Betaglycan Gene (TGFBR3) Polymorphism Is Associated with Increased Risk of Endometrial Cancer
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Population
2.2. Ethical Approval
2.3. Lifestyle Risk Factors
2.4. Genomic DNA Isolation
2.5. SNP Selection and Genotyping
2.6. Expression of the TGFBR3 Gene
2.7. Statistical Analysis
2.8. Bioinformatic Analysis
3. Results
3.1. SNPs Association with EC
3.2. Association of the TGFBR3 Gene SNPs and Betaglycan mRNA Expression—Genotype-Phenotype Analysis
4. Discussion
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Ferlay, J.; Colombet, M.; Soerjomataram, I.; Mathers, C.; Parkin, D.M.; Piñeros, M.; Znaor, A.; Bray, F. Estimating the global cancer incidence and mortality in 2018: GLOBOCAN sources and methods. Int. J. Cancer 2019, 144, 1941–1953. [Google Scholar] [CrossRef]
- Didkowska, J.; Wojciechowska, U.; Czaderny, K.; Olasek, P.; Ciuba, A. Cancer in Poland in 2017; Polish National Cancer Registry: Warsaw, Poland, 2019. [Google Scholar]
- Bokhman, J.V. Two pathogenetic types of endometrial carcinoma. Gynecol. Oncol. 1983, 15, 10–17. [Google Scholar] [CrossRef]
- Webb, P.M. Environmental (nongenetic) factors in gynecological cancers: Update and future perspectives. Future Oncol. 2015, 11, 295–307. [Google Scholar] [CrossRef] [PubMed]
- Murali, R.; Soslow, R.A.; Weigelt, B. Classification of endometrial carcinoma: More than two types. Lancet Oncol. 2014, 15, e268–e278. [Google Scholar] [CrossRef]
- Yen, T.T.; Wang, T.L.; Fader, A.N.; Shih, I.M.; Gaillard, S. Molecular Classification and Emerging Targeted Therapy in Endometrial Cancer. Int. J. Gynecol. Pathol. 2020, 39, 26–35. [Google Scholar] [CrossRef]
- Lee, Y.C.; Lheureux, S.; Oza, A.M. Treatment strategies for endometrial cancer: Current practice and perspective. Curr. Opin. Obstet. Gynecol. 2017, 29, 47–58. [Google Scholar] [CrossRef]
- Getz, G.; Gabriel, S.B.; Cibulskis, K.; Lander, E.; Sivachenko, A.; Sougnez, C.; Lawrence, M.; Kandoth, C.; Dooling, D.; Fulton, R.; et al. Integrated genomic characterization of endometrial carcinoma. Nature 2013, 497, 67–73. [Google Scholar] [CrossRef]
- Bell, D.W.; Ellenson, L.H. Molecular Genetics of Endometrial Carcinoma. Annu. Rev. Pathol. Mech. Dis. 2019, 14, 339–367. [Google Scholar] [CrossRef]
- Rossi, L.; Le Frere-Belda, M.A.; Laurent-Puig, P.; Buecher, B.; De Pauw, A.; Stoppa-Lyonnet, D.; Canlorbe, G.; Caron, O.; Borghese, B.; Colas, C.; et al. Clinicopathologic characteristics of endometrial cancer in lynch syndrome A French multicenter study. Int. J. Gynecol. Cancer 2017, 27, 953–960. [Google Scholar] [CrossRef]
- Parekh, T.V.; Gama, P.; Wen, X.; Demopoulos, R.; Munger, J.S.; Carcangiu, M.L.; Reiss, M.; Gold, L.I. Transforming growth factor β signaling is disabled early in human endometrial carcinogenesis concomitant with loss of growth inhibition. Cancer Res. 2002, 62, 2778–2790. [Google Scholar]
- Piestrzeniewicz-Ulanska, D.; Brys, M.; Semczuk, A.; Jakowicki, J.A.; Krajewska, W.M. Expression of TGF-β type I and II receptors in normal and cancerous human endometrium. Cancer Lett. 2002, 186, 231–239. [Google Scholar] [CrossRef]
- Piestrzeniewicz-Ulanska, D.; Brys, M.; Semczuk, A.; Rechberger, T.; Jakowicki, J.A.; Krajewska, W.M. TGF-β signaling is disrupted in endometrioid-type endometrial carcinomas. Gynecol. Oncol. 2004, 95, 173–180. [Google Scholar] [CrossRef] [PubMed]
- Drabsch, Y.; Ten Dijke, P. TGF-β signalling and its role in cancer progression and metastasis. Cancer Metastasis Rev. 2012, 31, 553–568. [Google Scholar] [CrossRef] [PubMed]
- Gordon, K.J.; Blobe, G.C. Role of transforming growth factor-β superfamily signaling pathways in human disease. Biochim. Biophys. Acta Mol. Basis Dis. 2008, 1782, 197–228. [Google Scholar] [CrossRef]
- Cheifetz, S.; Andres, J.L.; Massagué, J. The transforming growth factor-beta receptor type III is a membrane proteoglycan. Domain structure of the receptor. J. Biol. Chem. 1988, 263, 16984–16991. [Google Scholar]
- Bae, H.J.; Eun, J.W.; Noh, J.H.; Kim, J.K.; Jung, K.H.; Xie, H.J.; Park, W.S.; Lee, J.Y.; Nam, S.W. Down-regulation of transforming growth factor β receptor type III in hepatocellular carcinoma is not directly associated with genetic alterations or loss of heterozygosity. Oncol. Rep. 2009, 22, 475–480. [Google Scholar] [CrossRef]
- Bilandzic, M.; Chu, S.; Farnworth, P.G.; Harrison, C.; Nicholls, P.; Wang, Y.; Escalona, R.M.; Fuller, P.J.; Findlay, J.K.; Stenvers, K.L. Loss of betaglycan contributes to the malignant properties of human granulosa tumor cells. Mol. Endocrinol. 2009, 23, 539–548. [Google Scholar] [CrossRef]
- Copland, J.A.; Luxon, B.A.; Ajani, L.; Maity, T.; Campagnaro, E.; Guo, H.; LeGrand, S.N.; Tamboli, P.; Wood, C.G. Genomic profiling identifies alterations in TGFβ signaling through loss of TGFβ receptor expression in human renal cell carcinogenesis and progression. Oncogene 2003, 22, 8053–8062. [Google Scholar] [CrossRef]
- Dong, M.; How, T.; Kirkbride, K.C.; Gordon, K.J.; Lee, J.D.; Hempel, N.; Kelly, P.; Moeller, B.J.; Marks, J.R.; Blobe, G.C. The type III TGF-β receptor suppresses breast cancer progression. J. Clin. Investig. 2007, 117, 206–217. [Google Scholar] [CrossRef]
- Finger, E.C.; Turley, R.S.; Dong, M.; How, T.; Fields, T.A.; Blobe, G.C. TβRIII suppresses non-small cell lung cancer invasiveness and tumorigenicity. Carcinogenesis 2008, 29, 528–535. [Google Scholar] [CrossRef]
- Gordon, K.J.; Dong, M.; Chislock, E.M.; Fields, T.A.; Blobe, G.C. Loss of type III transforming growth factor beta receptor expression increases motility and invasiveness associated with epithelial to mesenchymal transition during pancreatic cancer progression. Carcinogenesis 2008, 29, 252–262. [Google Scholar] [CrossRef] [PubMed]
- Hempel, N.; How, T.; Dong, M.; Murphy, S.K.; Fields, T.A.; Blobe, G.C. Loss of betaglycan expression in ovarian cancer: Role in motility and invasion. Cancer Res. 2007, 67, 5231–5238. [Google Scholar] [CrossRef] [PubMed]
- Iolascon, A.; Giordani, L.; Borriello, A.; Carbone, R.; Izzo, A.; Tonini, G.P.; Gambini, C.; Della Ragione, F. Reduced expression of transforming growth factor-beta receptor type III in high stage neuroblastomas. Br. J. Cancer 2000, 82, 1171–1176. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.L.; Xiao, K.; Xue, B.; Yang, D.; Lei, Z.; Shan, Y.; Zhang, H.T. Dual role of TGFBR3 in bladder cancer. Oncol. Rep. 2013, 30, 1301–1308. [Google Scholar] [CrossRef]
- Sharifi, N.; Hurt, E.M.; Kawasaki, B.T.; Farrar, W.L. TGFBR3 loss and consequences in prostate cancer. Prostate 2007, 67, 301–311. [Google Scholar] [CrossRef]
- Turley, R.S.; Finger, E.C.; Hempel, N.; How, T.; Fields, T.A.; Blobe, G.C. The type III transforming growth factor-beta receptor as a novel tumor suppressor gene in prostate cancer. Cancer Res. 2007, 67, 1090–1098. [Google Scholar] [CrossRef]
- Zakrzewski, P.K.; Mokrosinski, J.; Cygankiewicz, A.I.; Semczuk, A.; Rechberger, T.; Skomra, D.; Krajewska, W.M. Dysregulation of betaglycan expression in primary human endometrial carcinomas. Cancer Investig. 2011, 29, 137–142. [Google Scholar] [CrossRef]
- Shi, Y.Y.; He, L. SHEsis, a powerful software platform for analyses of linkage disequilibrium, haplotype construction, and genetic association at polymorphism loci. Cell Res. 2005, 15, 97–98. [Google Scholar] [CrossRef]
- Boyle, A.P.; Hong, E.L.; Hariharan, M.; Cheng, Y.; Schaub, M.A.; Kasowski, M.; Karczewski, K.J.; Park, J.; Hitz, B.C.; Weng, S.; et al. Annotation of functional variation in personal genomes using RegulomeDB. Genome Res. 2012, 22, 1790–1797. [Google Scholar] [CrossRef]
- Akhurst, R.J. Targeting TGF-β Signaling for Therapeutic Gain. Cold Spring Harb. Perspect. Biol. 2017, 9, a022301. [Google Scholar] [CrossRef]
- Zhang, Y.; Alexander, P.B.; Wang, X.F. TGF-β family signaling in the control of cell proliferation and survival. Cold Spring Harb. Perspect. Biol. 2017, 9, a022145. [Google Scholar] [CrossRef] [PubMed]
- Xie, F.; Ling, L.; Van Dam, H.; Zhou, F.; Zhang, L. TGF-β signaling in cancer metastasis. Acta Biochim. Biophys. Sin. 2018, 50, 121–132. [Google Scholar] [CrossRef] [PubMed]
- Jenabi, E.; Poorolajal, J. The effect of body mass index on endometrial cancer: A meta-analysis. Public Health 2015, 129, 872–880. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Dai, X.; Chen, L.; Lee, A.C.; Tong, M.; Wise, M.; Chen, Q. Body Mass Index Is Positively Associated with Endometrial Cancer in Chinese Women, Especially Prior to Menopause. J. Cancer 2016, 7, 1169–1173. [Google Scholar] [CrossRef]
- Althubiti, M. Mutation frequencies in endometrial cancer patients of different ethnicities and tumor grades: An analytical study. Saudi J. Med. Med. Sci. 2019, 7, 16. [Google Scholar] [CrossRef]
- Park, S.L.; Goodman, M.T.; Zhang, Z.F.; Kolonel, L.N.; Henderson, B.E.; Setiawan, V.W. Body size, adult BMI gain and endometrial cancer risk: The multiethnic cohort. Int. J. Cancer 2010, 126, 490–499. [Google Scholar] [CrossRef]
- Garg, K.; Soslow, R.A. Endometrial carcinoma in women aged 40 years and younger. Arch. Pathol. Lab. Med. 2014, 138, 335–342. [Google Scholar] [CrossRef]
- O’Mara, T.A.; Glubb, D.M.; Kho, P.F.; Thompson, D.J.; Spurdle, A.B. Genome-Wide Association Studies of Endometrial Cancer: Latest Developments and Future Directions. Cancer Epidemiol. Biomark. Prev. 2019, 28, 1095–1102. [Google Scholar] [CrossRef]
- Chen, Q.; Tong, M.; Guo, F.; Lau, S.; Zhao, M. Parity correlates with the timing of developing endometrial cancer, but not subtype of endometrial cancer. J. Cancer 2015, 6, 1087–1092. [Google Scholar] [CrossRef]
- Karageorgi, S.; Hankinson, S.E.; Kraft, P.; De Vivo, I. Reproductive factors and postmenopausal hormone use in relation to endometrial cancer risk in the Nurses’ Health Study cohort 1976–2004. Int. J. Cancer 2010, 126, 208–216. [Google Scholar] [CrossRef]
- Dossus, L.; Allen, N.; Kaaks, R.; Bakken, K.; Lund, E.; Tjonneland, A.; Olsen, A.; Overvad, K.; Clavel-Chapelon, F.; Fournier, A.; et al. Reproductive risk factors and endometrial cancer: The European prospective investigation into cancer and nutrition. Int. J. Cancer 2010, 127, 442–451. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.-H.; Jong, S.; Park, B.-L.; Sub, H.; Yu, S.J.; Cheong, H.S.; Pasaje, C.F.A.; Bae, J.S.; Lee, H.-S.; Shin, H.D.; et al. TGFBR3 Polymorphisms and Its Haplotypes Associated with Chronic Hepatitis B Virus Infection and Age of Hepatocellular Carcinoma. Dig. Dis. 2011, 744, 278–283. [Google Scholar] [CrossRef]
- Charbonneau, B.; Moysich, K.B.; Kalli, K.R.; Oberg, A.L.; Vierkant, R.A.; Fogarty, Z.C.; Block, M.S.; Maurer, M.J.; Goergen, K.M.; Fridley, B.L.; et al. Large-scale evaluation of common variation in regulatory T cell-related genes and ovarian cancer outcome. Cancer Immunol. Res. 2014, 2, 332–340. [Google Scholar] [CrossRef] [PubMed]
- Xin, Z.; Zhang, W.; Xu, A.; Zhang, L.; Yan, T.; Li, Z.; Wu, X.; Zhu, X.; Ma, J.; Li, K.; et al. Polymorphisms in the potential functional regions of the TGF-β 1 and TGF-β receptor genes and disease susceptibility in HBV-related hepatocellular carcinoma patients. Mol. Carcinog. 2012, 51, E123–E131. [Google Scholar] [CrossRef] [PubMed]
- Chand, A.L.; Robertson, D.M.; Shelling, A.N.; Harrison, C.A. Mutational analysis of betaglycan/TGF-βRIII in premature ovarian failure. Fertil. Steril. 2007, 87, 210–212. [Google Scholar] [CrossRef]
- Dalgaard, M.D.; Weinhold, N.; Edsgärd, D.; Silver, J.D.; Pers, T.H.; Nielsen, J.E.; Jørgensen, N.; Juul, A.; Gerds, T.A.; Giwercman, A.; et al. A genome-wide association study of men with symptoms of testicular dysgenesis syndrome and its network biology interpretation. J. Med. Genet. 2012, 49, 58–65. [Google Scholar] [CrossRef]
- Elliott, L.; Ashley-Koch, A.E.; De Castro, L.; Jonassaint, J.; Price, J.; Ataga, K.I.; Levesque, M.C.; Brice Weinberg, J.; Eckman, J.R.; Orringer, E.P.; et al. Genetic polymorphisms associated with priapism in sickle cell disease. Br. J. Haematol. 2007, 137, 262–267. [Google Scholar] [CrossRef]
- Flanagan, J.M.; Frohlich, D.M.; Howard, T.A.; Schultz, W.H.; Driscoll, C.; Nagasubramanian, R.; Mortier, N.A.; Kimble, A.C.; Aygun, B.; Adams, R.J.; et al. Genetic predictors for stroke in children with sickle cell anemia. Blood 2011, 117, 6681–6684. [Google Scholar] [CrossRef]
- Hersh, C.P.; Hansel, N.N.; Barnes, K.C.; Lomas, D.A.; Pillai, S.G.; Coxson, H.O.; Mathias, R.A.; Rafaels, N.M.; Wise, R.A.; Connett, J.E.; et al. Transforming growth factor-β receptor-3 is associated with pulmonary emphysema. Am. J. Respir. Cell Mol. Biol. 2009, 41, 324–331. [Google Scholar] [CrossRef]
- Li, Z.; Allingham, R.R.; Nakano, M.; Jia, L.; Chen, Y.; Ikeda, Y.; Mani, B.; Chen, L.J.; Kee, C.; Garway-Heath, D.F.; et al. A common variant near TGFBR3 is associated with primary open angle glaucoma. Hum. Mol. Genet. 2015, 24, 3880–3892. [Google Scholar] [CrossRef]
- Duncan, E.L.; Danoy, P.; Kemp, J.P.; Leo, P.J.; McCloskey, E.; Nicholson, G.C.; Eastell, R.; Prince, R.L.; Eisman, J.A.; Jones, G.; et al. Genome-wide association study using extreme truncate selection identifies novel genes affecting bone mineral density and fracture risk. PLoS Genet. 2011, 7, e1001372. [Google Scholar] [CrossRef] [PubMed]
- Khor, C.C.; Ramdas, W.D.; Vithana, E.N.; Cornes, B.K.; Sim, X.; Tay, W.T.; Saw, S.M.; Lavanya, Y.Z.; Wu, R.; Wang, J.J.; et al. Genome-wide association studies in Asians confirm the involvement of ATOH7 and TGFBR3, and further identify CARD10 as a novel locus influencing optic discarea. Hum. Mol. Genet. 2011, 20, 1864–1872. [Google Scholar] [CrossRef] [PubMed]
- Xiong, D.H.; Liu, X.G.; Guo, Y.F.; Tan, L.J.; Wang, L.; Sha, B.Y.; Tang, Z.H.; Pan, F.; Yang, T.L.; Chen, X.D.; et al. Genome-wide Association and Follow-Up Replication Studies Identified ADAMTS18 and TGFBR3 as Bone Mass Candidate Genes in Different Ethnic Groups. Am. J. Hum. Genet. 2009, 84, 388–398. [Google Scholar] [CrossRef] [PubMed]
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: The next generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef] [PubMed]
- Makker, A.; Goel, M.M. Tumor progression, metastasis, and modulators of epithelial-mesenchymal transition in endometrioid endometrial carcinoma: An update. Endocr. Relat. Cancer 2016, 23, R85–R111. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Tian, X.-J.; Xing, J. Signal Transduction Pathways of EMT Induced by TGF-β, SHH, and WNT and Their Crosstalks. J. Clin. Med. 2016, 5, 41. [Google Scholar] [CrossRef]
- Zakrzewski, P.K.; Nowacka-Zawisza, M.; Semczuk, A.; Rechberger, T.; Gałczyński, K.; Krajewska, W.M. Significance of TGFBR3 allelic loss in the deregulation of TGF signaling in primary human endometrial carcinomas. Oncol. Rep. 2016, 35, 932–938. [Google Scholar] [CrossRef]
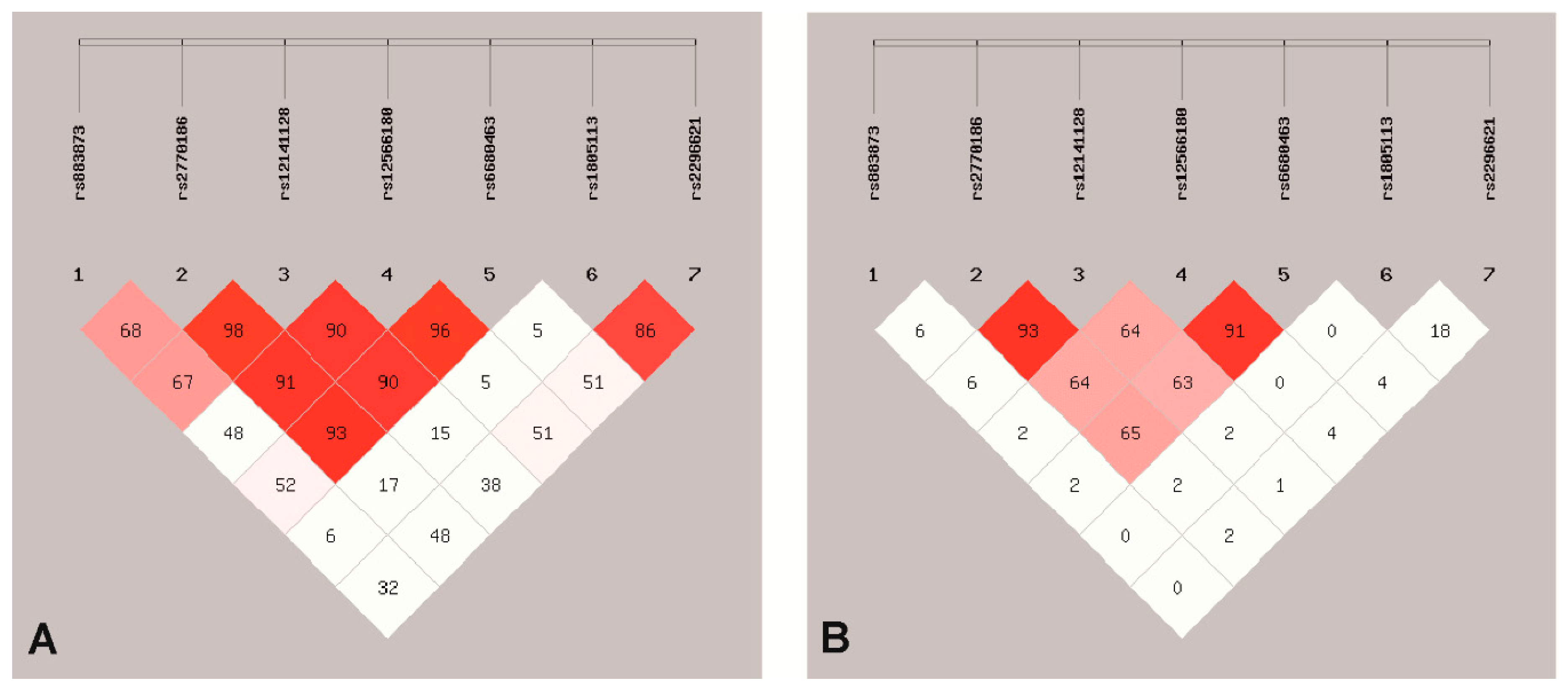
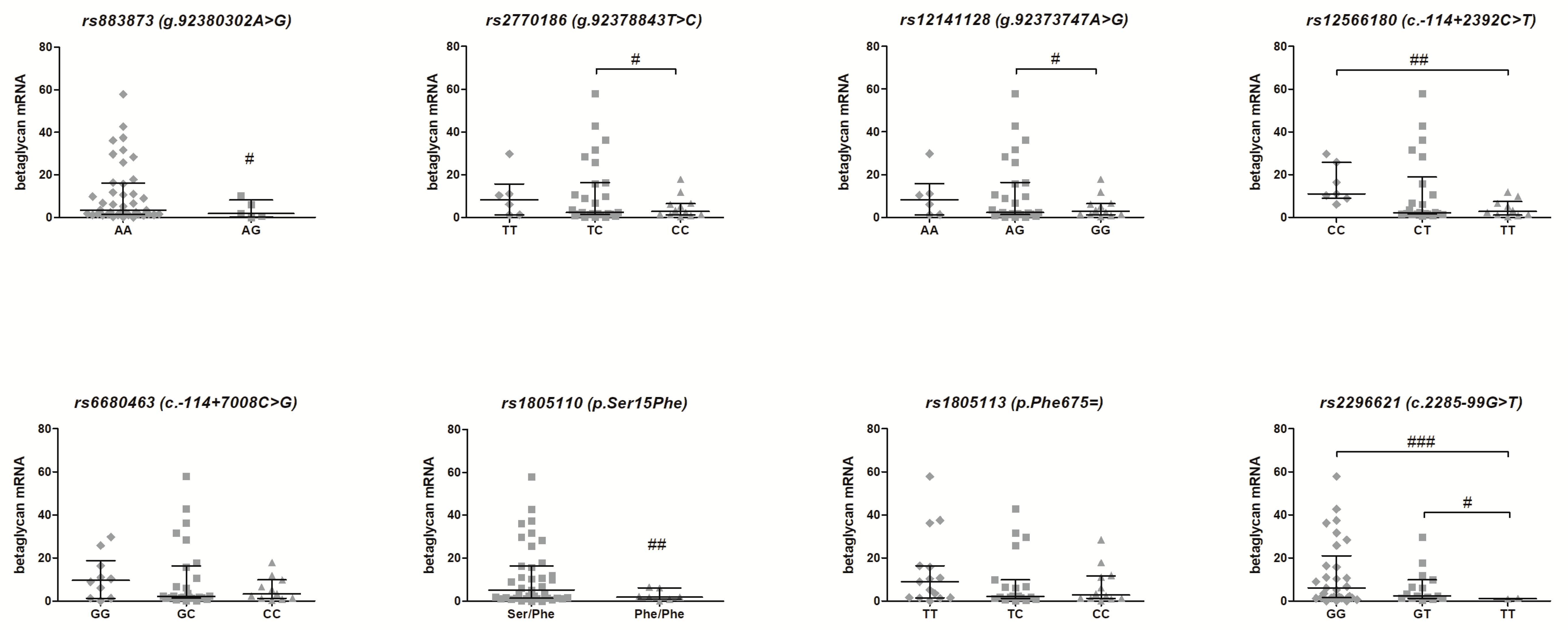
| Cases (n = 153) | Controls (n = 248) | p | |
|---|---|---|---|
| Socio-Demographic Characteristics | |||
| AGE (years) a | 62.1 ± 8.6 | 63.4 ± 9.0 | 0.1896 |
| min | 35 | 35 | |
| max | 85 | 85 | |
| 35–44 b | 2 (1.3) | 10 (4.0) | |
| 45–54 b | 25 (16.3) | 36 (14.5) | |
| 55–64 b | 75 (49.0) | 84 (33.9) | |
| 65–74 b | 38 (24.8) | 78 (31.5) | |
| 74–85 b | 13 (8.5) | 40 (16.1) | 0.009 |
| BMI (kg/m2) a | 28.5 ± 6.6 | 24.9 ± 4.0 | <0.001 |
| obesity (BMI > 30 kg/m2) b | 56 (36.6) | 11 (4.4) | <0.001 |
| Menarche (years) a | 13.8 ± 2.0 | 11.9 ± 1.6 | <0.001 |
| Parity (childbirths) a | 1.9 ± 1.4 | 1.1 ± 0.9 | <0.001 |
| Clinico-Pathological Characteristics | |||
| Histological Diagnosis | endometrial adenocarcinoma | ||
| Tumor Stage c | |||
| I b | 89 (58.2) | ||
| II b | 38 (24.8) | ||
| III b | 18 (11.8) | ||
| IV b | 8 (5.2) | ||
| Histological Grade d | |||
| G1 b | 37 (24.2) | ||
| G2 b | 94 (61.4) | ||
| G3 b | 22 (14.4) | ||
| Depth OF Myometrial Invasion | |||
| <1/2 b | 82 (53.6) | ||
| >1/2 b | 71 (46.4) | ||
| Vascular Space Invasion | |||
| not present b | 85 (55.6) | ||
| present b | 23 (15.0) | ||
| data not available b | 45 (29.4) |
| rs Number | Polymorphism | Localization | Maf |
|---|---|---|---|
| rs883873 | g.92380302A > G | 5′ regulatory region | 0.1394 |
| rs2770186 | g.92378843T > C | 5′ regulatory region | 0.4730 |
| rs12141128 | g.92373747A > G | 5′ regulatory region | 0.4736 |
| rs12566180 | c.−114 + 2392C > T | intron | 0.4209 |
| rs6680463 | c. −114 + 7008C > G | intron | 0.4687 |
| rs1805110 | c.44C > T (p.Ser15Phe) | exon | 0.1859 |
| rs1805113 | c.2025T > C (p.Phe675=) | exon | 0.2798 |
| rs2296621 | c.2285 − 99G > T | intron | 0.1050 |
| SNP Genotype/Allele | Cancer (n = 123) | Control (n = 248) | OR a | 95% CI | p value | OR b | 95% CI | p value | HWE c | ||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Number | Frequency (%) | Number | Frequency (%) | ||||||||
| rs883873 (g.92380302A > G) | |||||||||||
| AA | 107 | 69.9 | 200 | 80.6 | 1.00 | (ref.) | 1.00 | (ref.) | 0.3110 | ||
| AG | 45 | 29.4 | 47 | 19.0 | 1.79 | 1.12–2.87 | 0.0149 | 1.26 | 0.68–2.33 | 0.4607 | |
| GG | 1 | 0.7 | 1 | 0.4 | 1.87 | 0.12–30.18 | 1 | - | - | 1 | |
| χ2 = 6.049; p = 0.0486 | |||||||||||
| AG or GG vs. AA d | 46 | 48 | 1.79 | 1.12–2.86 | 0.0139 | 1.28 | 0.70–2.37 | 0.4242 | |||
| AG or AA vs. GG e | 152 | 247 | 0.62 | 0.04–9.91 | 1 | - | - | - | |||
| A | 259 | 84.6 | 447 | 90.1 | 0.60 | 0.39–0.93 | 0.0203 | 0.77 | 0.42–1.40 | 0.3846 | |
| G | 47 | 15.4 | 49 | 9.9 | 1.66 | 1.08–2.54 | 0.0203 | 1.31 | 0.72–2.39 | 0.3846 | |
| rs2770186 (g.92378843T > C) | |||||||||||
| TT | 22 | 14.4 | 46 | 18.5 | 1.00 | (ref.) | 1.00 | (ref.) | 0.8572 | ||
| TC | 89 | 58.2 | 120 | 48.4 | 1.55 | 0.87–2.76 | 0.1345 | 1.54 | 0.74–3.22 | 0.2469 | |
| CC | 42 | 27.5 | 82 | 33.1 | 1.07 | 0.57–2.01 | 0.8231 | 1.09 | 0.49–2.41 | 0.8286 | |
| χ2 = 3.672; p = 0.1595 | |||||||||||
| TC or CC vs. TT d | 131 | 202 | 1.36 | 0.78–2.36 | 0.2794 | 1.27 | 0.73–2.23 | 0.3961 | |||
| TC or TT vs. CC e | 111 | 166 | 1.31 | 0.84–2.03 | 0.2367 | 1.35 | 0.67–2.73 | 0.3930 | |||
| T | 133 | 43.5 | 212 | 42.7 | 1.03 | 0.77–1.37 | 0.8415 | 1.02 | 0.70–1.50 | 0.9100 | |
| C | 173 | 56.5 | 284 | 57.3 | 0.97 | 0.73–1.29 | 0.8415 | 0.98 | 0.67–1.44 | 0.9100 | |
| rs12141128 (g.92373747A > G) | |||||||||||
| AA | 23 | 15.0 | 49 | 19.8 | 1.00 | (ref.) | 1.00 | (ref.) | 0.6113 | ||
| AG | 88 | 57.5 | 118 | 47.6 | 1.59 | 0.90–2.80 | 0.1082 | 1.65 | 0.80–3.38 | 0.1742 | |
| GG | 42 | 27.5 | 81 | 32.7 | 1.10 | 0.59–2.05 | 0.7518 | 1.14 | 0.52–2.47 | 0.7418 | |
| χ2= 3.833; p = 0.1471 | |||||||||||
| AG or GG vs. AA d | 130 | 199 | 1.39 | 0.81–2.39 | 0.2318 | 1.43 | 0.72–2.83 | 0.3018 | |||
| AG or AA vs. GG e | 111 | 167 | 1.28 | 0.82–2.00 | 0.2713 | 1.27 | 0.72–2.23 | 0.4023 | |||
| A | 134 | 43.8 | 216 | 43.5 | 1.01 | 0.76–1.35 | 1.0000 | 1.00 | 0.68–1.46 | 0.9920 | |
| G | 172 | 56.2 | 280 | 56.5 | 0.99 | 0.74–1.32 | 1.0000 | 1.00 | 0.69–1.46 | 0.9920 | |
| rs12566180 (c.−114 + 2392C > T) | |||||||||||
| CC | 30 | 19.6 | 68 | 27.4 | 1.00 | (ref.) | 1.00 | (ref.) | 0.0570 | ||
| CT | 85 | 55.6 | 109 | 44.0 | 1.77 | 1.06–2.96 | 0.0393 | 2.22 | 1.15–4.30 | 0.0177 | |
| TT | 38 | 24.8 | 71 | 28.6 | 1.21 | 0.68–2.17 | 0.5169 | 1.18 | 0.58–2.41 | 0.6484 | |
| χ2 = 5.497; p = 0.0640 | |||||||||||
| CT or TT vs. CC d | 123 | 180 | 1.55 | 0.95–2.52 | 0.0769 | 1.73 | 0.94–3.17 | 0.0775 | |||
| CT or CC vs. TT e | 115 | 177 | 1.21 | 0.77–1.92 | 0.4062 | 1.42 | 0.80–2.51 | 0.2256 | |||
| C | 145 | 47.4 | 245 | 49.4 | 0.92 | 0.69–1.23 | 0.5777 | 0.95 | 0.67–1.35 | 0.7761 | |
| T | 161 | 52.6 | 251 | 50.6 | 1.08 | 0.81–1.44 | 0.5777 | 1.05 | 0.74–1.50 | 0.7761 | |
| rs6680463 (c.−114 + 7008C > G) | |||||||||||
| GG | 29 | 19.0 | 68 | 27.4 | 1.00 | (ref.) | 1.00 | (ref.) | 0.1275 | ||
| GC | 89 | 58.2 | 112 | 45.2 | 1.86 | 1.11–3.12 | 0.0174 | 2.34 | 1.20–4.53 | 0.0120 | |
| CC | 35 | 22.9 | 68 | 27.4 | 1.21 | 0.67–2.19 | 0.5376 | 1.09 | 0.53–2.24 | 0.8243 | |
| χ2 = 6.758; p = 0.0341 | |||||||||||
| GC or CC vs. GG d | 124 | 180 | 1.61 | 0.99–2.64 | 0.0544 | 1.74 | 0.95–3.20 | 0.0749 | |||
| GC or GG vs. CC e | 118 | 180 | 1.27 | 0.80–2.04 | 0.3125 | 1.60 | 0.90–2.86 | 0.1114 | |||
| G | 147 | 48.0 | 248 | 50.0 | 0.92 | 0.70–1.23 | 0.5902 | 0.99 | 0.69–1.41 | 0.9470 | |
| C | 159 | 52.0 | 248 | 50.0 | 1.08 | 0.81–1.44 | 0.5902 | 1.01 | 0.71–1.45 | 0.9470 | |
| rs1805110 c.44C > T (p.Ser15Phe) | |||||||||||
| CC | 0 | 0.0 | 0 | 0.0 | - | - | - | - | - | - | <0.001 |
| CT | 112 | 73.2 | 212 | 85.5 | 0.46 | 0.28–0.77 | 0.0024 | 0.79 | 0.42–1.50 | 0.4771 | |
| TT | 41 | 26.8 | 36 | 14.5 | 2.16 | 1.30–3.56 | 0.0024 | 1.26 | 0.66–2.39 | 0.4771 | |
| χ2 = 9.199; p = 0.0100 | |||||||||||
| CT or TT vs. CC d | 153 | 248 | - | - | - | - | - | - | |||
| CT or CC vs. TT e | 112 | 212 | 0.46 | 0.28–0.77 | 0.0024 | 0.79 | 0.42–1.50 | 0.4771 | |||
| C | 112 | 36.6 | 212 | 42.7 | 0.77 | 0.58–1.04 | 0.0853 | 0.79 | 0.42–1.50 | 0.4771 | |
| T | 194 | 63.4 | 284 | 57.3 | 1.29 | 0.96–1.73 | 0.0853 | 1.26 | 0.66–2.39 | 0.4771 | |
| rs1805113 c.2025T > C (p.Phe675=) | |||||||||||
| TT | 56 | 36.6 | 87 | 35.1 | 1.00 | (ref.) | 1.00 | (ref.) | 0.6117 | ||
| TC | 68 | 44.4 | 123 | 49.6 | 0.86 | 0.55–1.34 | 0.5055 | 0.62 | 0.34–1.12 | 0.1115 | |
| CC | 29 | 19.0 | 38 | 15.3 | 1.19 | 0.66–2.14 | 0.5707 | 0.93 | 0.44–1.96 | 0.8513 | |
| χ2 = 1.336; p = 0.5127 | |||||||||||
| TC or CC vs. TT d | 97 | 161 | 0.94 | 0.62–1.42 | 0.7518 | 0.70 | 0.40–1.21 | 0.2001 | |||
| TC or TT vs. CC e | 124 | 210 | 0.77 | 0.45–1.32 | 0.3438 | 0.81 | 0.42–1.57 | 0.5405 | |||
| T | 180 | 58.8 | 297 | 59.9 | 0.96 | 0.72–1.28 | 0.7642 | 1.10 | 0.77–1.58 | 0.6083 | |
| C | 126 | 41.2 | 199 | 40.1 | 1.04 | 0.78–1.40 | 0.7642 | 0.91 | 0.63–1.31 | 0.6083 | |
| rs2296621 c.2285 − 99G > T | |||||||||||
| GG | 103 | 67.3 | 178 | 71.8 | 1.00 | (ref.) | 1.00 | (ref.) | 0.0988 | ||
| GT | 43 | 28.1 | 68 | 27.4 | 1.09 | 0.70–1.72 | 0.6985 | 0.87 | 0.49–1.53 | 0.6226 | |
| TT | 7 | 4.6 | 2 | 0.8 | 6.05 | 1.23–29.66 | 0.0178 | 6.40 | 1.18–34.84 | 0.0317 | |
| χ2 = 6.272; p = 0.0435 | |||||||||||
| GT or TT vs. GG d | 50 | 70 | 1.23 | 0.80–1.91 | 0.3428 | 1.04 | 0.60–1.80 | 0.8951 | |||
| GT or GG vs. TT e | 146 | 246 | 0.17 | 0.03–0.83 | 0.0178 | 0.15 | 0.03–0.80 | 0.0267 | |||
| G | 249 | 81.4 | 424 | 85.5 | 0.74 | 0.51–1.09 | 0.1237 | 0.80 | 0.50–1.30 | 0.3703 | |
| T | 57 | 18.6 | 72 | 14.5 | 1.35 | 0.92–1.97 | 0.1237 | 1.24 | 0.77–2.00 | 0.3703 | |
| Combined Haplotype | Cancer | Control | OR | 95% CI | p Value (Bonferroni Corrected) | ||
|---|---|---|---|---|---|---|---|
| Number | Frequency (%) | Number | Frequency (%) | ||||
| rs2770186/rs12141128 | |||||||
| C/A | 1 | 0.003 | 6.02 | 0.012 | - | - | - |
| C/G | 172 | 0.562 | 277.98 | 0.560 | 0.98 | 0.73–1.30 | 1.0000 |
| T/A | 133 | 0.435 | 209.98 | 0.423 | 1.02 | 0.77–1.37 | 1.0000 |
| T/G | 0 | 0.000 | 2.02 | 0.004 | - | - | - |
| rs12566180/rs6680463 | |||||||
| C/C | 11.69 | 0.038 | 4.05 | 0.008 | 4.82 | 1.54–15.07 | 0.0116 |
| C/G | 133.31 | 0.436 | 240.95 | 0.486 | 0.82 | 0.61–1.09 | 0.6676 |
| T/C | 147.31 | 0.481 | 243.95 | 0.492 | 0.96 | 0.72–1.28 | 1.0000 |
| T/G | 13.69 | 0.045 | 7.05 | 0.014 | 3.25 | 1.29–8.15 | 0.0328 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zakrzewski, P.K.; Forma, E.; Cygankiewicz, A.I.; Bryś, M.; Wójcik-Krowiranda, K.; Bieńkiewicz, A.; Semczuk, A.; Krajewska, W.M. Betaglycan Gene (TGFBR3) Polymorphism Is Associated with Increased Risk of Endometrial Cancer. J. Clin. Med. 2020, 9, 3082. https://doi.org/10.3390/jcm9103082
Zakrzewski PK, Forma E, Cygankiewicz AI, Bryś M, Wójcik-Krowiranda K, Bieńkiewicz A, Semczuk A, Krajewska WM. Betaglycan Gene (TGFBR3) Polymorphism Is Associated with Increased Risk of Endometrial Cancer. Journal of Clinical Medicine. 2020; 9(10):3082. https://doi.org/10.3390/jcm9103082
Chicago/Turabian StyleZakrzewski, Piotr K., Ewa Forma, Adam I. Cygankiewicz, Magdalena Bryś, Katarzyna Wójcik-Krowiranda, Andrzej Bieńkiewicz, Andrzej Semczuk, and Wanda M. Krajewska. 2020. "Betaglycan Gene (TGFBR3) Polymorphism Is Associated with Increased Risk of Endometrial Cancer" Journal of Clinical Medicine 9, no. 10: 3082. https://doi.org/10.3390/jcm9103082
APA StyleZakrzewski, P. K., Forma, E., Cygankiewicz, A. I., Bryś, M., Wójcik-Krowiranda, K., Bieńkiewicz, A., Semczuk, A., & Krajewska, W. M. (2020). Betaglycan Gene (TGFBR3) Polymorphism Is Associated with Increased Risk of Endometrial Cancer. Journal of Clinical Medicine, 9(10), 3082. https://doi.org/10.3390/jcm9103082






