Dynamic Changes of Inhibitory Killer-Immunoglobulin-Like Receptors on NK Cells after Allogeneic Hematopoietic Stem Cell Transplantation: An Initial Study
Abstract
1. Introduction
2. Methods
2.1. Donor/Patient Samples and Data
2.2. NK Cell Isolation and Culture
2.3. Flow Cytometry
2.4. KIR Genotyping
2.5. Data Evaluation
3. Results
3.1. Expression of Inhibitory KIRs and Their Correlation with Clinical Data
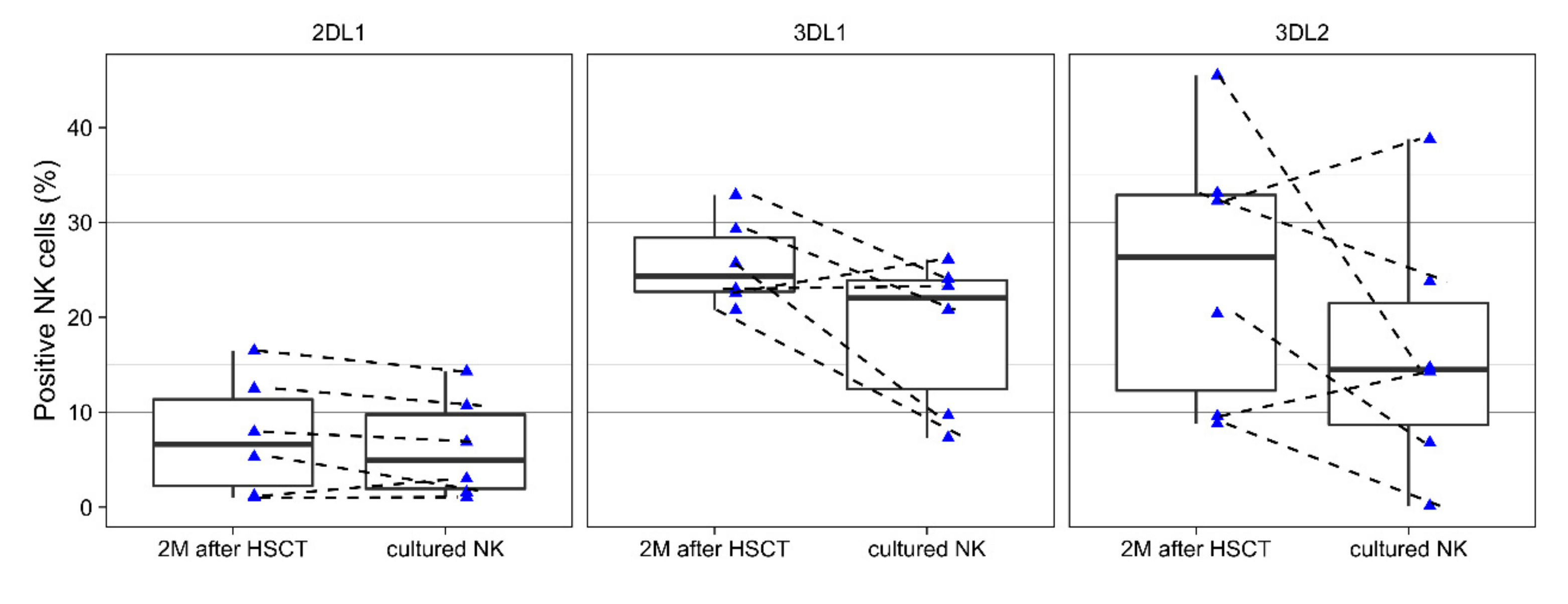
3.2. Changes of KIRs During Cell Culture
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Karre, K. Immunology. A perfect mismatch. Science 2002, 295, 2029–2031. [Google Scholar] [CrossRef]
- Caligiuri, M.A.; Velardi, A.; Scheinberg, D.A.; Borrello, I.M. Immunotherapeutic approaches for hematologic malignancies. Hematol. Am. Soc. Hematol. Educ. Program 2004, 1, 337–353. [Google Scholar] [CrossRef] [PubMed]
- Stern, L.; McGuire, H.; Avdic, S.; Rizzetto, S.; Fazekas de St Groth, B.; Luciani, F.; Slobedman, B.; Blyth, E. Mass Cytometry for the Assessment of Immune Reconstitution After Hematopoietic Stem Cell Transplantation. Front. Immunol. 2018, 9, 1672. [Google Scholar] [CrossRef]
- Della Chiesa, M.; Muccio, L.; Moretta, A. CMV induces rapid NK cell maturation in HSCT recipients. Immunol. Lett. 2013, 155, 11–13. [Google Scholar] [CrossRef]
- Vitale, C.; Chiossone, L.; Cantoni, C.; Morreale, G.; Cottalasso, F.; Moretti, S.; Pistorio, A.; Haupt, R.; Lanino, E.; Dini, G.; et al. The corticosteroid-induced inhibitory effect on NK cell function reflects down-regulation and/or dysfunction of triggering receptors involved in natural cytotoxicity. Eur. J. Immunol. 2004, 34, 3028–3038. [Google Scholar] [CrossRef]
- Paul, S.; Lal, G. The Molecular Mechanism of Natural Killer Cells Function and Its Importance in Cancer Immunotherapy. Front. Immunol. 2017, 8, 1124. [Google Scholar] [CrossRef]
- Horton, N.C.; Mathew, P.A. NKp44 and Natural Cytotoxicity Receptors as Damage-Associated Molecular Pattern Recognition Receptors. Front. Immunol. 2015, 6, 31. [Google Scholar] [CrossRef] [PubMed]
- Moretta, A.; Locatelli, F.; Moretta, L. Human NK cells: From HLA class I-specific killer Ig-like receptors to the therapy of acute leukemias. Immunol. Rev. 2008, 224, 58–69. [Google Scholar] [CrossRef] [PubMed]
- Middleton, D.; Gonzelez, F. The extensive polymorphism of KIR genes. Immunology 2010, 129, 8–19. [Google Scholar] [CrossRef] [PubMed]
- Shilling, H.G.; Guethlein, L.A.; Cheng, N.W.; Gardiner, C.M.; Rodriguez, R.; Tyan, D.; Parham, P. Allelic polymorphism synergizes with variable gene content to individualize human KIR genotype. J. Immunol. 2002, 168, 2307–2315. [Google Scholar] [CrossRef] [PubMed]
- Impola, U.; Turpeinen, H.; Alakulppi, N.; Linjama, T.; Volin, L.; Niittyvuopio, R.; Partanen, J.; Koskela, S. Donor Haplotype B of NK KIR Receptor Reduces the Relapse Risk in HLA-Identical Sibling Hematopoietic Stem Cell Transplantation of AML Patients. Front. Immunol. 2014, 5, 405. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Beziat, V.; Traherne, J.A.; Liu, L.L.; Jayaraman, J.; Enqvist, M.; Larsson, S.; Trowsdale, J.; Malmberg, K.J. Influence of KIR gene copy number on natural killer cell education. Blood 2013, 121, 4703–4707. [Google Scholar] [CrossRef] [PubMed]
- Koutsakos, M.; McWilliam, H.E.G.; Aktepe, T.E.; Fritzlar, S.; Illing, P.T.; Mifsud, N.A.; Purcell, A.W.; Rockman, S.; Reading, P.C.; Vivian, J.P.; et al. Downregulation of MHC Class I Expression by Influenza A and B Viruses. Front. Immunol. 2019, 10, 1158. [Google Scholar] [CrossRef]
- Gaafar, A.; Sheereen, A.; Almohareb, F.; Eldali, A.; Chaudhri, N.; Mohamed, S.Y.; Hanbali, A.; Shaheen, M.; Alfraih, F.; El Fakih, R.; et al. Prognostic role of KIR genes and HLA-C after hematopoietic stem cell transplantation in a patient cohort with acute myeloid leukemia from a consanguineous community. Bone Marrow Transplant. 2018, 53, 1170–1179. [Google Scholar] [CrossRef]
- Boudreau, J.E.; Hsu, K.C. Natural killer cell education in human health and disease. Curr. Opin. Immunol. 2018, 50, 102–111. [Google Scholar] [CrossRef] [PubMed]
- Ewen, E.M.; Pahl, J.H.W.; Miller, M.; Watzl, C.; Cerwenka, A. KIR downregulation by IL-12/15/18 unleashes human NK cells from KIR/HLA-I inhibition and enhances killing of tumor cells. Eur. J. Immunol. 2018, 48, 355–365. [Google Scholar] [CrossRef] [PubMed]
- Ullah, M.A.; Hill, G.R.; Tey, S.K. Functional Reconstitution of Natural Killer Cells in Allogeneic Hematopoietic Stem Cell Transplantation. Front. Immunol. 2016, 7, 144. [Google Scholar] [CrossRef]
- Daher, M.; Rezvani, K. Next generation natural killer cells for cancer immunotherapy: The promise of genetic engineering. Curr. Opin. Immunol. 2018, 51, 146–153. [Google Scholar] [CrossRef]
- Maniangou, B.; Legrand, N.; Alizadeh, M.; Guyet, U.; Willem, C.; David, G.; Charpentier, E.; Walencik, A.; Retiere, C.; Gagne, K. Killer Immunoglobulin-Like Receptor Allele Determination Using Next-Generation Sequencing Technology. Front. Immunol. 2017, 8, 547. [Google Scholar] [CrossRef]
- Wagner, I.; Schefzyk, D.; Pruschke, J.; Schofl, G.; Schone, B.; Gruber, N.; Lang, K.; Hofmann, J.; Gnahm, C.; Heyn, B.; et al. Allele-Level KIR Genotyping of More Than a Million Samples: Workflow, Algorithm, and Observations. Front. Immunol. 2018, 9, 2843. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing, Vienna, Austria. 2018. Available online: https://www.R-project.org/ (accessed on 12 March 2020).
- Wickham, H. ggplot2: Elegant Graphics for Data Analysis; Springer: New York, NY, USA, 2016. [Google Scholar]
- Pende, D.; Falco, M.; Vitale, M.; Cantoni, C.; Vitale, C.; Munari, E.; Bertaina, A.; Moretta, F.; del Zotto, G.; Pietra, G.; et al. Killer Ig-Like Receptors (KIRs): Their Role in NK Cell Modulation and Developments Leading to Their Clinical Exploitation. Front. Immunol. 2019, 10, 1179. [Google Scholar] [CrossRef] [PubMed]
- Falco, M.; Moretta, L.; Moretta, A.; Bottino, C. KIR and KIR ligand polymorphism: A new area for clinical applications? Tissue Antigens 2013, 82, 363–373. [Google Scholar] [CrossRef] [PubMed]
- Solloch, U.V.; Schefzyk, D.; Schäfer, G.; Massalski, C.; Kohler, M.; Pruschke, J.; Heidl, A.; Schetelig, J.; Schmidt, A.H.; Lange, V.; et al. Estimation of German KIR Allele Group Haplotype Frequencies. Front. Immunol. 2020, 11, 1–429. [Google Scholar] [CrossRef] [PubMed]
- Vierra-Green, C.; Roe, D.; Hou, L.; Hurley, C.K.; Rajalingam, R.; Reed, E.; Lebedeva, T.; Yu, N.; Stewart, M.; Noreen, H.; et al. Allele-level haplotype frequencies and pairwise linkage disequilibrium for 14 KIR loci in 506 European-American individuals. PLoS ONE 2012, 7, e47491. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Wang, T.; Hu, X.; Zhang, H.; Chen, L.; Bao, X.; He, J. Study of KIR gene expression at the mRNA level in specific donor-derived NK cells after allogeneic HSCT. Immunogenetics 2020, 72, 135–141. [Google Scholar] [CrossRef]
- Nguyen, S.; Achour, A.; Souchet, L.; Vigouroux, S.; Chevallier, P.; Furst, S.; Sirvent, A.; Bay, J.O.; Socié, G.; Ceballos, P.; et al. Clinical impact of NK-cell reconstitution after reduced intensity conditioned unrelated cord blood transplantation in patients with acute myeloid leukemia: Analysis of a prospective phase II multicenter trial on behalf of the Societe Francaise de Greffe de Moelle Osseuse et Therapie Cellulaire and Eurocord. Bone Marrow Transplant. 2017, 52, 1428–1435. [Google Scholar]
- Giebel, S.; Dziaczkowska, J.; Czerw, T.; Wojnar, J.; Krawczyk-Kulis, M.; Nowak, I.; Holowiecka, A.; Segatti, A.; Kyrcz-Krzemien, S.; Kusnierczyk, P.; et al. Sequential recovery of NK cell receptor repertoire after allogeneic hematopoietic SCT. Bone Marrow Transplant. 2010, 45, 1022–1030. [Google Scholar] [CrossRef][Green Version]
- Nutalai, R.; Gaudieri, S.; Jumnainsong, A.; Leelayuwat, C. Regulation of KIR3DL3 Expression via Mirna. Genes 2019, 10, 603. [Google Scholar] [CrossRef]
- Charoudeh, H.N.; Terszowski, G.; Czaja, K.; Gonzalez, A.; Schmitter, K.; Stern, M. Modulation of the natural killer cell KIR repertoire by cytomegalovirus infection. Eur. J. Immunol. 2013, 43, 480–487. [Google Scholar] [CrossRef]
- Moesta, A.K.; Parham, P. Diverse functionality among human NK cell receptors for the C1 epitope of HLA-C: KIR2DS2, KIR2DL2, and KIR2DL3. Front. Immunol. 2012, 3, 336. [Google Scholar] [CrossRef]
- Jones, D.C.; Peacock, S.; Hughes, D.; Traherne, J.A.; Allen, R.L.; Barnardo, M.C.; Friend, P.; Taylor, C.J.; Fuggle, S.; Trowsdale, J.; et al. Killer immunoglobulin-like receptor gene repertoire influences viral load of primary human cytomegalovirus infection in renal transplant patients. Genes Immun. 2014, 15, 562–568. [Google Scholar] [CrossRef] [PubMed]
- Behrendt, C.E.; Nakamura, R.; Forman, S.J.; Zaia, J.A. Donor killer immunoglobulin-like receptor genes and reactivation of cytomegalovirus after HLA-matched hematopoietic stem-cell transplantation: HLA-C allotype is an essential cofactor. Front. Immunol. 2013, 4, 36. [Google Scholar] [CrossRef] [PubMed]
- Cisneros, E.; Moraru, M.; Gomez-Lozano, N.; Lopez-Botet, M.; Vilches, C. KIR2DL5: An Orphan Inhibitory Receptor Displaying Complex Patterns of Polymorphism and Expression. Front. Immunol. 2012, 3, 289. [Google Scholar] [CrossRef]
- El-Serafi, I.; Abedi-Valugerdi, M.; Potacova, Z.; Afsharian, P.; Mattsson, J.; Moshfegh, A.; Hassan, M. Cyclophosphamide alters the gene expression profile in patients treated with high doses prior to stem cell transplantation. PLoS ONE 2014, 9, e86619. [Google Scholar] [CrossRef]
- Neuchel, C.; Furst, D.; Niederwieser, D.; Bunjes, D.; Tsamadou, C.; Wulf, G.; Pfreundschuh, M.; Wagner, E.; Stuhler, G.; Einsele, H.; et al. Impact of Donor Activating KIR Genes on HSCT Outcome in C1-Ligand Negative Myeloid Disease Patients Transplanted with Unrelated Donors-A Retrospective Study. PLoS ONE 2017, 12, e0169512. [Google Scholar] [CrossRef] [PubMed]
- Verheyden, S.; Demanet, C. Susceptibility to myeloid and lymphoid leukemia is mediated by distinct inhibitory KIR-HLA ligand interactions. Leukemia 2006, 20, 1437–1438. [Google Scholar] [CrossRef] [PubMed]
- Vey, N.; Karlin, L.; Sadot-Lebouvier, S.; Broussais, F.; Berton-Rigaud, D.; Rey, J.; Charbonnier, A.; Marie, D.; Andre, P.; Paturel, C.; et al. A phase 1 study of lirilumab (antibody against killer immunoglobulin-like receptor antibody KIR2D.; IPH2102) in patients with solid tumors and hematologic malignancies. Oncotarget 2018, 9, 17675–17688. [Google Scholar] [CrossRef]
- Holubova, M.; Leba, M.; Gmucova, H.; Caputo, V.S.; Jindra, P.; Lysak, D. Improving the Clinical Application of Natural Killer Cells by Modulating Signals Signal from Target Cells. Int. J. Mol. Sci. 2019, 20, 3472. [Google Scholar] [CrossRef]

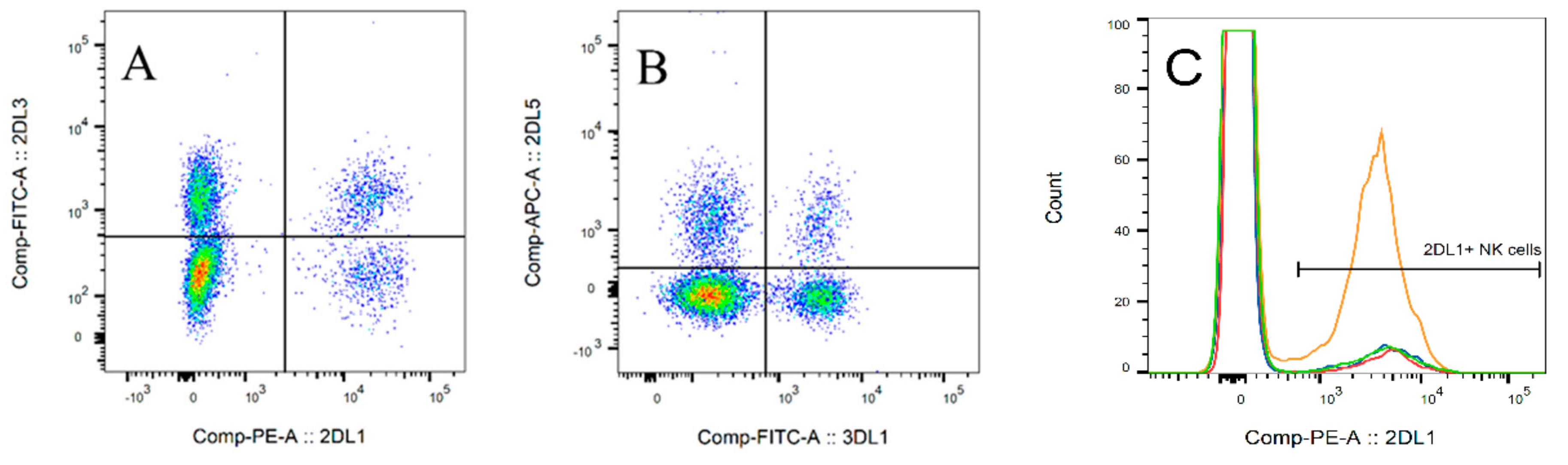
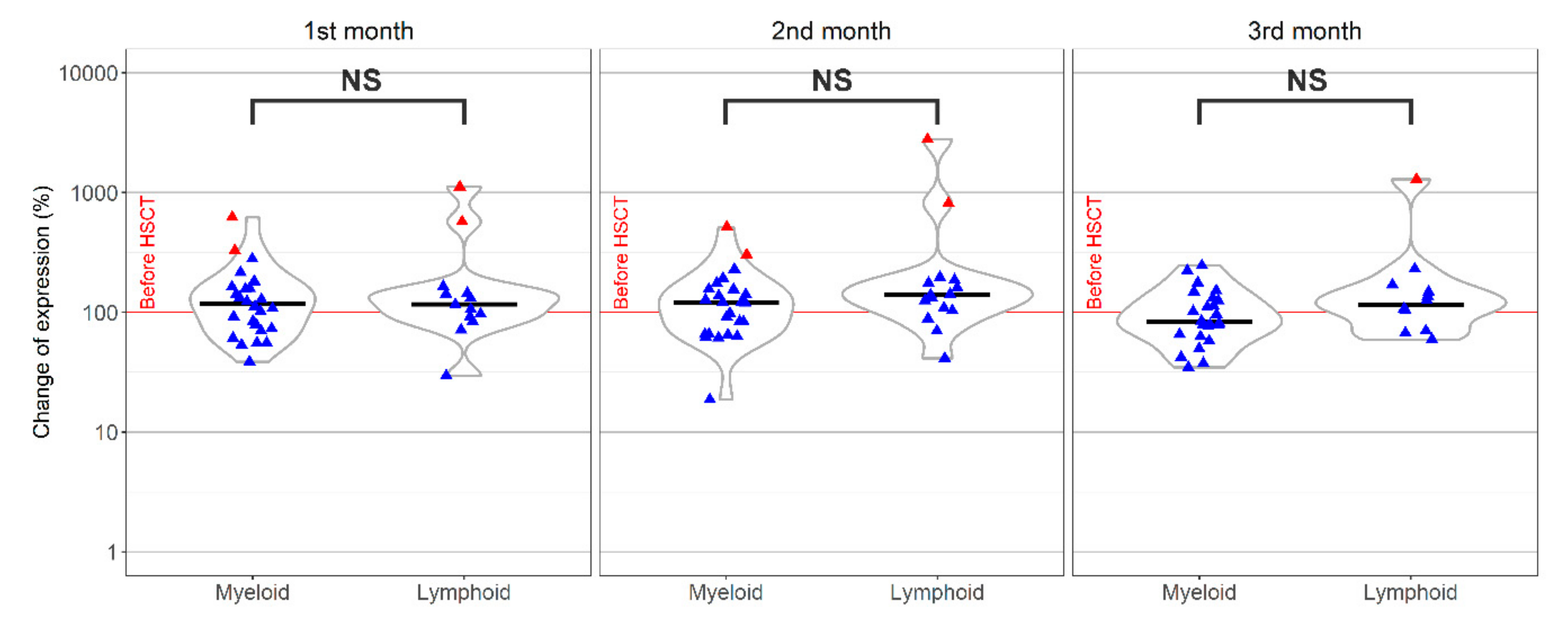
 . Median,
. Median,  .
.
 . Median,
. Median,  .
.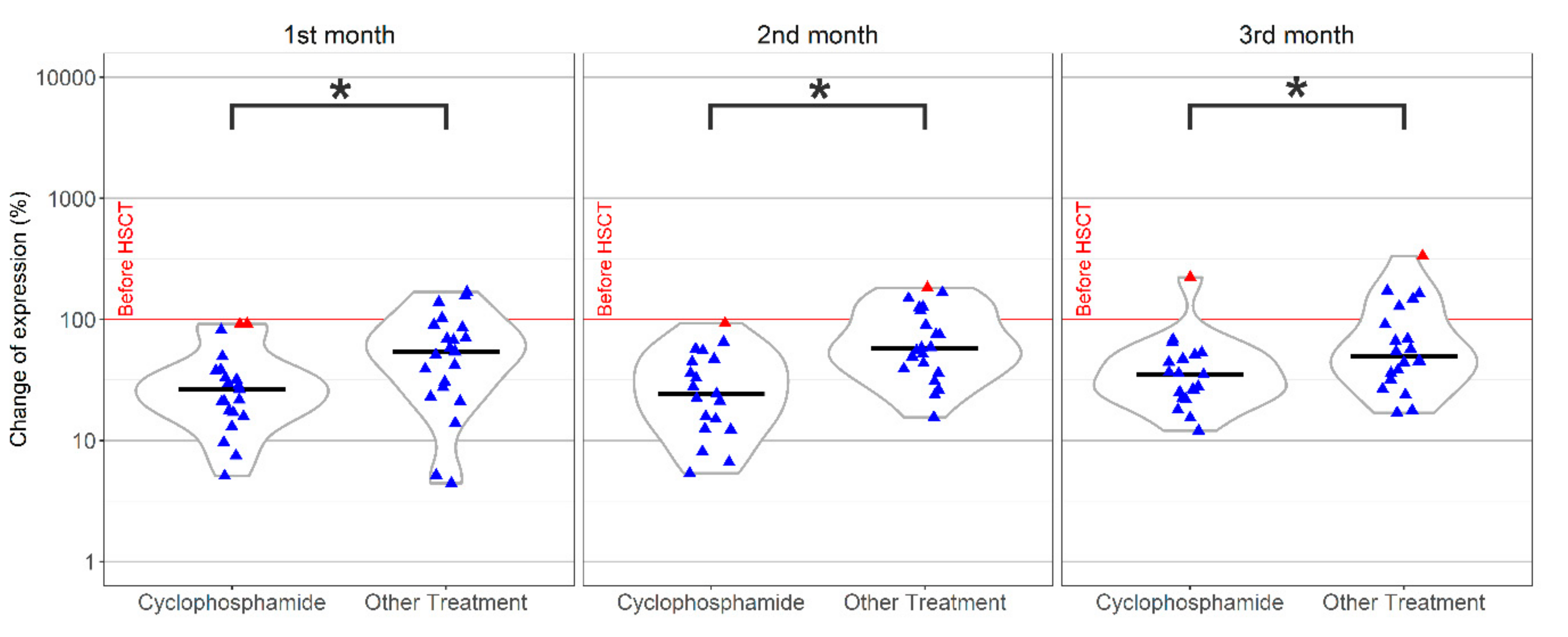
 ; Median,
; Median,  .
.
 ; Median,
; Median,  .
.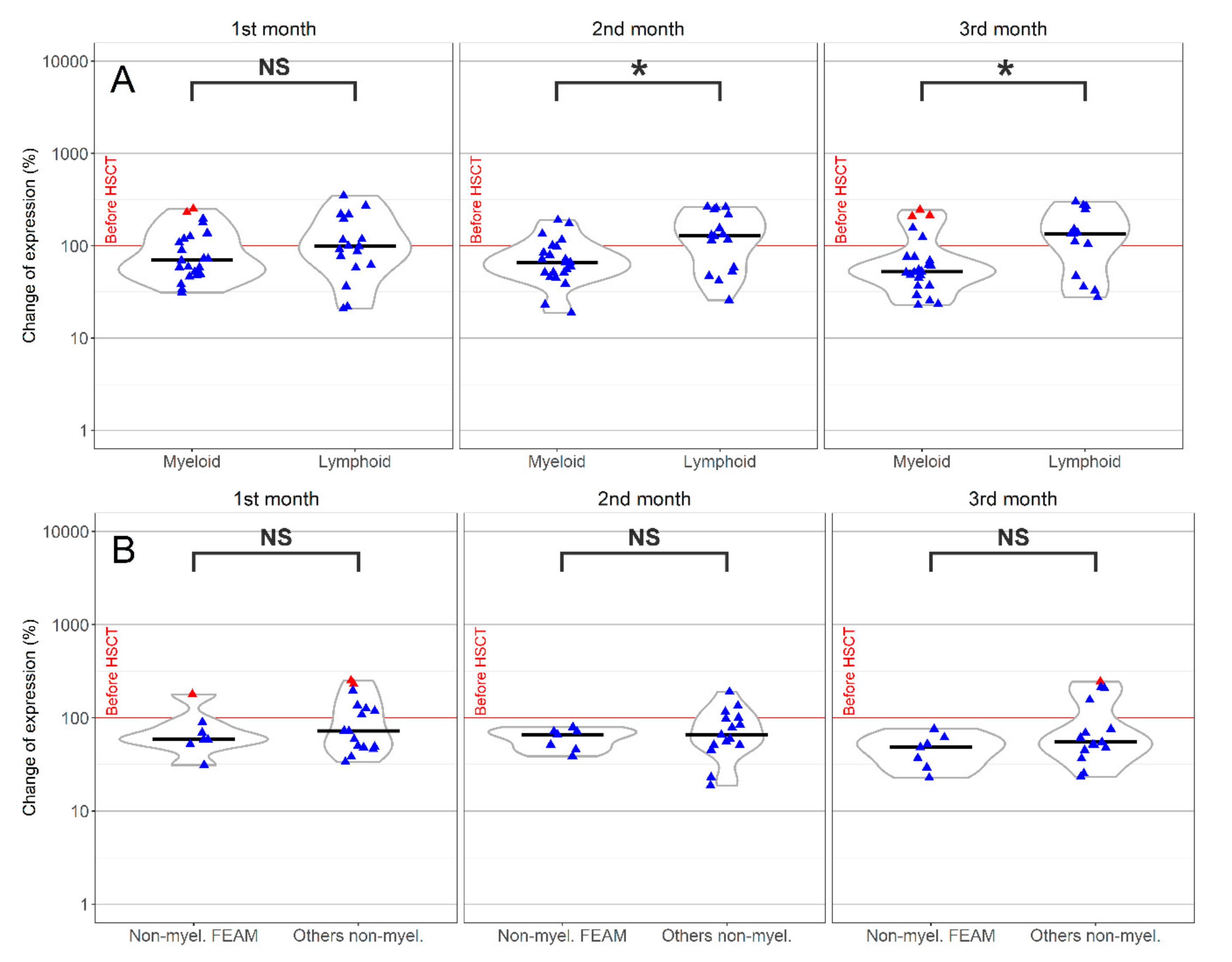
 ; Median,
; Median,  .
.
 ; Median,
; Median,  .
.
| Characteristic | Type | No. of Individuals | ||
|---|---|---|---|---|
| Diagnosis | Myeloid | 27 | ||
| Lymphoid | 17 | |||
| Sex (patients) | Male | 15 | ||
| Female | 29 | |||
| Age (patients) | Median: 50 | Range: 20–74 | ||
| Sex (donors) | Male | 28 | ||
| Female | 16 | |||
| Age (donors) | Median: 37.5 | Range: 20–60 | ||
| HSCT type | Matched | 19 | ||
| Mismatched | 25 | |||
| Disease status | CR | 25 | ||
| (at time of HSCT) | Non CR | 19 | ||
| Conditioning | Myeloablative | 5 | ||
| Non-myeloablative | 39 | |||
| Post-transplant treatment | Cyclophosphamide | 22 | ||
| Others | 22 |
| Time Point | 3DL1 | 3DL2+/3DL1− | 3DL2+/3DL1+ | 2DL5 | 2DL3 | 2DL1 | 2DL2+/2DL3+ | 2DL2+/2DL3− | 2DL1+/2DL3+ | |
|---|---|---|---|---|---|---|---|---|---|---|
| Before HSCT | Median | 17.1 | 1.6 | 21.5 | 0.2 | 15.8 | 16.6 | 33.8 | 14.4 | 3.7 |
| (Range) | (0–47.9) | (0–23.7) | (0.2–50.8) | (0–4.8) | (0.2–74.2) | (0.2–46) | (16.2–76) | (1.9–50.2) | (0–35) | |
| 1 month * | Median | 18.5 | 4.2 | 24.2 | 0.2 | 17.5 | 5.0 | 27.2 | 7.1 | 2.1 |
| (Range) | (0–46.9) | (0.5–63.5) | (0.7–77.1) | (0–9.3) | (0–40.6) | (0.4–37.9) | (3.9–58.1) | (0.6–49.5) | (0–15) | |
| 2 months * | Median | 20.5 | 2.7 | 25.4 | 0.2 | 17.5 | 5.8 | 27.0 | 6.6 | 2.7 |
| (Range) | (0–46) | (0.1–51.2) | (0.2–74.2) | (0–6.6) | (0.1–36.9) | (0.1–40.7) | (7.1–52.6) | (0.2–41.3) | (0–16.6) | |
| 3 months | Median | 18.0 | 2.3 | 20.7 | 0.2 | 16.7 | 7.8 * | 25.0 * | 9.9 | 2.7 |
| (Range) | (0–60) | (0–24.7) | (0.5–62.5) | (0–10.9) | (0.6–37.5) | (1.2–40.4) | (10.4–58) | (0.5–38.9) | (0.2–21.6) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dekojová, T.; Houdová, L.; Fatka, J.; Pitule, P.; Ostašov, P.; Caputo, V.S.; Gmucová, H.; Lysák, D.; Jindra, P.; Holubová, M. Dynamic Changes of Inhibitory Killer-Immunoglobulin-Like Receptors on NK Cells after Allogeneic Hematopoietic Stem Cell Transplantation: An Initial Study. J. Clin. Med. 2020, 9, 3502. https://doi.org/10.3390/jcm9113502
Dekojová T, Houdová L, Fatka J, Pitule P, Ostašov P, Caputo VS, Gmucová H, Lysák D, Jindra P, Holubová M. Dynamic Changes of Inhibitory Killer-Immunoglobulin-Like Receptors on NK Cells after Allogeneic Hematopoietic Stem Cell Transplantation: An Initial Study. Journal of Clinical Medicine. 2020; 9(11):3502. https://doi.org/10.3390/jcm9113502
Chicago/Turabian StyleDekojová, Tereza, Lucie Houdová, Jiří Fatka, Pavel Pitule, Pavel Ostašov, Valentina S. Caputo, Hana Gmucová, Daniel Lysák, Pavel Jindra, and Monika Holubová. 2020. "Dynamic Changes of Inhibitory Killer-Immunoglobulin-Like Receptors on NK Cells after Allogeneic Hematopoietic Stem Cell Transplantation: An Initial Study" Journal of Clinical Medicine 9, no. 11: 3502. https://doi.org/10.3390/jcm9113502
APA StyleDekojová, T., Houdová, L., Fatka, J., Pitule, P., Ostašov, P., Caputo, V. S., Gmucová, H., Lysák, D., Jindra, P., & Holubová, M. (2020). Dynamic Changes of Inhibitory Killer-Immunoglobulin-Like Receptors on NK Cells after Allogeneic Hematopoietic Stem Cell Transplantation: An Initial Study. Journal of Clinical Medicine, 9(11), 3502. https://doi.org/10.3390/jcm9113502





