associated with Shorter Survival in Patients with Advanced Non–Small Cell Lung Cancer. Clin. Cancer Res. 2014, 2, 4647–4660. [Google Scholar] [CrossRef]
- Poeta, M.L.; Manola, J.; Goldwasser, M.A.; Forastiere, A.; Benoit, N.; Califano, J.A.; Ridge, J.A.; Goodwin, J.; Kenady, D.; Saunders, J.; et al. TP53 mutations and survival in squamous-cell carcinoma of the head and neck. N. Engl. J. Med. 2007, 357, 2552–2561. [Google Scholar] [CrossRef]
- La Fleur, L.; Falk-Sörqvist, E.; Smeds, P.; Berglund, A.; Sundström, M.; Mattsson, J.S.; Brandén, E.; Koyi, H.; Isaksson, J.; Brunnström, H.; et al. Mutation patterns in a population-based non-small cell lung cancer cohort and prognostic impact of concomitant mutations in KRAS and TP53 or STK11. Lung Cancer 2019, 130, 50–58. [Google Scholar] [CrossRef]
- Zhao, J.; Han, Y.; Li, J.; Chai, R.; Bai, C. Prognostic value of KRAS/TP53/PIK3CA in non-small cell lung cancer. Oncol Lett. 2019, 17, 3233–3240. [Google Scholar] [CrossRef]
- Volckmar, A.-L.; Leichsenring, J.; Kirchner, M.; Christopoulos, P.; Neumann, O.; Budczies, J.; De Oliveira, C.M.M.; Rempel, E.; Buchhalter, I.; Brandt, R.; et al. Combined targeted DNA and RNA sequencing of advanced NSCLC in routine molecular diagnostics: Analysis of the first 3,000 Heidelberg cases. Int. J. Cancer 2019, 145, 649–661. [Google Scholar] [CrossRef]
- Jiao, X.-D.; Qin, B.-D.; You, P.; Cai, J.; Zang, Y.-S. The prognostic value of TP53 and its correlation with EGFR mutation in advanced non-small cell lung cancer, an analysis based on cBioPortal data base. Lung Cancer 2018, 123, 70–75. [Google Scholar] [CrossRef]
- Gu, J.; Zhou, Y.; Huang, L.; Ou, W.; Wu, J.; Li, S.; Xu, J.; Feng, J.; Liu, B. TP53 mutation is associated with a poor clinical outcome for non-small cell lung cancer: Evidence from a meta-analysis. Mol. Clin. Oncol. 2016, 5, 705–713. [Google Scholar] [CrossRef]
- Aggarwal, C.; Davis, C.W.; Mick, R.; Thompson, J.C.; Ahmed, S.; Jeffries, S.; Bagley, S.; Gabriel, P.; Evans, T.L.; Bauml, J.M.; et al. Influence of TP53 Mutation on Survival in Patients With Advanced EGFR-Mutant Non–Small-Cell Lung Cancer. JCO Precis. Oncol. 2018, 1–29. [Google Scholar] [CrossRef]
- Yu, H.A.; Arcila, M.E.; Rekhtman, N.; Sima, C.S.; Zakowski, M.F.; Pao, W.; Kris, M.; Miller, V.A.; Ladanyi, M.; Riely, G.J. Analysis of tumor specimens at the time of acquired resistance to EGFR-TKI therapy in 155 patients with EGFR-mutant lung cancers. Clin. Cancer Res. 2013, 19, 2240–2247. [Google Scholar] [CrossRef]
- Hou, H.; Qin, K.; Liang, Y.; Zhang, C.; Liu, N.; Jiang, H.; Liu, K.; Zhu, J.; Lv, H.; Li, T.; et al. Concurrent TP53 mutations predict poor outcomes of EGFR-TKI treatments in Chinese patients with advanced NSCLC. Cancer Manag. Res. 2019, 11, 5665–5675. [Google Scholar] [CrossRef]
- Jin, Y.; Shi, X.; Zhao, J.; He, Q.; Chen, M.; Yan, J.; Ou, Q.; Wu, X.; Shao, Y.W.; Yu, X. Mechanisms of primary resistance to EGFR targeted therapy in advanced lung adenocarcinomas. Lung Cancer 2018, 124, 110–116. [Google Scholar] [CrossRef]
- Chen, M.; Xu, Y.; Zhao, J.; Zhong, W.; Zhang, L.; Bi, Y.; Wang, M. Concurrent Driver Gene Mutations as Negative Predictive Factors in Epidermal Growth Factor Receptor-Positive Non-Small Cell Lung Cancer. EBioMedicine 2019, 42, 304–310. [Google Scholar] [CrossRef]
- Comprehensive molecular characterization of gastric adenocarcinoma. Nature 2014, 513, 202–209. [CrossRef] [PubMed]
- Corrigendum: Comprehensive molecular profiling of lung adenocarcinoma. Nature 2014, 514, 262. [CrossRef]
- Shi, J.; Hua, X.; Zhu, B.; Ravichandran, S.; Wang, M.; Nguyen, C.; Brodie, S.A.; Palleschi, A.; Alloisio, M.; Pariscenti, G.; et al. Somatic Genomics and Clinical Features of Lung Adenocarcinoma: A Retrospective Study. PLoS Med. 2016, 13, 1–24. [Google Scholar] [CrossRef] [PubMed]
- Jamal-Hanjani, M.; Wilson, G.A.; McGranahan, N.; Birkbak, N.J.; Watkins, T.B.; Veeriah, S.; Shafi, S.; Johnson, D.H.; Mitter, R.; Rosenthal, R.; et al. Tracking the Evolution of Non-Small-Cell Lung Cancer. New Engl. J. Med. 2017, 376, 2109–2121. [Google Scholar] [CrossRef]
- Brosh, R.; Rotter, V. When mutants gain new powers: news from the mutant p53 field. Nat. Rev. Cancer 2009, 9, 701. [Google Scholar] [CrossRef]
- Muller, P.A.J.; Vousden, K.H. P53 mutations in cancer. Nat. Cell Biol. 2013, 15, 2–8. [Google Scholar] [CrossRef]
- Liu, Y.; Xu, F.; Wang, Y.; Wu, Q.; Wang, B.; Yao, Y.; Zhang, Y.; Han-Zhang, H.; Ye, J.; Zhang, L.; et al. Mutations in exon 8 of TP53 are associated with shorter survival in patients with advanced lung cancer. Oncol. Lett. 2019, 18, 3159–3169. [Google Scholar] [CrossRef]
- Labbé, C.; Cabanero, M.; Korpanty, G.J.; Tomasini, P.; Doherty, M.K.; Mascaux, C.; Leighl, N.B. Lung Cancer Prognostic and predictive e ff ects of TP53 co-mutation in patients with EGFR-mutated non-small cell lung cancer (NSCLC). Lung Cancer 2017, 111, 23–29. [Google Scholar] [CrossRef]
- Xu, Y.; Tong, X.; Yan, J.; Wu, X.; Shao, Y.W.; Fan, Y. Short-Term Responders of Non–Small Cell Lung Cancer Patients to EGFR Tyrosine Kinase Inhibitors Display High Prevalence of TP53 Mutations and Primary Resistance Mechanisms. Transl. Oncol. 2018, 11, 1364–1369. [Google Scholar] [CrossRef]
- Leonetti, A.; Sharma, S.; Minari, R.; Perego, P.; Giovannetti, E.; Tiseo, M. Resistance mechanisms to osimertinib in EGFR-mutated non-small cell lung cancer. Br. J. Cancer 2019, 121, 725–737. [Google Scholar] [CrossRef]

| Characteristic | n | (%) |
|---|---|---|
| Gender | ||
| Female | 86 | (62.2) |
| Male | 50 | (36.8) |
| Age at first line TKI | ||
| Mean ± SD | 67.6 ± 11.2 | |
| Smoking habit † | ||
| Never smoker | 60 | (50.9) |
| Former smoker | 34 | (28.8) |
| Current smoker | 24 | (20.3) |
| Type of EGFR mutation | ||
| Exon 19 deletion | 73 | (53.7) |
| Exon 21 L858R | 48 | (35.3) |
| Other uncommon mutations | 15 | (11.0) |
| Type of EGFR exon 19 deletion | ||
| No exon 19 deletion | 63 | (46.3) |
| Deletion starts at codon 746 | 62 | (45.6) |
| Deletion starts at codon 747 | 11 | (8.1) |
| Type of TKI received in first line setting | ||
| Erlotinib * | 57 | (41.9) |
| Gefitinib | 42 | (30.9) |
| Afatinib | 37 | (27.2) |
| TP53 mutation | ||
| Wild type | 94 | (69.1) |
| Exon 5 | 12 | (8.8) |
| Exon 6 | 6 | (4.4) |
| Exon 7 | 13 | (9.6) |
| Exon 8 | 11 | (8.1) |
| Type of TP53 mutation | ||
| Wild type | 94 | (69.1) |
| Disruptive | 11 | (8.1) |
| Non-disruptive | 31 | (22.8) |
| All EGFR Mutations (n = 136) | Exon 19 Deletion (n = 73) | Exon 21 L858R (n = 48) | Other EGFR Mutations (n = 15) | p | |||||
|---|---|---|---|---|---|---|---|---|---|
| n | (%) | n | (%) | n | (%) | n | (%) | ||
| Best response † | 0.026 | ||||||||
| CR | 13 | (9.9) | 8 | (11.3) | 4 | (8.5) | 1 | (7.1) | |
| PR | 76 | (57.6) | 47 | (66.2) | 22 | (46.8) | 7 | (50.0) | |
| SD | 29 | (22.0) | 7 | (9.9) | 17 | (36.2) | 5 | (35.7) | |
| PD | 14 | (10.6) | 9 | (12.7) | 4 | (8.5) | 1 | (7.1) | |
| ORR | 89 | (67.4) | 55 | (77.5) | 26 | (55.3) | 6 | (54.6) | 0.029 |
| DCR | 118 | (89.4) | 62 | (87.3) | 43 | (91.5) | 11 | (100.0) | 0.844 |
| Duration of response | 0.260 | ||||||||
| Short-term responders | 36 | (26.4) | 19 | (26.0) | 10 | (20.8) | 7 | (46.7) | |
| Medium-term responders | 87 | (64.0) | 45 | (61.6) | 35 | (72.9) | 7 | (46.7) | |
| Long-term responders | 13 | (9.6) | 9 | (12.3) | 2 | (6.3) | 1 | (6.6) | |
| PFS | |||
|---|---|---|---|
| HR | 95% CI | p | |
| Exon 19 deletion | |||
| No | 1 | ||
| Yes | |||
| 0–6 months | 0.56 | (0.35–0.89) | 0.014 |
| 6–12 months | 0.67 | (0.40–1.12) | 0.123 |
| >12 months | 1.27 | (0.80–2.03) | 0.314 |
| TP53 exon 8 mutations | |||
| Wild-type TP53 | 1 | ||
| Non-Exon 8 mutations | 1.02 | 0.73–1.42 | 0.905 |
| Exon 8 mutations | 1.81 | 1.13–2.92 | 0.014 |
| PFS | OS | |||||
|---|---|---|---|---|---|---|
| HR | (95% CI) | p | HR | (95% CI) | p | |
| TP53 Exon 8 | ||||||
| Wild type | 1 | 1 | ||||
| Non-Exon 8 mutations | 1.15 | (0.37–3.59) | 0.811 | 1.55 | (0.42–5.76) | 0.514 |
| Exon 8 mutations | 2.39 | (0.77–7.45) | 0.134 | 4.86 | (1.25–18.90) | 0.023 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Canale, M.; Petracci, E.; Delmonte, A.; Bronte, G.; Chiadini, E.; Ludovini, V.; Dubini, A.; Papi, M.; Baglivo, S.; De Luigi, N.; et al. Concomitant TP53 Mutation Confers Worse Prognosis in EGFR-Mutated Non-Small Cell Lung Cancer Patients Treated with TKIs. J. Clin. Med. 2020, 9, 1047. https://doi.org/10.3390/jcm9041047
Canale M, Petracci E, Delmonte A, Bronte G, Chiadini E, Ludovini V, Dubini A, Papi M, Baglivo S, De Luigi N, et al. Concomitant TP53 Mutation Confers Worse Prognosis in EGFR-Mutated Non-Small Cell Lung Cancer Patients Treated with TKIs. Journal of Clinical Medicine. 2020; 9(4):1047. https://doi.org/10.3390/jcm9041047
Chicago/Turabian StyleCanale, Matteo, Elisabetta Petracci, Angelo Delmonte, Giuseppe Bronte, Elisa Chiadini, Vienna Ludovini, Alessandra Dubini, Maximilian Papi, Sara Baglivo, Nicoletta De Luigi, and et al. 2020. "Concomitant TP53 Mutation Confers Worse Prognosis in EGFR-Mutated Non-Small Cell Lung Cancer Patients Treated with TKIs" Journal of Clinical Medicine 9, no. 4: 1047. https://doi.org/10.3390/jcm9041047
APA StyleCanale, M., Petracci, E., Delmonte, A., Bronte, G., Chiadini, E., Ludovini, V., Dubini, A., Papi, M., Baglivo, S., De Luigi, N., Verlicchi, A., Chiari, R., Landi, L., Metro, G., Burgio, M. A., Crinò, L., & Ulivi, P. (2020). Concomitant TP53 Mutation Confers Worse Prognosis in EGFR-Mutated Non-Small Cell Lung Cancer Patients Treated with TKIs. Journal of Clinical Medicine, 9(4), 1047. https://doi.org/10.3390/jcm9041047






