Meta-Analysis of Differential miRNA Expression after Bariatric Surgery
Abstract
1. Introduction
2. Methods
2.1. Search Strategies
2.2. Study Selection
2.3. Data Collection Process
2.4. Synthesis of Results
3. Results
3.1. Selected Studies for the Meta-Analysis
3.2. Differential Expression of miRNA before and after Surgery
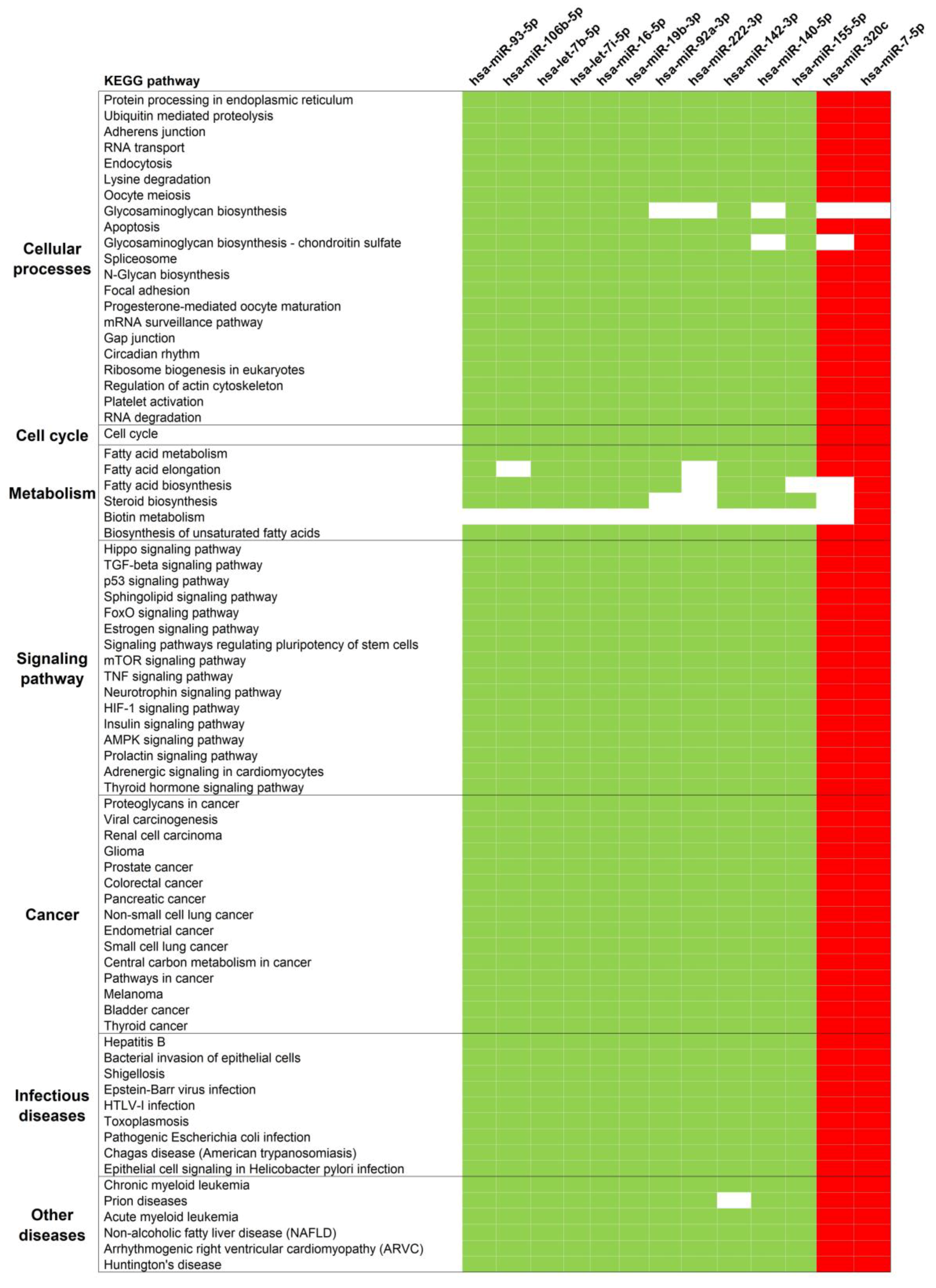
3.3. Pathway Analysis
4. Discussion
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Moshiri, M.; Osman, S.; Robinson, T.J.; Khandelwal, S.; Bhargava, P.; Rohrmann, C.A. Evolution of Bariatric Surgery: A Historical Perspective. Am. J. Roentgenol. 2013, 201, W40–W48. [Google Scholar] [CrossRef] [PubMed]
- Arterburn, D.E.; Courcoulas, A.P. Bariatric Surgery for Obesity and Metabolic Conditions in Adults. BMJ 2014, 349, g3961. [Google Scholar] [CrossRef] [PubMed]
- Maciejewski, M.L.; Arterburn, D.E.; Van Scoyoc, L.; Smith, V.A.; Yancy, W.S.; Weidenbacher, H.J.; Livingston, E.H.; Olsen, M.K. Bariatric Surgery and Long-Term Durability of Weight Loss. Jama Surg. 2016, 151, 1046. [Google Scholar] [CrossRef] [PubMed]
- Courcoulas, A.P.; King, W.C.; Belle, S.H.; Berk, P.; Flum, D.R.; Garcia, L.; Gourash, W.; Horlick, M.; Mitchell, J.E.; Pomp, A.; et al. Seven-Year Weight Trajectories and Health Outcomes in the Longitudinal Assessment of Bariatric Surgery (LABS) Study. Jama Surg. 2018, 153, 427. [Google Scholar] [CrossRef] [PubMed]
- Inge, T.H.; Courcoulas, A.P.; Jenkins, T.M.; Michalsky, M.P.; Helmrath, M.A.; Brandt, M.L.; Harmon, C.M.; Zeller, M.H.; Chen, M.K.; Xanthakos, S.A.; et al. Weight Loss and Health Status 3 Years after Bariatric Surgery in Adolescents. N. Engl. J. Med. 2016, 374, 113–123. [Google Scholar] [CrossRef] [PubMed]
- ASMBS. Estimate of Bariatric Surgery Numbers, 2011–2017 | American Society for Metabolic and Bariatric Surgery. Available online: https://asmbs.org/resources/estimate-of-bariatric-surgery-numbers (accessed on 29 July 2019).
- Angrisani, L.; Santonicola, A.; Iovino, P.; Formisano, G.; Buchwald, H.; Scopinaro, N. Bariatric Surgery Worldwide 2013. Obes. Surg. 2015, 25, 1822–1832. [Google Scholar] [CrossRef]
- Janik, M.R.; Stanowski, E.; Paśnik, K. Present Status of Bariatric Surgery in Poland. Videosurgery Miniinvasive Tech. 2016, 1, 22–25. [Google Scholar] [CrossRef]
- Chang, S.-H.; Stoll, C.R.T.; Song, J.; Varela, J.E.; Eagon, C.J.; Colditz, G.A. The Effectiveness and Risks of Bariatric Surgery: An Updated Systematic Review and Meta-Analysis, 2003–2012. Jama Surg. 2014, 149, 275. [Google Scholar] [CrossRef]
- Inge, T.H.; Jenkins, T.M.; Xanthakos, S.A.; Dixon, J.B.; Daniels, S.R.; Zeller, M.H.; Helmrath, M.A. Long-Term Outcomes of Bariatric Surgery in Adolescents with Severe Obesity (FABS-5+): A Prospective Follow-up Analysis. Lancet Diabetes Endocrinol. 2017, 5, 165–173. [Google Scholar] [CrossRef]
- Olbers, T.; Beamish, A.J.; Gronowitz, E.; Flodmark, C.-E.; Dahlgren, J.; Bruze, G.; Ekbom, K.; Friberg, P.; Göthberg, G.; Järvholm, K.; et al. Laparoscopic Roux-En-Y Gastric Bypass in Adolescents with Severe Obesity (AMOS): A Prospective, 5-Year, Swedish Nationwide Study. Lancet Diabetes Endocrinol. 2017, 5, 174–183. [Google Scholar] [CrossRef]
- Ibrahim, A.M.; Thumma, J.R.; Dimick, J.B. Reoperation and Medicare Expenditures After Laparoscopic Gastric Band Surgery. Jama Surg. 2017, 152, 835. [Google Scholar] [CrossRef]
- Lee, W.-J.; Chong, K.; Aung, L.; Chen, S.-C.; Ser, K.-H.; Lee, Y.-C. Metabolic Surgery for Diabetes Treatment: Sleeve Gastrectomy or Gastric Bypass? World J. Surg. 2017, 41, 216–223. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Lai, D.; Wu, D. Laparoscopic Roux-En-Y Gastric Bypass Versus Laparoscopic Sleeve Gastrectomy to Treat Morbid Obesity-Related Comorbidities: A Systematic Review and Meta-Analysis. Obes. Surg. 2016, 26, 429–442. [Google Scholar] [CrossRef] [PubMed]
- Sjöström, L.; Lindroos, A.-K.; Peltonen, M.; Torgerson, J.; Bouchard, C.; Carlsson, B.; Dahlgren, S.; Larsson, B.; Narbro, K.; Sjöström, C.D.; et al. Lifestyle, Diabetes, and Cardiovascular Risk Factors 10 Years after Bariatric Surgery. N. Engl. J. Med. 2004, 351, 2683–2693. [Google Scholar] [CrossRef] [PubMed]
- Poirier, P.; Cornier, M.-A.; Mazzone, T.; Stiles, S.; Cummings, S.; Klein, S.; McCullough, P.A.; Ren Fielding, C.; Franklin, B.A. Bariatric Surgery and Cardiovascular Risk Factors: A Scientific Statement from the American Heart Association. Circulation 2011, 123, 1683–1701. [Google Scholar] [CrossRef] [PubMed]
- Schauer, D.P.; Feigelson, H.S.; Koebnick, C.; Caan, B.; Weinmann, S.; Leonard, A.C.; Powers, J.D.; Yenumula, P.R.; Arterburn, D.E. Bariatric Surgery and the Risk of Cancer in a Large Multisite Cohort. Ann. Surg. 2019, 269, 95–101. [Google Scholar] [CrossRef]
- Dixon, J.B.; Zimmet, P.; Alberti, K.G.; Rubino, F. Bariatric Surgery: An IDF Statement for Obese Type 2 Diabetes. Diabet. Med. 2011, 28, 628–642. [Google Scholar] [CrossRef]
- Courcoulas, A.P.; Belle, S.H.; Neiberg, R.H.; Pierson, S.K.; Eagleton, J.K.; Kalarchian, M.A.; DeLany, J.P.; Lang, W.; Jakicic, J.M. Three-Year Outcomes of Bariatric Surgery vs. Lifestyle Intervention for Type 2 Diabetes Mellitus Treatment: A Randomized Clinical Trial. Jama Surg. 2015, 150, 931. [Google Scholar] [CrossRef]
- Cummings, D.E.; Arterburn, D.E.; Westbrook, E.O.; Kuzma, J.N.; Stewart, S.D.; Chan, C.P.; Bock, S.N.; Landers, J.T.; Kratz, M.; Foster-Schubert, K.E.; et al. Gastric Bypass Surgery vs Intensive Lifestyle and Medical Intervention for Type 2 Diabetes: The CROSSROADS Randomised Controlled Trial. Diabetologia 2016, 59, 945–953. [Google Scholar] [CrossRef]
- Ikramuddin, S.; Korner, J.; Lee, W.-J.; Connett, J.E.; Inabnet, W.B.; Billington, C.J.; Thomas, A.J.; Leslie, D.B.; Chong, K.; Jeffery, R.W.; et al. Roux-En-Y Gastric Bypass vs Intensive Medical Management for the Control of Type 2 Diabetes, Hypertension, and Hyperlipidemia: The Diabetes Surgery Study Randomized Clinical Trial. JAMA 2013, 309, 2240. [Google Scholar] [CrossRef]
- Mingrone, G.; Panunzi, S.; De Gaetano, A.; Guidone, C.; Iaconelli, A.; Nanni, G.; Castagneto, M.; Bornstein, S.; Rubino, F. Bariatric–Metabolic Surgery versus Conventional Medical Treatment in Obese Patients with Type 2 Diabetes: 5 Year Follow-up of an Open-Label, Single-Centre, Randomised Controlled Trial. Lancet 2015, 386, 964–973. [Google Scholar] [CrossRef]
- Rubino, F.; Nathan, D.M.; Eckel, R.H.; Schauer, P.R.; Alberti, K.G.M.M.; Zimmet, P.Z.; Del Prato, S.; Ji, L.; Sadikot, S.M.; Herman, W.H.; et al. Metabolic Surgery in the Treatment Algorithm for Type 2 Diabetes: A Joint Statement by International Diabetes Organizations. Diabetes Care 2016, 39, 861–877. [Google Scholar] [CrossRef] [PubMed]
- Aminian, A.; Brethauer, S.A.; Andalib, A.; Nowacki, A.S.; Jimenez, A.; Corcelles, R.; Hanipah, Z.N.; Punchai, S.; Bhatt, D.L.; Kashyap, S.R.; et al. Individualized Metabolic Surgery Score: Procedure Selection Based on Diabetes Severity. Ann. Surg. 2017, 266, 650–657. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.-C.; Hsu, N.-Y.; Lee, W.-J.; Chen, S.-C.; Ser, K.-H.; Lee, Y.-C. Prediction of Type 2 Diabetes Remission after Metabolic Surgery: A Comparison of the Individualized Metabolic Surgery Score and the ABCD Score. Surg. Obes. Relat. Dis. 2018, 14, 640–645. [Google Scholar] [CrossRef] [PubMed]
- Ashrafian, H.; Bueter, M.; Ahmed, K.; Suliman, A.; Bloom, S.R.; Darzi, A.; Athanasiou, T. Metabolic Surgery: An Evolution through Bariatric Animal Models: Metabolic and Bariatric Surgery Animal Models. Obes. Rev. 2010, 11, 907–920. [Google Scholar] [CrossRef] [PubMed]
- Li, J.V.; Ashrafian, H.; Bueter, M.; Kinross, J.; Sands, C.; le Roux, C.W.; Bloom, S.R.; Darzi, A.; Athanasiou, T.; Marchesi, J.R.; et al. Metabolic Surgery Profoundly Influences Gut Microbial-Host Metabolic Cross-Talk. Gut 2011, 60, 1214–1223. [Google Scholar] [CrossRef] [PubMed]
- Izquierdo, A.G.; Crujeiras, A.B. Obesity-Related Epigenetic Changes After Bariatric Surgery. Front. Endocrinol. 2019, 10, 232. [Google Scholar] [CrossRef]
- Ortega, F.J.; Mercader, J.M.; Catalan, V.; Moreno-Navarrete, J.M.; Pueyo, N.; Sabater, M.; Gomez-Ambrosi, J.; Anglada, R.; Fernandez-Formoso, J.A.; Ricart, W.; et al. Targeting the Circulating MicroRNA Signature of Obesity. Clin. Chem. 2013, 59, 781–792. [Google Scholar] [CrossRef]
- Alkandari, A.; Ashrafian, H.; Sathyapalan, T.; Sedman, P.; Darzi, A.; Holmes, E.; Athanasiou, T.; Atkin, S.L.; Gooderham, N.J. Improved Physiology and Metabolic Flux after Roux-En-Y Gastric Bypass Is Associated with Temporal Changes in the Circulating MicroRNAome: A Longitudinal Study in Humans. Bmc Obes. 2018, 5, 20. [Google Scholar] [CrossRef]
- Atkin, S.L.; Ramachandran, V.; Yousri, N.A.; Benurwar, M.; Simper, S.C.; McKinlay, R.; Adams, T.D.; Najafi-Shoushtari, S.H.; Hunt, S.C. Changes in Blood MicroRNA Expression and Early Metabolic Responsiveness 21 Days Following Bariatric Surgery. Front. Endocrinol. 2019, 9, 773. [Google Scholar] [CrossRef]
- Bae, Y.; Kim, Y.; Lee, H.; Kim, H.; Jeon, J.S.; Noh, H.; Han, D.C.; Ryu, S.; Kwon, S.H. Bariatric Surgery Alters MicroRNA Content of Circulating Exosomes in Patients with Obesity. Obesity 2019, 27, 264–271. [Google Scholar] [CrossRef] [PubMed]
- Blum, A.; Yehuda, H.; Geron, N.; Meerson, A. Elevated Levels of MiR-122 in Serum May Contribute to Improved Endothelial Function and Lower Oncologic Risk Following Bariatric Surgery. Isr. Med. Assoc. J. Imaj 2017, 19, 620–624. [Google Scholar] [PubMed]
- Guo, W.; Han, H.; Wang, Y.; Zhang, X.; Liu, S.; Zhang, G.; Hu, S. MiR-200a Regulates Rheb-Mediated Amelioration of Insulin Resistance after Duodenal–Jejunal Bypass. Int. J. Obes. 2016, 40, 1222–1232. [Google Scholar] [CrossRef] [PubMed]
- Wei, G.; Yi, S.; Yong, D.; Shaozhuang, L.; Guangyong, Z.; Sanyuan, H. MiR-320 Mediates Diabetes Amelioration after Duodenal-Jejunal Bypass via Targeting AdipoR1. Surg. Obes. Relat. Dis. 2018, 14, 960–971. [Google Scholar] [CrossRef] [PubMed]
- Hohensinner, P.J.; Kaun, C.; Ebenbauer, B.; Hackl, M.; Demyanets, S.; Richter, D.; Prager, M.; Wojta, J.; Rega-Kaun, G. Reduction of Premature Aging Markers After Gastric Bypass Surgery in Morbidly Obese Patients. Obes. Surg. 2018, 28, 2804–2810. [Google Scholar] [CrossRef] [PubMed]
- Hubal, M.J.; Nadler, E.P.; Ferrante, S.C.; Barberio, M.D.; Suh, J.-H.; Wang, J.; Dohm, G.L.; Pories, W.J.; Mietus-Snyder, M.; Freishtat, R.J. Circulating Adipocyte-Derived Exosomal MicroRNAs Associated with Decreased Insulin Resistance after Gastric Bypass: Gastric Bypass Alters Exosomal MicroRNAs. Obesity 2017, 25, 102–110. [Google Scholar] [CrossRef] [PubMed]
- Hulsmans, M.; Sinnaeve, P.; Van der Schueren, B.; Mathieu, C.; Janssens, S.; Holvoet, P. Decreased MiR-181a Expression in Monocytes of Obese Patients Is Associated with the Occurrence of Metabolic Syndrome and Coronary Artery Disease. J. Clin. Endocrinol. Metab. 2012, 97, E1213–E1218. [Google Scholar] [CrossRef] [PubMed]
- Kwon, I.G.; Ha, T.K.; Ryu, S.-W.; Ha, E. Roux-En-Y Gastric Bypass Stimulates Hypothalamic MiR-122 and Inhibits Cardiac and Hepatic MiR-122 Expressions. J. Surg. Res. 2015, 199, 371–377. [Google Scholar] [CrossRef]
- Lirun, K.; Sewe, M.; Yong, W. A Pilot Study: The Effect of Roux-En-Y Gastric Bypass on the Serum MicroRNAs of the Type 2 Diabetes Patient. Obes. Surg. 2015, 25, 2386–2392. [Google Scholar] [CrossRef]
- Mysore, R.; Ortega, F.J.; Latorre, J.; Ahonen, M.; Savolainen-Peltonen, H.; Fischer-Posovszky, P.; Wabitsch, M.; Olkkonen, V.M.; Fernández-Real, J.M.; Haridas, P.A.N. MicroRNA-221-3p Regulates Angiopoietin-Like 8 (ANGPTL8) Expression in Adipocytes. J. Clin. Endocrinol. Metab. 2017, 102, 4001–4012. [Google Scholar] [CrossRef]
- Ortega, F.J.; Mercader, J.M.; Moreno-Navarrete, J.M.; Nonell, L.; Puigdecanet, E.; Rodriquez-Hermosa, J.I.; Rovira, O.; Xifra, G.; Guerra, E.; Moreno, M.; et al. Surgery-Induced Weight Loss Is Associated With the Downregulation of Genes Targeted by MicroRNAs in Adipose Tissue. J. Clin. Endocrinol. Metab. 2015, 100, E1467–E1476. [Google Scholar] [CrossRef] [PubMed]
- Ortega, F.J.; Moreno, M.; Mercader, J.M.; Moreno-Navarrete, J.M.; Fuentes-Batllevell, N.; Sabater, M.; Ricart, W.; Fernández-Real, J.M. Inflammation Triggers Specific MicroRNA Profiles in Human Adipocytes and Macrophages and in Their Supernatants. Clin. Epigenetics 2015, 7, 49. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Wang, D.-S.; Cheng, Y.-S.; Jia, B.-L.; Yu, G.; Yin, X.-Q.; Wang, Y. Expression of MicroRNA-448 and SIRT1 and Prognosis of Obese Type 2 Diabetic Mellitus Patients After Laparoscopic Bariatric Surgery. Cell. Physiol. Biochem. 2018, 45, 935–950. [Google Scholar] [CrossRef] [PubMed]
- Wu, Q.; Li, J.V.; Seyfried, F.; le Roux, C.W.; Ashrafian, H.; Athanasiou, T.; Fenske, W.; Darzi, A.; Nicholson, J.K.; Holmes, E.; et al. Metabolic Phenotype-MicroRNA Data Fusion Analysis of the Systemic Consequences of Roux-En-Y Gastric Bypass Surgery. Int. J. Obes. 2015, 39, 1126–1134. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.; Kim, M.; Han, J.; Yeom, K.-H.; Lee, S.; Baek, S.H.; Kim, V.N. MicroRNA Genes Are Transcribed by RNA Polymerase II. Embo J. 2004, 23, 4051–4060. [Google Scholar] [CrossRef] [PubMed]
- Bartel, D.P. MicroRNAs. Cell 2004, 116, 281–297. [Google Scholar] [CrossRef]
- Kim, V.N.; Han, J.; Siomi, M.C. Biogenesis of Small RNAs in Animals. Nat. Rev. Mol. Cell Biol. 2009, 10, 126–139. [Google Scholar] [CrossRef]
- Krol, J.; Loedige, I.; Filipowicz, W. The Widespread Regulation of MicroRNA Biogenesis, Function and Decay. Nat. Rev. Genet. 2010, 11, 597–610. [Google Scholar] [CrossRef]
- Lim, L.P.; Lau, N.C.; Garrett-Engele, P.; Grimson, A.; Schelter, J.M.; Castle, J.; Bartel, D.P.; Linsley, P.S.; Johnson, J.M. Microarray Analysis Shows That Some MicroRNAs Downregulate Large Numbers of Target MRNAs. Nature 2005, 433, 769–773. [Google Scholar] [CrossRef]
- Hausser, J.; Zavolan, M. Identification and Consequences of MiRNA–Target Interactions—Beyond Repression of Gene Expression. Nat. Rev. Genet. 2014, 15, 599–612. [Google Scholar] [CrossRef]
- Ebert, M.S.; Sharp, P.A. Roles for MicroRNAs in Conferring Robustness to Biological Processes. Cell 2012, 149, 515–524. [Google Scholar] [CrossRef] [PubMed]
- Karbiener, M.; Fischer, C.; Nowitsch, S.; Opriessnig, P.; Papak, C.; Ailhaud, G.; Dani, C.; Amri, E.-Z.; Scheideler, M. MicroRNA MiR-27b Impairs Human Adipocyte Differentiation and Targets PPARγ. Biochem. Biophys. Res. Commun. 2009, 390, 247–251. [Google Scholar] [CrossRef] [PubMed]
- Plaisance, V.; Abderrahmani, A.; Perret-Menoud, V.; Jacquemin, P.; Lemaigre, F.; Regazzi, R. MicroRNA-9 Controls the Expression of Granuphilin/Slp4 and the Secretory Response of Insulin-Producing Cells. J. Biol. Chem. 2006, 281, 26932–26942. [Google Scholar] [CrossRef] [PubMed]
- Lu, H.; Buchan, R.J.; Cook, S.A. MicroRNA-223 Regulates Glut4 Expression and Cardiomyocyte Glucose Metabolism. Cardiovasc. Res. 2010, 86, 410–420. [Google Scholar] [CrossRef] [PubMed]
- Esau, C.; Davis, S.; Murray, S.F.; Yu, X.X.; Pandey, S.K.; Pear, M.; Watts, L.; Booten, S.L.; Graham, M.; McKay, R.; et al. MiR-122 Regulation of Lipid Metabolism Revealed by in Vivo Antisense Targeting. Cell Metab. 2006, 3, 87–98. [Google Scholar] [CrossRef] [PubMed]
- Oger, F.; Gheeraert, C.; Mogilenko, D.; Benomar, Y.; Molendi-Coste, O.; Bouchaert, E.; Caron, S.; Dombrowicz, D.; Pattou, F.; Duez, H.; et al. Cell-Specific Dysregulation of MicroRNA Expression in Obese White Adipose Tissue. J. Clin. Endocrinol. Metab. 2014, 99, 2821–2833. [Google Scholar] [CrossRef]
- Hilton, C.; Neville, M.J.; Karpe, F. MicroRNAs in Adipose Tissue: Their Role in Adipogenesis and Obesity. Int. J. Obes. 2013, 37, 325–332. [Google Scholar] [CrossRef]
- Trajkovski, M.; Hausser, J.; Soutschek, J.; Bhat, B.; Akin, A.; Zavolan, M.; Heim, M.H.; Stoffel, M. MicroRNAs 103 and 107 Regulate Insulin Sensitivity. Nature 2011, 474, 649–653. [Google Scholar] [CrossRef]
- Arner, P.; Kulyté, A. MicroRNA Regulatory Networks in Human Adipose Tissue and Obesity. Nat. Rev. Endocrinol. 2015, 11, 276–288. [Google Scholar] [CrossRef]
- Guay, C.; Regazzi, R. Circulating MicroRNAs as Novel Biomarkers for Diabetes Mellitus. Nat. Rev. Endocrinol. 2013, 9, 513–521. [Google Scholar] [CrossRef]
- Rottiers, V.; Näär, A.M. MicroRNAs in Metabolism and Metabolic Disorders. Nat. Rev. Mol. Cell Biol. 2012, 13, 239–250. [Google Scholar] [CrossRef] [PubMed]
- Xie, H.; Lim, B.; Lodish, H.F. MicroRNAs Induced During Adipogenesis That Accelerate Fat Cell Development Are Downregulated in Obesity. Diabetes 2009, 58, 1050–1057. [Google Scholar] [CrossRef] [PubMed]
- Hulsmans, M.; De Keyzer, D.; Holvoet, P. MicroRNAs Regulating Oxidative Stress and Inflammation in Relation to Obesity and Atherosclerosis. Faseb J. 2011, 25, 2515–2527. [Google Scholar] [CrossRef] [PubMed]
- Bramer, W.M.; Rethlefsen, M.L.; Kleijnen, J.; Franco, O.H. Optimal Database Combinations for Literature Searches in Systematic Reviews: A Prospective Exploratory Study. Syst. Rev. 2017, 6, 245. [Google Scholar] [CrossRef] [PubMed]
- Zhu, H.; Leung, S. Identification of Potential MicroRNA Biomarkers by Meta-Analysis. In Computational Drug Discovery and Design; Gore, M., Jagtap, U.B., Eds.; Springer: New York, NY, USA, 2018; Volume 1762, pp. 473–484. [Google Scholar] [CrossRef]
- Vlachos, I.S.; Zagganas, K.; Paraskevopoulou, M.D.; Georgakilas, G.; Karagkouni, D.; Vergoulis, T.; Dalamagas, T.; Hatzigeorgiou, A.G. DIANA-MiRPath v3.0: Deciphering MicroRNA Function with Experimental Support. Nucleic Acids Res. 2015, 43, W460–W466. [Google Scholar] [CrossRef] [PubMed]
- The FANTOM Consortium; de Rie, D.; Abugessaisa, I.; Alam, T.; Arner, E.; Arner, P.; Ashoor, H.; Åström, G.; Babina, M.; Bertin, N.; et al. An Integrated Expression Atlas of MiRNAs and Their Promoters in Human and Mouse. Nat. Biotechnol. 2017, 35, 872–878. [Google Scholar] [CrossRef]
- Martín-Núñez, G.M.; Cabrera-Mulero, A.; Alcaide-Torres, J.; García-Fuentes, E.; Tinahones, F.J.; Morcillo, S. No Effect of Different Bariatric Surgery Procedures on LINE-1 DNA Methylation in Diabetic and Nondiabetic Morbidly Obese Patients. Surg. Obes. Relat. Dis. 2017, 13, 442–450. [Google Scholar] [CrossRef]
- Balasar, Ö.; Çakır, T.; Erkal, Ö.; Aslaner, A.; Çekiç, B.; Uyar, M.; Bülbüller, N.; Oruç, M.T. The Effect of Rs9939609 FTO Gene Polymorphism on Weight Loss after Laparoscopic Sleeve Gastrectomy. Surg. Endosc. 2016, 30, 121–125. [Google Scholar] [CrossRef]
- Käkelä, P.; Jääskeläinen, T.; Torpström, J.; Ilves, I.; Venesmaa, S.; Pääkkönen, M.; Gylling, H.; Paajanen, H.; Uusitupa, M.; Pihlajamäki, J. Genetic Risk Score Does Not Predict the Outcome of Obesity Surgery. Obes. Surg. 2014, 24, 128–133. [Google Scholar] [CrossRef]
- Zhang, W.; Lu, W.; Ananthan, S.; Suto, M.J.; Li, Y. Discovery of Novel Frizzled-7 Inhibitors by Targeting the Receptor’s Transmembrane Domain. Oncotarget 2017, 8. [Google Scholar] [CrossRef]
- Christodoulides, C.; Scarda, A.; Granzotto, M.; Milan, G.; Dalla Nora, E.; Keogh, J.; De Pergola, G.; Stirling, H.; Pannacciulli, N.; Sethi, J.K.; et al. WNT10B Mutations in Human Obesity. Diabetologia 2006, 49, 678–684. [Google Scholar] [CrossRef] [PubMed]
- Mayas, M.D.; Ortega, F.J.; Macías-González, M.; Bernal, R.; Gómez-Huelgas, R.; Fernández-Real, J.M.; Tinahones, F.J. Inverse Relation between FASN Expression in Human Adipose Tissue and the Insulin Resistance Level. Nutr. Metab. 2010, 7, 3. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Wang, D.; DuBois, R.N. Associations Between Obesity and Cancer: The Role of Fatty Acid Synthase. Jnci J. Natl. Cancer Inst. 2012, 104, 343–345. [Google Scholar] [CrossRef] [PubMed]
- O’Connell, R.M.; Rao, D.S.; Baltimore, D. MicroRNA Regulation of Inflammatory Responses. Annu. Rev. Immunol. 2012, 30, 295–312. [Google Scholar] [CrossRef] [PubMed]
- Moore, C.S.; Rao, V.T.S.; Durafourt, B.A.; Bedell, B.J.; Ludwin, S.K.; Bar-Or, A.; Antel, J.P. MiR-155 as a Multiple Sclerosis–Relevant Regulator of Myeloid Cell Polarization. Ann. Neurol. 2013, 74, 709–720. [Google Scholar] [CrossRef]
- Faraoni, I.; Antonetti, F.R.; Cardone, J.; Bonmassar, E. MiR-155 Gene: A Typical Multifunctional MicroRNA. Biochim. Biophys. Acta Bba Mol. Basis Dis. 2009, 1792, 497–505. [Google Scholar] [CrossRef]
- Ding, S.; Huang, H.; Xu, Y.; Zhu, H.; Zhong, C. MiR-222 in Cardiovascular Diseases: Physiology and Pathology. Biomed Res. Int. 2017, 2017. [Google Scholar] [CrossRef]
- Li, Y.; Kowdley, K.V. Method for MicroRNA Isolation from Clinical Serum Samples. Anal. Biochem. 2012, 431, 69–75. [Google Scholar] [CrossRef]
- Kroh, E.M.; Parkin, R.K.; Mitchell, P.S.; Tewari, M. Analysis of Circulating MicroRNA Biomarkers in Plasma and Serum Using Quantitative Reverse Transcription-PCR (QRT-PCR). Methods 2010, 50, 298–301. [Google Scholar] [CrossRef]
- Chugh, P.; Dittmer, D.P. Potential Pitfalls in MicroRNA Profiling. Wiley Interdiscip. Rev. Rna 2012, 3, 601–616. [Google Scholar] [CrossRef]
- Git, A.; Dvinge, H.; Salmon-Divon, M.; Osborne, M.; Kutter, C.; Hadfield, J.; Bertone, P.; Caldas, C. Systematic Comparison of Microarray Profiling, Real-Time PCR, and next-Generation Sequencing Technologies for Measuring Differential MicroRNA Expression. RNA 2010, 16, 991–1006. [Google Scholar] [CrossRef] [PubMed]
- Sato, F.; Tsuchiya, S.; Terasawa, K.; Tsujimoto, G. Intra-Platform Repeatability and Inter-Platform Comparability of MicroRNA Microarray Technology. PLoS ONE 2009, 4, e5540. [Google Scholar] [CrossRef] [PubMed]
- Schwarzenbach, H.; da Silva, A.M.; Calin, G.; Pantel, K. Which Is the Accurate Data Normalization Strategy for MicroRNA Quantification? Clin. Chem. 2015, 61, 1333–1342. [Google Scholar] [CrossRef] [PubMed]
- Vandesompele, J.; De Preter, K.; Pattyn, F.; Poppe, B.; Van Roy, N.; De Paepe, A.; Speleman, F. Accurate Normalization of Real-Time Quantitative RT-PCR Data by Geometric Averaging of Multiple Internal Control Genes. Genome Biol. 2002, 3, RESEARCH0034. [Google Scholar] [CrossRef] [PubMed]
- Huang, R.S.; Gamazon, E.R.; Ziliak, D.; Wen, Y.; Im, H.K.; Zhang, W.; Wing, C.; Duan, S.; Bleibel, W.K.; Cox, N.J.; et al. Population Differences in MicroRNA Expression and Biological Implications. Rna Biol. 2011, 8, 692–701. [Google Scholar] [CrossRef] [PubMed]
- Rawlings-Goss, R.A.; Campbell, M.C.; Tishkoff, S.A. Global Population-Specific Variation in MiRNA Associated with Cancer Risk and Clinical Biomarkers. Bmc Med. Genom. 2014, 7, 53. [Google Scholar] [CrossRef] [PubMed]
- Cui, C.; Yang, W.; Shi, J.; Zhou, Y.; Yang, J.; Cui, Q.; Zhou, Y. Identification and Analysis of Human Sex-Biased MicroRNAs. Genom. Proteom. Bioinform. 2018, 16, 200–211. [Google Scholar] [CrossRef] [PubMed]
- Guo, L.; Zhang, Q.; Ma, X.; Wang, J.; Liang, T. MiRNA and MRNA Expression Analysis Reveals Potential Sex-Biased MiRNA Expression. Sci. Rep. 2017, 7, 39812. [Google Scholar] [CrossRef]
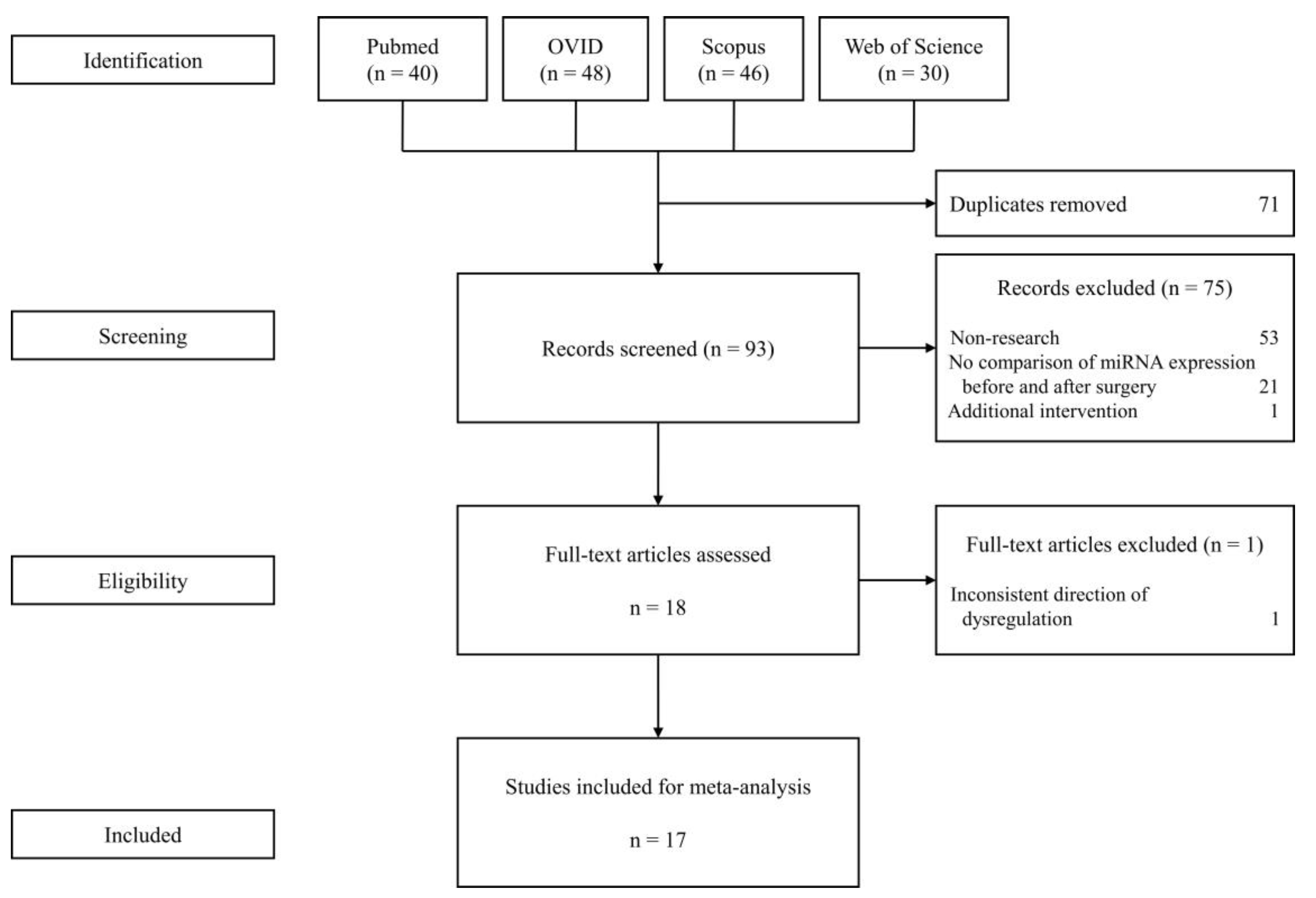
| Study | Year | Country | Sample Size | Sex (Males/Females) |
|---|---|---|---|---|
| Human studies (comparing before vs. after bariatric surgery) | ||||
| Ortega et al. [29] | 2013 | Spain | 22 | 5/17 |
| Alkandari et al. [30] | 2018 | UK | 9 | 4/5 |
| Atkin et al. [31] | 2019 | USA | 29 | 9/20 |
| Bae et al. [32] | 2019 | South Korea | 12 | Unspecified |
| Blum et al. [33] | 2017 | Israel | 21 | 14/7 |
| Hohensinner et al. [36] | 2018 | Austria | 58 | 17/41 |
| Hubal et al. [37] | 2017 | USA | 6 | 0/6 |
| Hulsmans et al. [38] | 2012 | Belgium | 21 | 7/14 |
| Lirun et al. [40] | 2015 | China | 18 | 4/11 |
| Mysore et al. [41] | 2017 | Spain | 22 | 0/22 |
| Ortega et al. [42] | 2015 | Spain | 25 | 0/25 |
| Ortega et al. [43] | 2015 | Spain | 9 | 0/9 |
| Wang et al. [44] | 2018 | China | 124 | 46/78 |
| Animal studies (comparing bariatric vs sham surgery) | ||||
| Guo et al. [34] | 2016 | China | 35 | 35/0 |
| Wei et al. [35] | 2018 | China | 45 | 45/0 |
| Kwon et al. [39] | 2015 | South Korea | 25 | 25/0 |
| Wu et al. [45] | 2015 | UK | 12 | 12/0 |
| Study | Tissue | Isolation | Platform | Normalization |
|---|---|---|---|---|
| Human studies | ||||
| Ortega et al. [29] | Plasma | mirVana PARIS Isolation Kit | TaqMan array miRNA cards in a subset and qPCR in the final sample | Geometric mean of six miRNAs (hsa-miR-106a-5p, hsa-miR-146a-5p, hsa-miR-19b-3p, hsa-miR-223-3p, hsa-miR-186-5p, hsa-miR-199a-3p) |
| Alkandari et al. [30] | Plasma | mirVana PARIS Isolation Kit | miRCURY qPCR panel | Four miRNAs (hsa-miR-223-3p, hsa-miR-26a-5p, hsa-miR-101-3p, and hsa-miR-19a-3p) |
| Atkin et al. [31] | Plasma | miRCURY RNA Isolation kit | qPCR and a FANTOM miRNA atlas [68] | Global mean |
| Bae et al. [32] | Exosome | miRNeasy Mini Kit | Small RNA sequencing | Relative log expression using DESeq2 |
| Blum et al. [33] | Serum | miRNeasy serum/plasma kit | RNA sequencing in a subset and qPCR in the final sample | hsa-miR-451a |
| Hohensinner et al. [36] | Plasma | miRNA tissue lysis kit | qPCR | RNA spike-in |
| Hubal et al. [37] | Exosome | mirVANA miRNA Isolation Kit | GeneChip miRNA 4.0 Array | RMA algorithm |
| Hulsmans et al. [38] | Monocytes | TRIzol reagent | qPCR | RNU5G |
| Lirun et al. [40] | Plasma | mirVana RNA Isolation Kit | GeneChip miRNA 3.0 Array | RMA algorithm |
| Mysore et al. [41] | Subcutaneous Adipose Tissue (SAT) | miRNeasy Mini kit | qPCR | RNU44 |
| Ortega et al. [42] | SAT | miRNeasy Mini Kit | GeneChip miRNA 3.0 array in a subset and qPCR in the final sample | RMA algorithm and RNU48 |
| Ortega et al. [43] | SAT | miRNeasy Mini Kit | qPCR | RNU6B |
| Wang et al. [44] | Circulating Endothelial Progenitor Cells | High Pure RNA kit | qPCR | RNU6 |
| Animal studies | ||||
| Guo et al. [34] | Liver | TRIzol reagent | miProfile Customized Rat qPCR arrays | 5S rRNA and RsnRNA U6 |
| Wei et al. [35] | Liver | TRIzol reagent | miProfile Customized Rat qPCR arrays | 5S rRNA, RsnRNA U6, rno-miR-25, and rno-miR-186 |
| Kwon et al. [39] | Hypothalamus, Heart, and Liver | Unspecified | Agilent Rat miRNA 8x15k microarray for hypothalamus and heart samples, then qPCR for liver and validation | Whole-array and RNU6 |
| Wu et al. [45] | Plasma and Liver | mirVANA PARIS RNA Isolation kit | TaqMan Array Rodent Card | RNU6-1, RNU6-2, rno-miR-16-5p, rno-miR-223-3p, mmu-miR-1937b |
| Study | Year | Bariatric Surgery Type | Time of Observation after Surgery |
|---|---|---|---|
| Human studies | |||
| Ortega et al. [29] | 2013 | RYGB | 12 months |
| Alkandari et al. [30] | 2018 | RYGB | 1, 3, 6, 9, and 12 months |
| Atkin et al. [31] | 2019 | RYGB | 21 days |
| Bae et al. [32] | 2019 | RYGB and SG | 6 months |
| Blum et al. [33] | 2017 | SG | 3 months |
| Hohensinner et al. [36] | 2018 | RYGB | 24 months |
| Hubal et al. [37] | 2017 | RYGB | 12 months |
| Hulsmans et al. [38] | 2012 | RYGB | 3 months |
| Lirun et al. [40] | 2015 | RYGB | 3 months |
| Mysore et al. [41] | 2017 | RYGB | 24 months |
| Ortega et al. [42] | 2015 | RYGB | 24 months |
| Ortega et al. [43] | 2015 | RYGB | 24 months |
| Wang et al. [44] | 2018 | Not specified | 3 months |
| Animal studies | |||
| Guo et al. [34] | 2016 | DJB and SG | 2, 4, 8 weeks |
| Wei et al. [35] | 2018 | DJB | 2, 4, 8 weeks |
| Kwon et al. [39] | 2015 | RYGB | 25 days |
| Wu et al. [45] | 2015 | RYGB | 53 days |
| miRNA | miRBase | References | Direction of Expression | No. of Subjects | Tissue | Time of Observation | |
|---|---|---|---|---|---|---|---|
| Group 1 miRNAs (same direction of expression after surgery in two or more studies) | |||||||
| 1 | hsa-miR-93-5p | MIMAT0000093 | Lirun [40] | − | 15 | Plasma | 3 months |
| Alkandari [30] | 9 | Plasma | 3 months | ||||
| 2 | hsa-miR-106b-5p | MIMAT0000680 | Lirun [40] | − | 15 | Plasma | 3 months |
| Alkandari [30] | 9 | Plasma | 3, 12 months | ||||
| 3 | hsa-let-7b-5p | MIMAT0000063 | Lirun [40] | − | 15 | Plasma | 3 months |
| Alkandari [30] | 9 | Plasma | 3 months | ||||
| 4 | hsa-let-7i-5p | MIMAT0000415 | Lirun [40] | − | 15 | Plasma | 3 months |
| Alkandari [30] | 9 | Plasma | 6, 9 months | ||||
| Atkin [31] | 29 | Plasma | 21 days | ||||
| 5 | hsa-miR-16-5p | MIMAT0000069 | Lirun [40] | − | 15 | Plasma | 3 months |
| Hubal [37] | 6 | Exosomes | 12 months | ||||
| 6 | hsa-miR-19b-3p | MIMAT0000074 | Ortega [43] | − | 9 | SAT | 24 months |
| Lirun [40] | 15 | Plasma | 3 months | ||||
| Ortega [29] | 22 | Plasma | 12 months | ||||
| 7 | hsa-miR-92a-3p | MIMAT0000092 | Lirun [40] | − | 15 | Plasma | 3 months |
| Alkandari [30] | 9 | Plasma | 9, 12 months | ||||
| 8 | hsa-miR-222-3p | MIMAT0000279 | Ortega [29] | − | 22 | Plasma | 12 months |
| Ortega [43] | 9 | SAT | 24 months | ||||
| 9 | hsa-miR-142-3p | MIMAT0000434 | Bae [32] | − | 12 | Exosome | 6 months |
| Ortega [29] | 22 | Plasma | 12 months | ||||
| 10 | hsa-miR-140-5p | MIMAT0000431 | Bae [32] | − | 12 | Exosome | 6 months |
| Ortega [29] | 22 | Plasma | 12 months | ||||
| 11 | hsa-miR-155-5p | MIMAT0000646 | Ortega [43] | − | 9 | SAT | 24 months |
| Ortega [42] | 25 | SAT | 24 months | ||||
| 12 | rno-miR-320-3p | MIMAT0000903 | Wu [45] | − | 4 | Plasma | 53 days |
| Wei [35] | 5 | liver | 2 months | ||||
| 13 | hsa-miR-320c | MIMAT0005793 | Atkin [31] | + | 29 | Plasma | 21 days |
| Lirun [40] | 15 | Plasma | 3 months | ||||
| 14 | hsa-miR-7-5p | MIMAT0000252 | Atkin [31] | + | 29 | Plasma | 21 days |
| Bae [32] | 12 | Exosome | 6 months | ||||
| Group 2 miRNAs (overall same direction of expression after surgery in two or more studies) | |||||||
| 1 | hsa-miR-125b-5p | MIMAT0000423 | Ortega [29] | − | 22 | Plasma | 12 months |
| Alkandari [30] | − | 9 | Plasma | 6, 9, 12 months | |||
| Hubal [37] | + | 6 | Exosomes | 12 months | |||
| 2 | hsa-miR-130b-3p | MIMAT0000691 | Ortega [42] | − | 25 | SAT | 24 months |
| Alkandari [30] | − | 9 | Plasma | 12 months | |||
| Ortega [29] | + | 22 | Plasma | 12 months | |||
| 3 | hsa-miR-221-3p | MIMAT0000278 | Ortega [43] | − | 9 | SAT | 24 months |
| Ortega [42] | − | 25 | SAT | 24 months | |||
| Mysore [41] | − | 22 | SAT | 24 months | |||
| Lirun [40] | − | 15 | Plasma | 3 months | |||
| Ortega [29] | + | 22 | Plasma | 12 months | |||
| 4 | rno-miR-122-5p | MIMAT0000827 | Kwon [39] | − | 25 | heart | 25 days |
| Kwon [39] | − | 25 | liver | 25 days | |||
| Wu [45] | − | 4 | Plasma | 53 days | |||
| Wu [45] | − | 8 | Liver | 53 days | |||
| Kwon [39] | + | 25 | hypothalamus | 25 days | |||
| 5 | hsa-miR-146a-5p | MIMAT0000449 | Lirun [40] | − | 15 | Plasma | 3 months |
| Ortega [43] | − | 9 | SAT | 24 months | |||
| Ortega [29] | + | 22 | Plasma | 12 months | |||
| 6 | rno-miR-503-5p | MIMAT0003213 | Kwon [39] | + | 25 | hypothalamus | 25 days |
| Kwon [39] | + | 25 | heart | 25 days | |||
| Wei [35] | − | 4 | liver | 2 months | |||
| Group 3 miRNAs (reported in at least two studies, but with no agreement in direction of expression) | |||||||
| 1 | hsa-miR-21-5p | MIMAT0000076 | Alkandari [30] | − | 9 | Plasma | 9, 12 months |
| Ortega [29] | + | 22 | Plasma | 12 months | |||
| 2 | hsa-miR-33a-5p | MIMAT0000091 | Bae [32] | − | 12 | Exosome | 6 months |
| Alkandari [30] | + | 9 | Plasma | 6 months | |||
| 3 | hsa-miR-320a-3p | MIMAT0000510 | Alkandari [30] | − | 9 | Plasma | 6, 9, 12 months |
| Lirun [40] | + | 15 | Plasma | 3 months | |||
| 4 | hsa-miR-320b | MIMAT0005792 | Alkandari [30] | − | 9 | Plasma | 9 months |
| Lirun [40] | + | 15 | Plasma | 3 months | |||
| 5 | hsa-miR-378a-3p | MIMAT0000732 | Alkandari [30] | − | 9 | Plasma | 6, 9, 12 months |
| Lirun [40] | + | 15 | Plasma | 3 months | |||
| 6 | hsa-miR-103-3p | MIMAT0000101 | Lirun [40] | − | 15 | Plasma | 3 months |
| Hubal [37] | + | 6 | Exosomes | 12 months | |||
| 7 | rno-miR-133b-3p | MIMAT0003126 | Wei [35] | − | 4 | liver | 2 months |
| Kwon [39] | + | 25 | hypothalamus | 25 days | |||
| 8 | rno-miR-194-5p | MIMAT0000869 | Kwon [39] | − | 25 | heart | 25 days |
| Guo [34] | + | 4 | liver | 2 months | |||
| 9 | hsa-miR-122-5p | MIMAT0000421 | Ortega [29] | − | 22 | Plasma | 12 months |
| Blum [33] | + | 21 | Serum | 3 months | |||
| 10 | rno-miR-146a-5p | MIMAT0000852 | Wu [45] | − | 4 | Plasma | 53 days |
| Kwon [39] | + | 25 | hypothalamus | 25 days | |||
| 11 | rno-miR-542-3p | MIMAT0003179 | Wei [35] | − | 4 | liver | 2 months |
| Kwon [39] | + | 25 | hypothalamus | 25 days | |||
| 12 | hsa-miR-191-5p | MIMAT0000440 | Lirun [40] | − | 15 | Plasma | 3 months |
| Bae [32] | + | 12 | Exosome | 6 months | |||
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Langi, G.; Szczerbinski, L.; Kretowski, A. Meta-Analysis of Differential miRNA Expression after Bariatric Surgery. J. Clin. Med. 2019, 8, 1220. https://doi.org/10.3390/jcm8081220
Langi G, Szczerbinski L, Kretowski A. Meta-Analysis of Differential miRNA Expression after Bariatric Surgery. Journal of Clinical Medicine. 2019; 8(8):1220. https://doi.org/10.3390/jcm8081220
Chicago/Turabian StyleLangi, Gladys, Lukasz Szczerbinski, and Adam Kretowski. 2019. "Meta-Analysis of Differential miRNA Expression after Bariatric Surgery" Journal of Clinical Medicine 8, no. 8: 1220. https://doi.org/10.3390/jcm8081220
APA StyleLangi, G., Szczerbinski, L., & Kretowski, A. (2019). Meta-Analysis of Differential miRNA Expression after Bariatric Surgery. Journal of Clinical Medicine, 8(8), 1220. https://doi.org/10.3390/jcm8081220





