Importance of Virulence Factors for the Persistence of Oral Bacteria in the Inflamed Gingival Crevice and in the Pathogenesis of Periodontal Disease
Abstract
1. Introduction
2. The Role of Micro-organisms in Periodontitis—A Historical Perspective
3. “Putative Periodontal Pathogens”
4. Periodontitis—An Inflammatory Disease or an Infectious Disease?
5. Factors that Promote Bacterial Colonization and Persistence
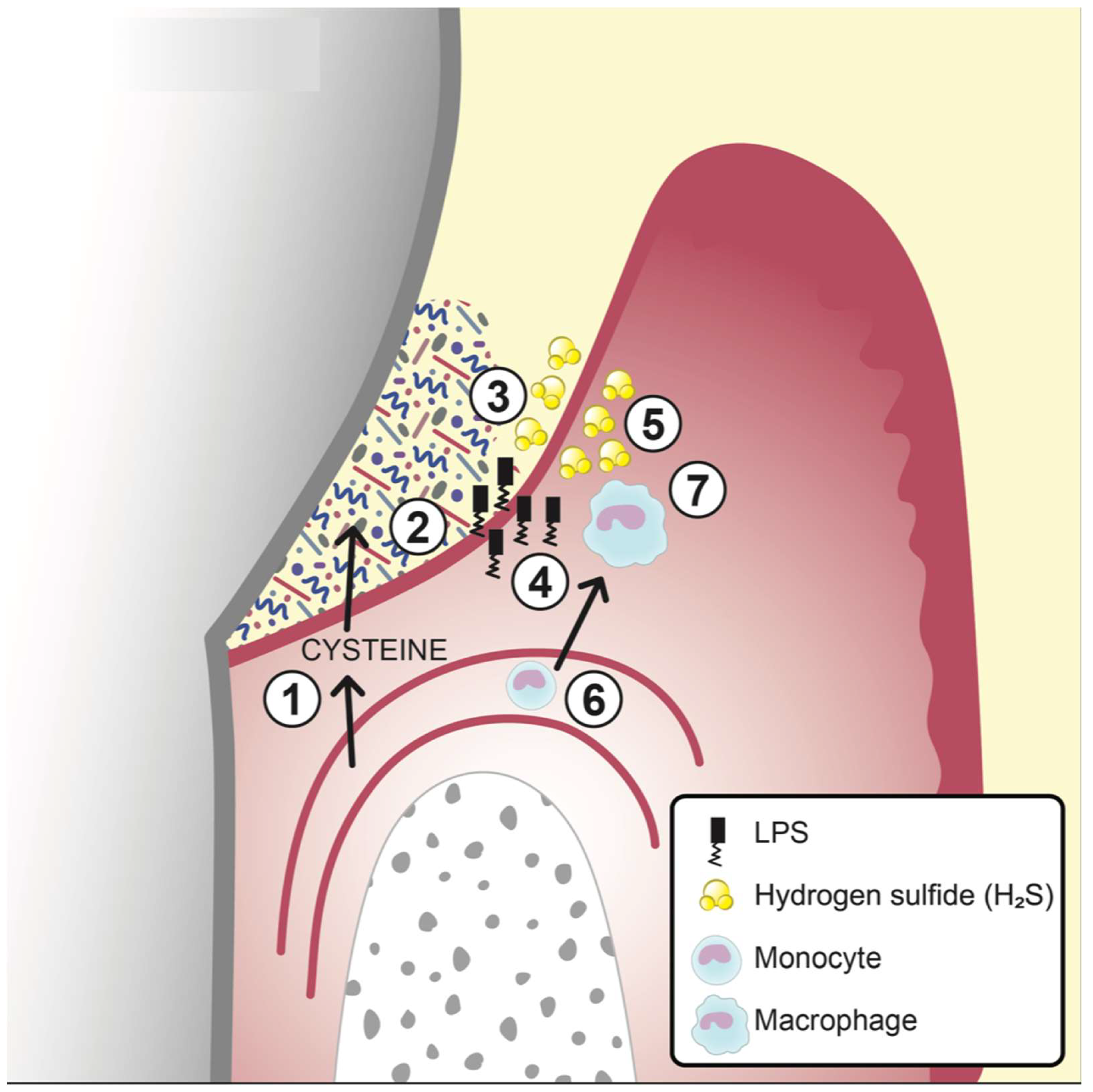
6. Factors Promoting Gingival Inflammation
6.1. Bacterial Metabolites as Pro-Inflammatory and/or Cytotoxic Agents
6.2. Bacterial Cell-Wall Constituents as Pro-inflammatory Factors
7. Invasion and Factors that Promote Tissue Degradation
7.1. The Intercellular Route
7.2. The Intracellular Route
7.3. Invasion by Trauma
7.4. Tissue Degrading Factors
8. Factors for Evasion of Host Defence
8.1. Exotoxins
8.2. Capsule Formation
9. Systemic Implications of Periodontal Bacteria
10. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Hashim, D.; Cionca, N.; Combescure, C.; Mombelli, A. The diagnosis of peri-implantitis: A systematic review on the predictive value of bleeding on probing. Clin. Oral Implants Res. 2018, 29 (Suppl. S16), 276–293. [Google Scholar] [CrossRef]
- Teles, R.; Teles, F.; Frias-Lopez, J.; Paster, B.; Haffajee, A. Lessons learned and unlearned in periodontal microbiology. Periodontology 2000 2013, 62, 95–162. [Google Scholar] [CrossRef] [PubMed]
- Baelum, V.; Lopez, R. Periodontal disease epidemiology—Learned and unlearned? Periodontology 2000 2013, 62, 37–58. [Google Scholar] [CrossRef] [PubMed]
- Kinane, D.F.; Stathopoulou, P.G.; Papapanou, P.N. Periodontal diseases. Nat. Rev. Dis. Primers 2017, 3, 17038. [Google Scholar] [CrossRef]
- Bui, F.Q.; Almeida-da-Silva, C.L.C.; Huynh, B.; Trinh, A.; Liu, J.; Woodward, J.; Asadi, H.; Ojcius, D.M. Association between periodontal pathogens and systemic disease. Biomed. J. 2019, 42, 27–35. [Google Scholar] [CrossRef] [PubMed]
- Loe, H.; Theilade, E.; Jensen, S.B. Experimental gingivitis in man. J. Periodontal. 1965, 36, 177–187. [Google Scholar] [CrossRef] [PubMed]
- Socransky, S.S.; Haffajee, A.D. Evidence of bacterial etiology: A historical perspective. Periodontology 2000 1994, 5, 7–25. [Google Scholar] [CrossRef] [PubMed]
- Socransky, S.S.; Haffajee, A.D. Periodontal microbial ecology. Periodontology 2000 2005, 38, 135–187. [Google Scholar] [CrossRef] [PubMed]
- Socransky, S.S. Criteria for the infectious agents in dental caries and periodontal disease. J. Clin. Periodontal. 1979, 6, 16–21. [Google Scholar] [CrossRef] [PubMed]
- Loesche, W.J. The therapeutic use of antimicrobial agents in patients with periodontal disease. Scand. J. Infect. Dis. Suppl. 1985, 46, 106–114. [Google Scholar] [PubMed]
- Page, R.C.; Kornman, K.S. The pathogenesis of human periodontitis: An introduction. Periodontology 2000 1997, 14, 9–11. [Google Scholar] [CrossRef] [PubMed]
- Meyle, J.; Chapple, I. Molecular aspects of the pathogenesis of periodontitis. Periodontology 2000 2015, 69, 7–17. [Google Scholar] [CrossRef] [PubMed]
- Haber, J.; Wattles, J.; Crowley, M.; Mandell, R.; Joshipura, K.; Kent, R.L. Evidence for cigarette smoking as a major risk factor for periodontitis. J. Periodontal. 1993, 64, 16–23. [Google Scholar] [CrossRef] [PubMed]
- Emrich, L.J.; Shlossman, M.; Genco, R.J. Periodontal disease in non-insulin-dependent diabetes mellitus. J. Periodontal. 1991, 62, 123–131. [Google Scholar] [CrossRef] [PubMed]
- Loos, B.G.; Papantonopoulos, G.; Jepsen, S.; Laine, M.L. What is the contribution of genetics to periodontal risk? Dent. Clin. 2015, 59, 761–780. [Google Scholar] [CrossRef]
- Nibali, L.; Bayliss-Chapman, J.; Almofareh, S.A.; Zhou, Y.; Divaris, K.; Vieira, A.R. What Is the heritability of periodontitis? A systematic review. J. Dent. Res. 2019, 98, 632–641. [Google Scholar] [CrossRef]
- Kassebaum, N.J.; Bernabe, E.; Dahiya, M.; Bhandari, B.; Murray, C.J.; Marcenes, W. Global burden of severe periodontitis in 1990–2010: A systematic review and meta-regression. J. Dent. Res. 2014, 93, 1045–1053. [Google Scholar] [CrossRef]
- Larsson, L. Current concepts of epigenetics and its role in periodontitis. Curr. Oral Health Rep. 2017, 4, 286–293. [Google Scholar] [CrossRef]
- Luo, Y.; Peng, X.; Duan, D.; Liu, C.; Xu, X.; Zhou, X. Epigenetic regulations in the pathogenesis of periodontitis. Curr. Stem Cell Res. Ther. 2018, 13, 144–150. [Google Scholar] [CrossRef]
- De-La-Torre, J.; Quindos, G.; Marcos-Arias, C.; Marichalar-Mendia, X.; Gainza, M.L.; Eraso, E.; Acha-Sagredo, A.; Aguirre-Urizar, J.M. Oral Candida colonization in patients with chronic periodontitis. Is there any relationship? Rev. Iberoam. Micol. 2018, 35, 134–139. [Google Scholar] [CrossRef]
- Slots, J. Periodontitis: Facts, fallacies and the future. Periodontology 2000 2017, 75, 7–23. [Google Scholar] [CrossRef] [PubMed]
- Rosier, B.T.; Marsh, P.D.; Mira, A. Resilience of the oral microbiota in health: Mechanisms that prevent dysbiosis. J. Dent. Res. 2018, 97, 371–380. [Google Scholar] [CrossRef] [PubMed]
- Lamont, R.J.; Koo, H.; Hajishengallis, G. The oral microbiota: Dynamic communities and host interactions. Nat. Rev. Microbiol. 2018, 16, 745–759. [Google Scholar] [CrossRef] [PubMed]
- Kilian, M.; Chapple, I.L.; Hannig, M.; Marsh, P.D.; Meuric, V.; Pedersen, A.M.; Tonetti, M.S.; Wade, W.G.; Zaura, E. The oral microbiome—An update for oral healthcare professionals. Br. Dent. J. 2016, 221, 657–666. [Google Scholar] [CrossRef] [PubMed]
- Lopez, R.; Hujoel, P.; Belibasakis, G.N. On putative periodontal pathogens: An epidemiological perspective. Virulence 2015, 6, 249–257. [Google Scholar] [CrossRef] [PubMed]
- Dahlen, G.; Manji, F.; Baelum, V.; Fejerskov, O. Putative periodontopathogens in “diseased” and “non-diseased” persons exhibiting poor oral hygiene. J. Clin. Periodontal. 1992, 19, 35–42. [Google Scholar] [CrossRef]
- Socransky, S.S.; Haffajee, A.D.; Cugini, M.A.; Smith, C.; Kent, R.L., Jr. Microbial complexes in subgingival plaque. J. Clin. Periodontal. 1998, 25, 134–144. [Google Scholar] [CrossRef]
- Lamont, R.J.; Jenkinson, H.F. Life below the gum line: Pathogenic mechanisms of Porphyromonas gingivalis. Microbiol. Mol. Biol. Rev. 1998, 62, 1244–1263. [Google Scholar]
- Holt, S.C.; Ebersole, J.L. Porphyromonas gingivalis, Treponema denticola, and Tannerella forsythia: The “red complex”, a prototype polybacterial pathogenic consortium in periodontitis. Periodontology 2000 2005, 38, 72–122. [Google Scholar] [CrossRef]
- Fine, D.H.; Patil, A.G.; Velusamy, S.K. Aggregatibacter actinomycetemcomitans (Aa) under the radar: Myths and misunderstandings of Aa and its role in aggressive periodontitis. Front. Immunol. 2019, 10, 728. [Google Scholar] [CrossRef]
- Listgarten, M.A.; Hellden, L. Relative distribution of bacteria at clinically healthy and periodontally diseased sites in humans. J. Clin. Periodontal. 1978, 5, 115–132. [Google Scholar] [CrossRef] [PubMed]
- Huo, Y.B.; Chan, Y.; Lacap-Bugler, D.C.; Mo, S.; Woo, P.C.Y.; Leung, W.K.; Watt, R.M. Multilocus sequence analysis of phylogroup 1 and 2 oral treponeme strains. Appl. Environ. Microbiol. 2017, 83, e02499-16. [Google Scholar] [CrossRef] [PubMed]
- You, M.; Mo, S.; Leung, W.K.; Watt, R.M. Comparative analysis of oral treponemes associated with periodontal health and disease. BMC Infect. Dis. 2013, 13, 174. [Google Scholar] [CrossRef] [PubMed]
- Haubek, D.; Ennibi, O.K.; Poulsen, K.; Vaeth, M.; Poulsen, S.; Kilian, M. Risk of aggressive periodontitis in adolescent carriers of the JP2 clone of Aggregatibacter (Actinobacillus) actinomycetemcomitans in Morocco: A prospective longitudinal cohort study. Lancet 2008, 371, 237–242. [Google Scholar] [CrossRef]
- Perez-Chaparro, P.J.; Goncalves, C.; Figueiredo, L.C.; Faveri, M.; Lobao, E.; Tamashiro, N.; Duarte, P.; Feres, M. Newly identified pathogens associated with periodontitis: A systematic review. J. Dent. Res. 2014, 93, 846–858. [Google Scholar] [CrossRef] [PubMed]
- Wade, W.G. Has the use of molecular methods for the characterization of the human oral microbiome changed our understanding of the role of bacteria in the pathogenesis of periodontal disease? J. Clin. Periodontal. 2011, 38 (Suppl. S11), 7–16. [Google Scholar] [CrossRef] [PubMed]
- Curtis, M.A. Periodontal microbiology—The lid’s off the box again. J. Dent. Res. 2014, 93, 840–842. [Google Scholar] [CrossRef]
- Charalampakis, G.; Dahlen, G.; Carlen, A.; Leonhardt, A. Bacterial markers vs. clinical markers to predict progression of chronic periodontitis: A 2-yr prospective observational study. Eur. J. Oral Sci. 2013, 121, 394–402. [Google Scholar] [CrossRef]
- Casadevall, A.; Pirofski, L.A. Virulence factors and their mechanisms of action: The view from a damage-response framework. J. Water Health 2009, 7 (Suppl. S1), S2–S18. [Google Scholar] [CrossRef]
- Isenberg, H.D. Pathogenicity and virulence: Another view. Clin. Microbiol. Rev. 1988, 1, 40–53. [Google Scholar] [CrossRef]
- Anonymous. Dorland´s Medical Dictionary; Saunders: Philadelphia, PA, USA, 2007. [Google Scholar]
- Manji, F.; Dahlen, G.; Fejerskov, O. Caries and periodontitis: Contesting the conventional wisdom on their aetiology. Caries Res. 2018, 52, 548–564. [Google Scholar] [CrossRef] [PubMed]
- Dahlen, G. Microbiology and treatment of dental abscesses and periodontal-endodontic lesions. Periodontology 2000 2002, 28, 206–239. [Google Scholar] [CrossRef] [PubMed]
- Lewis, M.A.; MacFarlane, T.W.; McGowan, D.A. Quantitative bacteriology of acute dento-alveolar abscesses. J. Med. Microbiol. 1986, 21, 101–104. [Google Scholar] [CrossRef] [PubMed]
- Charalampakis, G.; Belibasakis, G.N. Microbiome of peri-implant infections: Lessons from conventional, molecular and metagenomic analyses. Virulence 2015, 6, 183–187. [Google Scholar] [CrossRef] [PubMed]
- Berglundh, T.; Gislason, O.; Lekholm, U.; Sennerby, L.; Lindhe, J. Histopathological observations of human periimplantitis lesions. J. Clin. Periodontal. 2004, 31, 341–347. [Google Scholar] [CrossRef] [PubMed]
- Dahlen, G.; Fabricius, L.; Holm, S.E.; Moller, A. Interactions within a collection of eight bacterial strains isolated from a monkey dental root canal. Oral Microbiol. Immunol. 1987, 2, 164–170. [Google Scholar] [CrossRef] [PubMed]
- Project, C.H.M. Structure, function and diversity of the healthy human microbiome. Nature 2012, 486, 207–214. [Google Scholar] [CrossRef]
- Verma, D.; Garg, P.K.; Dubey, A.K. Insights into the human oral microbiome. Arch. Microbiol. 2018, 200, 525–540. [Google Scholar] [CrossRef] [PubMed]
- Marsh, P.; Lewis, M.; Williams, D.; Marsh, P.; Martin, M.; Lewis, M.; Williams, D. Oral Microbiology, 5th ed.; Churchill Livingstone: Edingburgh, UK, 2009. [Google Scholar]
- Carlsson, J. Growth and nutrition as ecological factors. In Oral Bacterial Ecology. The Molecular Basis; Kuramitsu, H.K., Ellen, R.P., Eds.; Horizon Scientific Press: Norfolk, UK, 2000; pp. 68–130. [Google Scholar]
- Devine, D.A.; Cosseau, C. Host defense peptides in the oral cavity. Adv. Appl. Microbiol. 2008, 63, 281–322. [Google Scholar] [CrossRef] [PubMed]
- Theilade, E.; Wright, W.H.; Jensen, S.B.; Loe, H. Experimental gingivitis in man. II. A longitudinal clinical and bacteriological investigation. J. Periodontal Res. 1966, 1, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Moore, L.V.; Moore, W.E.; Cato, E.P.; Smibert, R.M.; Burmeister, J.A.; Best, A.M.; Ranney, R.R. Bacteriology of human gingivitis. J. Dent. Res. 1987, 66, 989–995. [Google Scholar] [CrossRef] [PubMed]
- Slots, J. Subgingival microflora and periodontal disease. J. Clin. Periodontal. 1979, 6, 351–382. [Google Scholar] [CrossRef] [PubMed]
- Griffen, A.L.; Beall, C.J.; Campbell, J.H.; Firestone, N.D.; Kumar, P.S.; Yang, Z.K.; Podar, M.; Leys, E.J. Distinct and complex bacterial profiles in human periodontitis and health revealed by 16S pyrosequencing. ISME J. 2012, 6, 1176–1185. [Google Scholar] [CrossRef] [PubMed]
- Yost, S.; Duran-Pinedo, A.E.; Teles, R.; Krishnan, K.; Frias-Lopez, J. Functional signatures of oral dysbiosis during periodontitis progression revealed by microbial metatranscriptome analysis. Genome Med. 2015, 7, 27. [Google Scholar] [CrossRef]
- Takahashi, N. Oral microbiome metabolism: From “who are they?” to “what are they doing?”. J. Dent. Res. 2015, 94, 1628–1637. [Google Scholar] [CrossRef]
- Basic, A.; Alizadehgharib, S.; Dahlen, G.; Dahlgren, U. Hydrogen sulfide exposure induces NLRP3 inflammasome-dependent IL-1beta and IL-18 secretion in human mononuclear leukocytes in vitro. Clin. Exp. Dent. Res. 2017, 3, 115–120. [Google Scholar] [CrossRef]
- Zhang, J.H.; Dong, Z.; Chu, L. Hydrogen sulfide induces apoptosis in human periodontium cells. J. Periodontal Res. 2010, 45, 71–78. [Google Scholar] [CrossRef]
- Nakano, Y.; Yoshimura, M.; Koga, T. Methyl mercaptan production by periodontal bacteria. Int. Dent. J. 2002, 52 (Suppl. S3), 217–220. [Google Scholar] [CrossRef]
- Uematsu, H.; Sato, N.; Djais, A.; Hoshino, E. Degradation of arginine by Slackia exigua ATCC 700122 and Cryptobacterium curtum ATCC 700683. Oral Microbiol. Immunol. 2006, 21, 381–384. [Google Scholar] [CrossRef]
- Niederman, R.; Brunkhorst, B.; Smith, S.; Weinreb, R.N.; Ryder, M.I. Ammonia as a potential mediator of adult human periodontal infection: Inhibition of neutrophil function. Arch. Oral Biol. 1990, 35, S205–S209. [Google Scholar] [CrossRef]
- Uematsu, H.; Sato, N.; Hossain, M.Z.; Ikeda, T.; Hoshino, E. Degradation of arginine and other amino acids by butyrate-producing asaccharolytic anaerobic gram-positive rods in periodontal pockets. Arch. Oral Biol. 2003, 48, 423–429. [Google Scholar] [CrossRef]
- Niederman, R.; Zhang, J.; Kashket, S. Short-chain carboxylic-acid-stimulated, PMN-mediated gingival inflammation. Crit. Rev. Oral Biol. Med. 1997, 8, 269–290. [Google Scholar] [CrossRef] [PubMed]
- Niederman, R.; Buyle-Bodin, Y.; Lu, B.Y.; Robinson, P.; Naleway, C. Short-chain carboxylic acid concentration in human gingival crevicular fluid. J. Dent. Res. 1997, 76, 575–579. [Google Scholar] [CrossRef] [PubMed]
- Qiqiang, L.; Huanxin, M.; Xuejun, G. Longitudinal study of volatile fatty acids in the gingival crevicular fluid of patients with periodontitis before and after nonsurgical therapy. J. Periodontal Res. 2012, 47, 740–749. [Google Scholar] [CrossRef] [PubMed]
- Lu, R.; Meng, H.; Gao, X.; Xu, L.; Feng, X. Effect of non-surgical periodontal treatment on short chain fatty acid levels in gingival crevicular fluid of patients with generalized aggressive periodontitis. J. Periodontal Res. 2014, 49, 574–583. [Google Scholar] [CrossRef] [PubMed]
- Tsuda, H.; Ochiai, K.; Suzuki, N.; Otsuka, K. Butyrate, a bacterial metabolite, induces apoptosis and autophagic cell death in gingival epithelial cells. J. Periodontal Res. 2010, 45, 626–634. [Google Scholar] [CrossRef] [PubMed]
- Kurita-Ochiai, T.; Seto, S.; Suzuki, N.; Yamamoto, M.; Otsuka, K.; Abe, K.; Ochiai, K. Butyric acid induces apoptosis in inflamed fibroblasts. J. Dent. Res. 2008, 87, 51–55. [Google Scholar] [CrossRef]
- Chang, M.C.; Tsai, Y.L.; Chen, Y.W.; Chan, C.P.; Huang, C.F.; Lan, W.C.; Lin, C.C.; Lan, W.H.; Jeng, J.H. Butyrate induces reactive oxygen species production and affects cell cycle progression in human gingival fibroblasts. J. Periodontal Res. 2013, 48, 66–73. [Google Scholar] [CrossRef]
- Bjorkman, L.; Martensson, J.; Winther, M.; Gabl, M.; Holdfeldt, A.; Uhrbom, M.; Bylund, J.; Hojgaard Hansen, A.; Pandey, S.K.; Ulven, T.; et al. The Neutrophil response induced by an agonist for free fatty acid receptor 2 (GPR43) is primed by tumor necrosis factor alpha and by Receptor UNCOUPLING from the cytoskeleton but attenuated by tissue recruitment. Mol. Cell Biol. 2016, 36, 2583–2595. [Google Scholar] [CrossRef]
- Alvarez-Curto, E.; Milligan, G. Metabolism meets immunity: The role of free fatty acid receptors in the immune system. Biochem. Pharmacol. 2016, 114, 3–13. [Google Scholar] [CrossRef]
- Maslowski, K.M.; Vieira, A.T.; Ng, A.; Kranich, J.; Sierro, F.; Yu, D.; Schilter, H.C.; Rolph, M.S.; Mackay, F.; Artis, D.; et al. Regulation of inflammatory responses by gut microbiota and chemoattractant receptor GPR43. Nature 2009, 461, 1282–1286. [Google Scholar] [CrossRef] [PubMed]
- Barnes, V.M.; Teles, R.; Trivedi, H.M.; Devizio, W.; Xu, T.; Mitchell, M.W.; Milburn, M.V.; Guo, L. Acceleration of purine degradation by periodontal diseases. J. Dent. Res. 2009, 88, 851–855. [Google Scholar] [CrossRef] [PubMed]
- Enersen, M.; Nakano, K.; Amano, A. Porphyromonas gingivalis fimbriae. J. Oral Microbiol. 2013, 5, 20265. [Google Scholar] [CrossRef] [PubMed]
- Singhrao, S.K.; Olsen, I. Are Porphyromonas gingivalis outer membrane vesicles microbullets for sporadic Alzheimer’s disease manifestation? J. Alzheimers Dis. Rep. 2018, 2, 219–228. [Google Scholar] [CrossRef] [PubMed]
- Neyen, C.; Lemaitre, B. Sensing gram-negative bacteria: A phylogenetic perspective. Curr. Opin. Immunol. 2016, 38, 8–17. [Google Scholar] [CrossRef] [PubMed]
- Paramonov, N.; Aduse-Opoku, J.; Hashim, A.; Rangarajan, M.; Curtis, M.A. Identification of the linkage between A-polysaccharide and the core in the A-lipopolysaccharide of Porphyromonas gingivalis W50. J. Bacteriol. 2015, 197, 1735–1746. [Google Scholar] [CrossRef]
- Tribble, G.D.; Lamont, R.J. Bacterial invasion of epithelial cells and spreading in periodontal tissue. Periodontology 2000 2010, 52, 68–83. [Google Scholar] [CrossRef]
- Ji, S.; Choi, Y.S.; Choi, Y. Bacterial invasion and persistence: Critical events in the pathogenesis of periodontitis? J. Periodontal Res. 2015, 50, 570–585. [Google Scholar] [CrossRef]
- Lux, R.; Miller, J.N.; Park, N.H.; Shi, W. Motility and chemotaxis in tissue penetration of oral epithelial cell layers by Treponema denticola. Infect. Immun. 2001, 69, 6276–6283. [Google Scholar] [CrossRef]
- Katz, J.; Yang, Q.B.; Zhang, P.; Potempa, J.; Travis, J.; Michalek, S.M.; Balkovetz, D.F. Hydrolysis of epithelial junctional proteins by Porphyromonas gingivalis gingipains. Infect. Immun. 2002, 70, 2512–2518. [Google Scholar] [CrossRef]
- Listgarten, M.A. Electron microscopic observations on the bacterial flora of acute necrotizing Ulcerative gingivitis. J. Periodontal. 1965, 36, 328–339. [Google Scholar] [CrossRef] [PubMed]
- Listgarten, M.A.; Socransky, S.S. Ultrastructural characteristics of a spirochete in the lesion of acute necrotizing Ulcerative gingivostomatitis (Vincent’s infection). Arch. Oral Biol. 1964, 9, 95–96. [Google Scholar] [CrossRef]
- Berglundh, T.; Zitzmann, N.U.; Donati, M. Are peri-implantitis lesions different from periodontitis lesions? J. Clin. Periodontol. 2011, 38 (Suppl. S11), 188–202. [Google Scholar] [CrossRef]
- Rudney, J.D.; Chen, R.; Zhang, G. Streptococci dominate the diverse flora within buccal cells. J. Dent. Res. 2005, 84, 1165–1171. [Google Scholar] [CrossRef] [PubMed]
- Rudney, J.D.; Chen, R.; Sedgewick, G.J. Actinobacillus actinomycetemcomitans, Porphyromonas gingivalis, and Tannerella forsythensis are components of a polymicrobial intracellular flora within human buccal cells. J. Dent. Res. 2005, 84, 59–63. [Google Scholar] [CrossRef] [PubMed]
- Dibart, S.; Skobe, Z.; Snapp, K.R.; Socransky, S.S.; Smith, C.M.; Kent, R. Identification of bacterial species on or in crevicular epithelial cells from healthy and periodontally diseased patients using DNA-DNA hybridization. Oral Microbiol. Immunol. 1998, 13, 30–35. [Google Scholar] [CrossRef] [PubMed]
- Madianos, P.N.; Papapanou, P.N.; Nannmark, U.; Dahlen, G.; Sandros, J. Porphyromonas gingivalis FDC381 multiplies and persists within human oral epithelial cells in vitro. Infect. Immun. 1996, 64, 660–664. [Google Scholar]
- Amano, A.; Kuboniwa, M.; Nakagawa, I.; Akiyama, S.; Morisaki, I.; Hamada, S. Prevalence of specific genotypes of Porphyromonas gingivalis fimA and periodontal health status. J. Dent. Res. 2000, 79, 1664–1668. [Google Scholar] [CrossRef]
- Jotwani, R.; Cutler, C.W. Fimbriated Porphyromonas gingivalis is more efficient than fimbria-deficient P. gingivalis in entering human dendritic cells in vitro and induces an inflammatory Th1 effector response. Infect. Immun. 2004, 72, 1725–1732. [Google Scholar] [CrossRef]
- Wang, M.; Liang, S.; Hosur, K.B.; Domon, H.; Yoshimura, F.; Amano, A.; Hajishengallis, G. Differential virulence and innate immune interactions of Type I and II fimbrial genotypes of Porphyromonas gingivalis. Oral Microbiol. Immunol. 2009, 24, 478–484. [Google Scholar] [CrossRef]
- Burnett, G.W.; Scherp, H.W. Oral Microbiology and Infectious Disease, 3rd ed.; The William & Wilkins Company: Baltimore, MD, USA, 1968; p. 982. [Google Scholar]
- Attstrom, R.; Schroeder, H.E. Effect of experimental neutropenia on initial gingivitis in dogs. Scand. J. Dent. Res. 1979, 87, 7–23. [Google Scholar] [CrossRef] [PubMed]
- Sundqvist, G.; Carlsson, J.; Herrmann, B.; Tarnvik, A. Degradation of human immunoglobulins G and M and complement factors C3 and C5 by black-pigmented bacteroides. J. Med. Microbiol. 1985, 19, 85–94. [Google Scholar] [CrossRef] [PubMed]
- Lina, G.; Piemont, Y.; Godail-Gamot, F.; Bes, M.; Peter, M.O.; Gauduchon, V.; Vandenesch, F.; Etienne, J. Involvement of Panton-Valentine leukocidin-producing Staphylococcus aureus in primary skin infections and pneumonia. Clin. Infect. Dis. 1999, 29, 1128–1132. [Google Scholar] [CrossRef]
- Tadepalli, S.; Stewart, G.C.; Nagaraja, T.G.; Narayanan, S.K. Human Fusobacterium necrophorum strains have a leukotoxin gene and exhibit leukotoxic activity. J. Med. Microbiol. 2008, 57, 225–231. [Google Scholar] [CrossRef][Green Version]
- Kachlany, S.C. Aggregatibacter actinomycetemcomitans leukotoxin: From threat to therapy. J. Dent. Res. 2010, 89, 561–570. [Google Scholar] [CrossRef] [PubMed]
- Johansson, A. Aggregatibacter actinomycetemcomitans leukotoxin: A powerful tool with capacity to cause imbalance in the host inflammatory response. Toxins 2011, 3, 242–259. [Google Scholar] [CrossRef] [PubMed]
- Hoglund Aberg, C.; Kwamin, F.; Claesson, R.; Dahlen, G.; Johansson, A.; Haubek, D. Progression of attachment loss is strongly associated with presence of the JP2 genotype of Aggregatibacter actinomycetemcomitans: A prospective cohort study of a young adolescent population. J. Clin. Periodontal. 2014, 41, 232–241. [Google Scholar] [CrossRef]
- Aberg, C.H.; Kelk, P.; Johansson, A. Aggregatibacter actinomycetemcomitans: Virulence of its leukotoxin and association with aggressive periodontitis. Virulence 2015, 6, 188–195. [Google Scholar] [CrossRef]
- Fortanier, A.C.; Venekamp, R.P.; Boonacker, C.W.; Hak, E.; Schilder, A.G.; Sanders, E.A.; Damoiseaux, R.A. Pneumococcal conjugate vaccines for preventing acute otitis media in children. Cochrane Database Syst. Rev. 2019, 5, Cd001480. [Google Scholar] [CrossRef]
- Wang, S.; Tafalla, M.; Hanssens, L.; Dolhain, J. A review of Haemophilus influenzae disease in Europe from 2000–2014: Challenges, successes and the contribution of hexavalent combination vaccines. Expert Rev. Vaccines 2017, 16, 1095–1105. [Google Scholar] [CrossRef]
- Laine, M.L.; van Winkelhoff, A.J. Virulence of six capsular serotypes of Porphyromonas gingivalis in a mouse model. Oral Microbiol. Immunol. 1998, 13, 322–325. [Google Scholar] [CrossRef] [PubMed]
- Gharbia, S.E.; Haapasalo, M.; Shah, H.N.; Kotiranta, A.; Lounatmaa, K.; Pearce, M.A.; Devine, D.A. Characterization of Prevotella intermedia and Prevotella nigrescens isolates from periodontic and endodontic infections. J. Periodontal. 1994, 65, 56–61. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.; Wyant, T.; Anaya-Bergman, C.; Aduse-Opoku, J.; Brunner, J.; Laine, M.L.; Curtis, M.A.; Lewis, J.P. The capsule of Porphyromonas gingivalis leads to a reduction in the host inflammatory response, evasion of phagocytosis, and increase in virulence. Infect. Immun. 2011, 79, 4533–4542. [Google Scholar] [CrossRef] [PubMed]
- Yoshino, T.; Laine, M.L.; van Winkelhoff, A.J.; Dahlen, G. Genotype variation and capsular serotypes of Porphyromonas gingivalis from chronic periodontitis and periodontal abscesses. FEMS Microbiol. Lett. 2007, 270, 75–81. [Google Scholar] [CrossRef] [PubMed]
- Slots, J. Focal infection of periodontal origin. Periodontology 2000 2019, 79, 233–235. [Google Scholar] [CrossRef] [PubMed]
- Jepsen, S.; Caton, J.G.; Albandar, J.M.; Bissada, N.F.; Bouchard, P.; Cortellini, P.; Demirel, K.; de Sanctis, M.; Ercoli, C.; Fan, J.; et al. Periodontal manifestations of systemic diseases and developmental and acquired conditions: Consensus report of workgroup 3 of the 2017 world workshop on the classification of periodontal and peri-implant diseases and conditions. J. Periodontal. 2018, 89 (Suppl. S1), S237–S248. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.K.; Wu, Y.T.; Chang, Y.C. Association between chronic periodontitis and the risk of Alzheimer’s disease: A retrospective, population-based, matched-cohort study. Alzheimers Res. Ther. 2017, 9, 56. [Google Scholar] [CrossRef]
- Potempa, J.; Mydel, P.; Koziel, J. The case for periodontitis in the pathogenesis of rheumatoid arthritis. Nat. Rev. Rheumatol. 2017, 13, 606–620. [Google Scholar] [CrossRef]
- Konig, M.F.; Abusleme, L.; Reinholdt, J.; Palmer, R.J.; Teles, R.P.; Sampson, K.; Rosen, A.; Nigrovic, P.A.; Sokolove, J.; Giles, J.T.; et al. Aggregatibacter actinomycetemcomitans-induced hypercitrullination links periodontal infection to autoimmunity in rheumatoid arthritis. Sci. Transl. Med. 2016, 8, 369ra176. [Google Scholar] [CrossRef]
- McGraw, W.T.; Potempa, J.; Farley, D.; Travis, J. Purification, characterization, and sequence analysis of a potential virulence factor from Porphyromonas gingivalis, peptidylarginine deiminase. Infect. Immun. 1999, 67, 3248–3256. [Google Scholar]

| “Putative Periodontal Pathogens” | Gram Stain | Main Metabolic Trait | Motility | Proteolytic Activity | Carbohydrate Fermentation | Major end Products | Factors of Significance | Subtyping |
|---|---|---|---|---|---|---|---|---|
| Aggregatibacter actinomycetemcomitans | Gramneg | Facultative anaerobic | No | Weak | Glycolytic | Lactic acid, Succinic acid Acetic acid, Propionic acid | Leukotoxin Cytodescen-ding (Cdt) toxin | Serotypes a-e, Non-serotypable isolates are frequent Specific genotypes (JP2) |
| Porphyromonas gingivalis | Gramneg | Anaerobic | No | Strong | Asaccharolytic | NH3 H2S Phenylacetic acid Indole | Gingipains (RgpA and Kgp) | FimA genotypes: I-V Arg-specific RgpA: A-C Lys-specific Kgp: I and II Capsular subtypes: K1-K6 |
| Tannerella forsythia | Gramneg | Anaerobic | No | Strong | Weak glycolytic | H2S (weak) Acetic acid Propionic acid | Trypsin-like and PrtH proteases | Variations in the leucine-rich repeat BspA protein are existing but no subtyping is presented |
| Treponema denticola | Gramneg | Anaerobic | Strong | Strong | Weak glycolytic | H2S (strong) | Spirochetes (spiral shaped) | Seven oral Treponema species identified but no subtyping is known Hundreds of spirochetal genotypes (OTU′s) found |
| Prevotella intermedia/nigrescens | Gramneg | Anaerobic | No | Strong | Glycolytic | NH3 H2S Succinic acid Acetic | Indole | Capsule is produced but no subtyping is known |
| Fusobacterium nucleatum | Gramneg | Anaerobic | No | Strong | Glycolytic | NH3 H2S (strong) Butyric acid | Fusiform Morphology (threadlike) | Three subspecies reported variation in the outer-membrane structure |
| Campylobacter rectus/gracilis | Gramneg | Microaerophilic | weak | Weak | Asaccharolytic | H2S Succinic acid | Nd | not known |
| Parvimonas micra | Grampos | Anaerobic | No | Strong | Glycolytic | NH3 H2S (weak), Acetic acid | Nd | not known |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dahlen, G.; Basic, A.; Bylund, J. Importance of Virulence Factors for the Persistence of Oral Bacteria in the Inflamed Gingival Crevice and in the Pathogenesis of Periodontal Disease. J. Clin. Med. 2019, 8, 1339. https://doi.org/10.3390/jcm8091339
Dahlen G, Basic A, Bylund J. Importance of Virulence Factors for the Persistence of Oral Bacteria in the Inflamed Gingival Crevice and in the Pathogenesis of Periodontal Disease. Journal of Clinical Medicine. 2019; 8(9):1339. https://doi.org/10.3390/jcm8091339
Chicago/Turabian StyleDahlen, Gunnar, Amina Basic, and Johan Bylund. 2019. "Importance of Virulence Factors for the Persistence of Oral Bacteria in the Inflamed Gingival Crevice and in the Pathogenesis of Periodontal Disease" Journal of Clinical Medicine 8, no. 9: 1339. https://doi.org/10.3390/jcm8091339
APA StyleDahlen, G., Basic, A., & Bylund, J. (2019). Importance of Virulence Factors for the Persistence of Oral Bacteria in the Inflamed Gingival Crevice and in the Pathogenesis of Periodontal Disease. Journal of Clinical Medicine, 8(9), 1339. https://doi.org/10.3390/jcm8091339




