Rapid Characterization of Virulence Determinants in Helicobacter pylori Isolated from Non-Atrophic Gastritis Patients by Next-Generation Sequencing
Abstract
:1. Introduction
2. Materials and Methods
2.1. Histology of Gastric Biopsy Specimens
2.2. Clinical H. pylori Isolates, Culture Methods and Phenotypic Drug Susceptibility Testing
2.3. DNA Extraction, Library Preparation and Sequencing of H. pylori Strains
2.4. Bioinformatic and Statistical Analysis
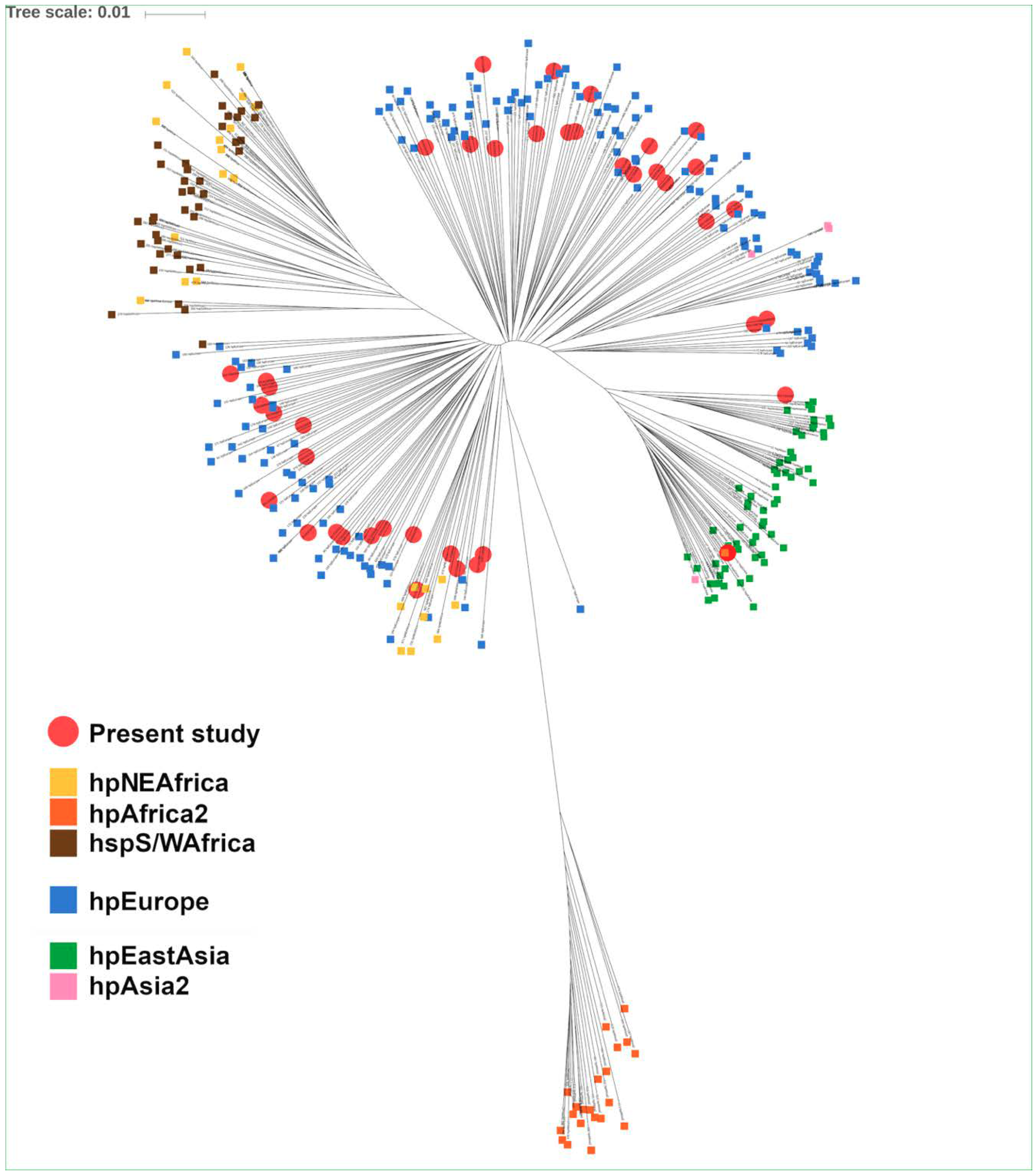
2.5. Genotyping and Phylogenetic Analysis
2.6. Ethics Approval, Consent to Participate and Consent for Publication
3. Results and Discussion
3.1. Patient Demographics and H. pylori Epidemiology
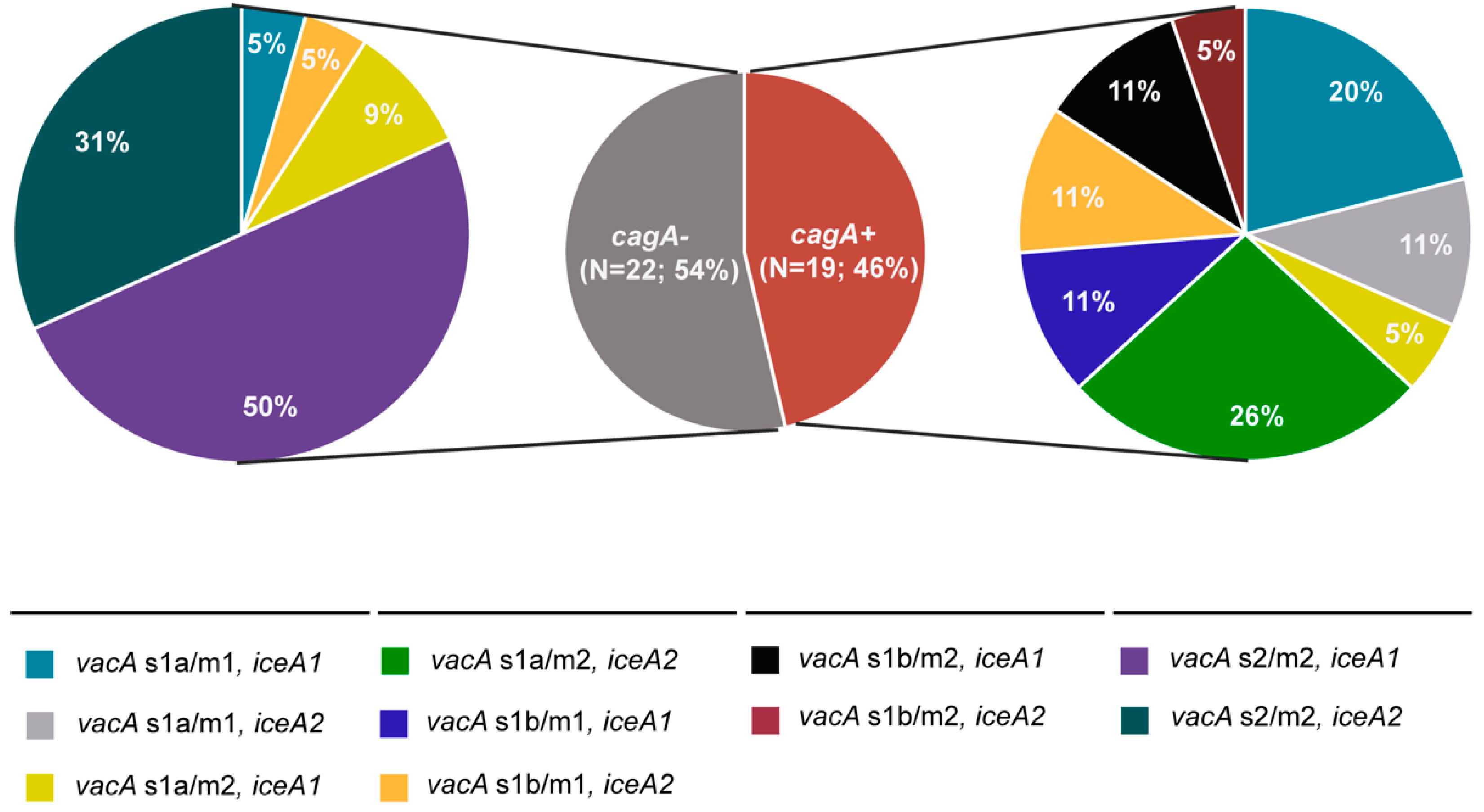
3.2. Presence of Virulence Determinants in H. pylori Isolates
3.3. Presence of Genes Encoding OMPs in H. pylori Isolates
3.4. Presence of Multiple Virulence Determinants and OMPs in H. pylori Isolates
4. Conclusion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Availability of Data and Material
Conflicts of Interest
References
- Wroblewski, L.E.; Peek, R.M.; Wilson, K.T. Helicobacter pylori and gastric cancer: Factors that modulate disease risk. Clin. Microbiol. Rev. 2010, 23, 713–739. [Google Scholar] [CrossRef] [PubMed]
- Jonaitis, L.; Pellicano, R.; Kupcinskas, L. Helicobacter pylori and nonmalignant upper gastrointestinal diseases. Helicobacter 2018. [Google Scholar] [CrossRef] [PubMed]
- Atherton, J.C.; Cao, P.; Peek, R.M.; Tummuru, M.K.; Blaser, M.J.; Cover, T.L. Mosaicism in vacuolating cytotoxin alleles of Helicobacter pylori association of specific vacA types with cytotoxin production and peptic ulceration. J. Biol. Chem. 1995, 270, 17771–17777. [Google Scholar] [CrossRef] [PubMed]
- Blaser, M.J.; Perez-Perez, G.I.; Kleanthous, H.; Cover, T.L.; Peek, R.M.; Chyou, P.; Stemmermann, G.N.; Nomura, A. Infection with Helicobacter pylori strains possessing cagA is associated with an increased risk of developing adenocarcinoma of the stomach. Cancer Res. 1995, 55, 2111–2115. [Google Scholar] [PubMed]
- Censini, S.; Lange, C.; Xiang, Z.; Crabtree, J.E.; Ghiara, P.; Borodovsky, M.; Rappuoli, R.; Covacci, A. Cag, a pathogenicity island of Helicobacter pylori, encodes type I-specific and disease-associated virulence factors. Proc. Natl. Acad. Sci. USA 1996, 93, 14648–14653. [Google Scholar] [CrossRef] [PubMed]
- Atherton, J.C. The pathogenesis of Helicobacter pylori induced gastro-duodenal diseases. Annu. Rev. Pathol. 2006, 1, 63–96. [Google Scholar] [CrossRef] [PubMed]
- Camilo, V.; Sugiyama, T.; Touati, E. Pathogenesis of Helicobacter pylori infection. Helicobacter 2017, 22. [Google Scholar] [CrossRef] [PubMed]
- Greenfield, L.K.; Jones, N.L. Modulation of autophagy by Helicobacter pylori and its role in gastric carcinogenesis. Trends Microbiol. 2013, 21, 602–612. [Google Scholar] [CrossRef] [PubMed]
- Atherton, J.; Peek, R., Jr.; Tham, K.; Cover, T.; Blaser, M. Clinical and pathological importance of heterogeneity in vacA, the vacuolating cytotoxin gene of Helicobacter pylori. Gastroenterology 1997, 112, 92–99. [Google Scholar] [CrossRef]
- Peek, J.R.; Thompson, S.A.; Donahue, J.P.; Tham, K.T.; Atherton, J.C.; Blaser, M.J.; Miller, G.G. Adherence to gastric epithelial cells induces expression of a Helicobacter pylori gene, iceA, that is associated with clinical outcome. Proc. Assoc. Am. Phys. 1998, 110, 531–544. [Google Scholar] [PubMed]
- van Doorn, L.J.; Figueiredo, C.; Sanna, R.; Plaisier, A.; Schneeberger, P.; de Boer, W.; Quint, W. Clinical relevance of the cagA, vacA, and iceA status of Helicobacter pylori. Gastroenterology 1998, 115, 58–66. [Google Scholar] [CrossRef]
- Hussein NR, Argent RH, Marx CK, Patel SR, Robinson K, Atherton JC: Helicobacter pylori dupA is polymorphic, and its active form induces proinflammatory cytokine secretion by mononuclear cells. J. Infect. Dis. 2010, 202, 261–269. [CrossRef] [PubMed]
- Yamaoka, Y.; Ojo, O.; Fujimoto, S.; Odenbreit, S.; Haas, R.; Gutierrez, O.; El-Zimaity, H.M.; Reddy, R.; Arnqvist, A.; Graham, D.Y. Helicobacter pylori outer membrane proteins and gastroduodenal disease. Gut 2006, 55, 775–781. [Google Scholar] [CrossRef] [PubMed]
- Ando, T.; Peek, R.M.; Lee, Y.C.; Krishna, U.; Kusugami, K.; Blaser, M.J. Host cell responses to genotypically similar Helicobacter pylori isolates from United States and Japan. Clin. Diagn. Lab. Immunol. 2002, 9, 167–175. [Google Scholar] [CrossRef] [PubMed]
- Lu, H.; Hsu, P.I.; Graham, D.Y.; Yamaoka, Y. Duodenal ulcer promoting gene of Helicobacter pylori. Gastroenterology 2005, 128, 833–848. [Google Scholar] [CrossRef]
- Abadi, A.T.B.; Taghvaei, T.; Wolfram, L.; Kusters, J.G. Infection with Helicobacter pylori strains lacking dupA is associated with an increased risk of gastric ulcer and gastric cancer development. J. Med. Microbiol. 2012, 61, 23–30. [Google Scholar] [CrossRef] [PubMed]
- Tomb, J.F.; White, O.; Kerlavage, A.R.; Clayton, R.A.; Sutton, G.G.; Fleischmann, R.D.; Ketchum, K.A.; Klenk, H.P.; Gill, S.; Dougherty, B.A. The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature 1997, 388, 539–547. [Google Scholar] [CrossRef] [PubMed]
- Alm, R.A.; Ling, L.S.L.; Moir, D.T.; King, B.L.; Brown, E.D.; Doig, P.C.; Smith, D.R.; Noonan, B.; Guild, B.C.; Carmel, G. Genomic-sequence comparison of two unrelated isolates of the human gastric pathogen Helicobacter pylori. Nature 1999, 397, 176–180. [Google Scholar] [CrossRef]
- Cover, T.L. Helicobacter pylori diversity and gastric cancer risk. MBio 2016, 7, e01869-15. [Google Scholar] [CrossRef]
- Pride, D.T.; Meinersmann, R.J.; Blaser, M.J. Allelic variation within Helicobacter pylori babA and babB. Infect. Immun. 2001, 69, 1160–1171. [Google Scholar] [CrossRef]
- Lauener, F.; Imkamp, F.; Lehours, P.; Buissonnière, A.; Benejat, L.; Zbinden, R.; Keller, P.; Wagner, K. Genetic determinants and prediction of antibiotic resistance phenotypes in Helicobacter pylori. J. Clin Med. 2019, 8, 53. [Google Scholar] [CrossRef] [PubMed]
- Dixon, M.F.; Genta, R.M.; Yardley, J.H.; Correa, P. Classification and grading of gastritis: The updated Sydney system. Am. J. Surg. Pathol. 1996, 20, 1161–1181. [Google Scholar] [CrossRef] [PubMed]
- McFarland, J. The nephelometer: An instrument for estimating the number of bacteria in suspensions used for calculating the opsonic index and for vaccines. JAMA 1907, 49, 1176–1178. [Google Scholar] [CrossRef]
- EUCAST. Breakpoint Tables for Interpretation of MICs and Zone Diameters, Version 9.0. 2019. Available online: http://www.eucast.org (accessed on 1 January 2019).
- Hays, C.; Burucoa, C.; Lehours, P.; Tran, C.T.; Leleu, A.; Raymond, J. Molecular characterization of Helicobacter pylori resistance to rifamycins. Helicobacter 2018, 23. [Google Scholar] [CrossRef] [PubMed]
- CASFM/EUCAST. Breakpoint Tables for Interpretation of MICs and Zone Diameters, Version 1.0. 2019. Available online: www.sfm-microbiologie.org (accessed on 3 January 2019).
- Hunt, M.; Mather, A.E.; Sánchez-Busó, L.; Page, A.J.; Parkhill, J.; Keane, J.A.; Harris, S.R. ARIBA: Rapid antimicrobial resistance genotyping directly from sequencing reads. Microb. Genom. 2017, 3. [Google Scholar] [CrossRef] [PubMed]
- R Development Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2018; ISBN 3-900051-07-0. Available online: http://www.R-project.org (accessed on 12 July 2019).
- Falush, D.; Wirth, T.; Linz, B.; Pritchard, J.K.; Stephens, M.; Kidd, M.; Blaser, M.J.; Graham, D.Y.; Vacher, S.; Perez-Perez, G.I. Traces of human migrations in Helicobacter pylori populations. Science 2003, 299, 1582–1585. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef] [PubMed]
- Pritchard, J.K.; Stephens, M.; Donnelly, P. Inference of population structure using multilocus genotype data. Genetics 2000, 155, 945–959. [Google Scholar] [PubMed]
- Falush, D.; Stephens, M.; Pritchard, J.K. Inference of population structure using multilocus genotype data: Linked loci and correlated allele frequencies. Genetics 2003, 164, 1567–1587. [Google Scholar] [PubMed]
- Malfertheiner, P.; Megraud, F.; O’morain, C.; Gisbert, J.; Kuipers, E.; Axon, A.; Bazzoli, F.; Gasbarrini, A.; Atherton, J.; Graham, D. Management of Helicobacter pylori infection—the Maastricht V/Florence consensus report. Gut 2017, 66, 6–30. [Google Scholar] [CrossRef] [PubMed]
- Wagner, K.; Imkamp, F.; Pires, V.; Keller, P. Evaluation of Lightmix Mycoplasma macrolide assay for detection of macrolide-resistant Mycoplasma pneumoniae in pneumonia patients. Clin. Microbiol. Infect. 2019, 25. [Google Scholar] [CrossRef] [PubMed]
- Oleastro, M.; Rocha, R.; Vale, F.F. Population genetic structure of Helicobacter pylori strains from Portuguese-speaking countries. Helicobacter 2017, 22. [Google Scholar] [CrossRef] [PubMed]
- Thorell, K.; Yahara, K.; Berthenet, E.; Lawson, D.J.; Mikhail, J.; Kato, I.; Mendez, A.; Rizzato, C.; Bravo, M.M.; Suzuki, R. Rapid evolution of distinct Helicobacter pylori subpopulations in the Americas. PLoS Genet. 2017, 13. [Google Scholar] [CrossRef] [PubMed]
- Figueiredo, C.; Van Doorn, L.J.; Nogueira, C.; Soares, J.; Pinho, C.; Figueira, P.; Quint, W.; Carneiro, F. Helicobacter pylori genotypes are associated with clinical outcome in Portuguese patients and show a high prevalence of infections with multiple strains. Scand. J. Gastroenterol. 2001, 36, 128–135. [Google Scholar] [CrossRef] [PubMed]
- Erzin, Y.; Koksal, V.; Altun, S.; Dobrucali, A.; Aslan, M.; Erdamar, S.; Dirican, A.; Kocazeybek, B. Prevalence of Helicobacter pylori vacA, cagA, cagE, iceA, babA2 genotypes and correlation with clinical outcome in Turkish patients with dyspepsia. Helicobacter 2006, 11, 574–580. [Google Scholar] [CrossRef] [PubMed]
- Miehlke, S.; Kirsch, C.; Agha-Amiri, K.; Günther, T.; Lehn, N.; Malfertheiner, P.; Stolte, M.; Ehninger, G.; Bayerdörffer, E. The Helicobacter pylori vacA s1, m1 genotype and cagA is associated with gastric carcinoma in Germany. Int. J. Cancer 2000, 87, 322–327. [Google Scholar] [CrossRef]
- Heikkinen, M.; Mayo, K.; Megraud, F.; Vornanen, M.; Marin, S.; Pikkarainen, P.; Julkunen, R. Association of CagA-positive and CagA-negative Helicobacter pylori strains with patients’ symptoms and gastritis in primary care patients with functional upper abdominal complaints. Scand. J. Gastroenterol. 1998, 33, 31–38. [Google Scholar]
- Audibert, C.; Janvier, B.; Grignon, B.; Salaüna, L.; Burucoa, C.; Lecron, J.C.; Fauchère, J.L. Correlation between IL-8 induction, cagA status and vacA genotypes in 153 French Helicobacter pylori isolates. Res. Microbiol. 2000, 15, 191–200. [Google Scholar] [CrossRef]
- Chiarini, A.; Calà, C.; Bonura, C.; Gullo, A.; Giuliana, G.; Peralta, S.; D’Arpa, F.; Giammanco, A. Prevalence of virulence-associated genotypes of Helicobacter pylori and correlation with severity of gastric pathology in patients from western Sicily, Italy. Eur. J. Clin. Microbiol. Infect. Dis. 2009, 28, 437. [Google Scholar] [CrossRef]
- Boyanova, L.; Markovska, R.; Yordanov, D.; Marina, M.; Ivanova, K.; Panayotov, S.; Gergova, G.; Mitov, I. High prevalence of virulent Helicobacter pylori strains in symptomatic Bulgarian patients. Diagn. Microbiol. Infect. Dis. 2009, 64, 374–380. [Google Scholar] [CrossRef]
- Andreson, H.; Loivukene, K.; Sillakivi, T.; Maaroos, H.I.; Ustav, M.; Peetsalu, A.; Mikelsaar, M. Association of cagA and vacA genotypes of Helicobacter pylori with gastric diseases in Estonia. J. Clin. Microbiol. 2002, 40, 298–300. [Google Scholar] [CrossRef] [PubMed]
- Van Doorn, L.J.; Figueiredo, C.; Mégraud, F.; Pena, S.; Midolo, P.; Queiroz, D.M.D.M.; Carneiro, F.; Vanderborght, B.; Maria Da Glória, F.P.; Sanna, R. Geographic distribution of vacA allelic types of Helicobacter pylori. Gastroenterology 1999, 116, 823–830. [Google Scholar] [CrossRef]
- Yamaoka, Y.; Kodama, T.; Kita, M.; Imanishi, J.; Kashima, K.; Graham, D.Y. Relationship of vacA genotypes of Helicobacter pylori to cagA status, cytotoxin production, and clinical outcome. Helicobacter 1998, 3, 241–253. [Google Scholar] [CrossRef] [PubMed]
- Letley, D.P.; Rhead, J.L.; Twells, R.J.; Dove, B.; Atherton, J.C. Determinants of non-toxicity in the gastric pathogen Helicobacter pylori. J. Biol. Chem. 2003, 278, 26734–26741. [Google Scholar] [CrossRef] [PubMed]
- Van Doorn, L.; Figueiredo, C.; Rossau, R.; Jannes, G.; Van Asbroeck, M.; Sousa, J.; Carneiro, F.; Quint, W. Typing of Helicobacter pylori vacA gene and detection of cagA gene by PCR and reverse hybridization. J. Clin. Microbiol. 1998, 36, 1271–1276. [Google Scholar] [PubMed]
- Figueiredo, C.; Machado, J.C.; Pharoah, P.; Seruca, R.; Sousa, S.; Carvalho, R.; Capelinha, A.F.; Quint, W.; Caldas, C.; van Doorn, L.J. Helicobacter pylori and interleukin 1 genotyping: An opportunity to identify high-risk individuals for gastric carcinoma. J. Natl. Cancer. Inst. 2002, 94, 1680–1687. [Google Scholar] [CrossRef] [PubMed]
- Kidd, M.; Lastovica, A.; Atherton, J.; Louw, J. Heterogeneity in the Helicobacter pylori vacA and cagA genes: Association with gastroduodenal disease in South Africa? Gut 1999, 45, 499–502. [Google Scholar] [CrossRef]
- Miehlke, S.; Yu, J.; Schuppler, M.; Frings, C.; Kirsch, C.; Negraszus, N.; Morgner, A.; Stolte, M.; Ehninger, G.; Bayerdörffer, E. Helicobacter pylori vacA, iceA, and cagA status and pattern of gastritis in patients with malignant and benign gastroduodenal disease. Am. J. Gastroenterol. 2001, 96, 1008–1013. [Google Scholar] [CrossRef]
- Boyanova, L.; Yordanov, D.; Gergova, G.; Markovska, R.; Mitov, I. Association of iceA and babA genotypes in Helicobacter pylori strains with patient and strain characteristics. Antonie Van Leeuwenhoek 2010, 98, 343–350. [Google Scholar] [CrossRef]
- Yamaoka, Y.; Kodama, T.; Gutierrez, O.; Kim, J.G.; Kashima, K.; Graham, D.Y. Relationship between Helicobacter pylori iceA, cagA, and vacA status and clinical outcome: Studies in four different countries. J. Clin. Microbiol. 1999, 37, 2274–2279. [Google Scholar]
- Nishiya, D.; Shimoyama, T.; Fukuda, S.; Yoshimura, T.; Tanaka, M.; Munakata, A. Evaluation of the clinical relevance of the iceA1 gene in patients with Helicobacter pylori infection in Japan. Scand. J. Gastroenterol. 2000, 35, 36–39. [Google Scholar] [PubMed]
- Yamaoka, Y.; Kikuchi, S.; El–Zimaity, H.M.; Gutierrez, O.; Osato, M.S.; Graham, D.Y. Importance of Helicobacter pylori oipA in clinical presentation, gastric inflammation, and mucosal interleukin 8 production. Gastroenterology 2002, 123, 414–424. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Zheng, Q.; Chen, X.; Xiao, S.; Liu, W.; Lu, H. The Helicobacter pylori duodenal ulcer promoting gene, dupA in China. BMC Gastroenterol. 2008, 8. [Google Scholar] [CrossRef] [PubMed]
- Pereira, W.N.; Ferraz, M.A.; Zabaglia, L.M.; de Labio, R.W.; Orcini, W.A.; Ximenez, J.P.B.; Neto, A.C.; Payão, S.L.M.; Rasmussen, L.T. Association among H. pylori virulence markers dupA, cagA and vacA in Brazilian patients. J. Venom Anim. Toxins Incl. Trop. Dis. 2014, 20. [Google Scholar] [CrossRef] [PubMed]
- Arachchi, H.J.; Kalra, V.; Lal, B.; Bhatia, V.; Baba, C.; Chakravarthy, S.; Rohatgi, S.; Sarma, P.M.; Mishra, V.; Das, B. Prevalence of duodenal ulcer-promoting gene (dupA) of Helicobacter pylori in patients with duodenal ulcer in North Indian population. Helicobacter 2007, 12, 591–597. [Google Scholar] [CrossRef] [PubMed]
- Gomes, L.I.; Rocha, G.A.; Rocha, A.M.; Soares, T.F.; Oliveira, C.A.; Bittencourt, P.F.; Queiroz, D.M. Lack of association between Helicobacter pylori infection with dupA-positive strains and gastroduodenal diseases in Brazilian patients. Int. J. Med. Microbiol. 2008, 298, 223–230. [Google Scholar] [CrossRef]
- Gerhard, M.; Lehn, N.; Neumayer, N.; Borén, T.; Rad, R.; Schepp, W.; Miehlke, S.; Classen, M.; Prinz, C. Clinical relevance of the Helicobacter pylori gene for blood-group antigen-binding adhesin. Proc. Natl. Acad. Sci. USA 1999, 96, 12778–12783. [Google Scholar] [CrossRef] [PubMed]
- Mahdavi, J.; Sondén, B.; Hurtig, M.; Olfat, F.O.; Forsberg, L.; Roche, N.; Ångström, J.; Larsson, T.; Teneberg, S.; Karlsson, K.A. Helicobacter pylori SabA adhesin in persistent infection and chronic inflammation. Science 2002, 297, 573–578. [Google Scholar] [CrossRef]
- Lehours, P.; Ménard, A.; Dupouy, S.; Bergey, B.; Richy, F.; Zerbib, F.; Ruskoné-Fourmestraux, A.; Delchier, J.C.; Mégraud, F. Evaluation of the association of nine Helicobacter pylori virulence factors with strains involved in low-grade gastric mucosa-associated lymphoid tissue lymphoma. Infect. Immun. 2004, 72, 880–888. [Google Scholar] [CrossRef]
- De Jonge, R.; Pot, R.G.; Loffeld, R.J.; Van Vliet, A.H.; Kuipers, E.J.; Kusters, J.G. The functional status of the Helicobacter pylori sabB adhesin gene as a putative marker for disease outcome. Helicobacter 2004, 9, 158–164. [Google Scholar] [CrossRef]
- Sheu, B.S.; Odenbreit, S.; Hung, K.H.; Liu, C.P.; Sheu, S.M.; Yang, H.B.; Wu, J.J. Interaction between host gastric Sialyl-Lewis X and H. pylori SabA enhances H. pylori density in patients lacking gastric Lewis B antigen. Am. J. Gastroenterol. 2006, 101, 36–44. [Google Scholar] [CrossRef] [PubMed]
- Kennemann, L.; Brenneke, B.; Andres, S.; Engstrand, L.; Meyer, T.F.; Aebischer, T.; Josenhans, C.; Suerbaum, S. In vivo sequence variation in HopZ, a phase-variable outer membrane protein of Helicobacter pylori. Infect. Immun. 2012, 80, 4364–4373. [Google Scholar] [CrossRef] [PubMed]
- Peck, B.; Ortkamp, M.; Diehl, K.D.; Hundt, E.; Knapp, B. Conservation, localization and expression of HopZ, a protein involved in adhesion of Helicobacter pylori. Nucleic Acid. Res. 1999, 27, 3325–3333. [Google Scholar] [CrossRef] [PubMed]
- Yamaoka, Y.; Kudo, T.; Lu, H.; Casola, A.; Brasier, A.R.; Graham, D.Y. Role of interferon-stimulated responsive element-like element in interleukin-8 promoter in Helicobacter pylori infection. Gastroenterology 2004, 126, 1030–1043. [Google Scholar] [CrossRef] [PubMed]
- Giannakis, M.; Backhed, H.K.; Chen, S.L.; Faith, J.J.; Wu, M.; Guruge, J.L.; Engstrand, L.; Gordon, J.I. The response of gastric epithelial progenitors to Helicobacter pylori isolates obtained from Swedish patients with chronic atrophic gastritis. J. Biol. Chem. 2009, 284, 30383–30394. [Google Scholar] [CrossRef] [PubMed]
- Yamaoka, Y.; Kita, M.; Kodama, T.; Imamura, S.; Ohno, T.; Sawai, N.; Ishimaru, A.; Imanishi, J.; Graham, D.Y. Helicobacter pylori infection in mice: Role of outer membrane proteins in colonization and inflammation. Gastroenterology 2002, 123, 1992–2004. [Google Scholar] [CrossRef] [PubMed]
- Yamaoka, Y.; Kwon, D.H.; Graham, D.Y. A Mr 34,000 proinflammatory outer membrane protein (oipA) of Helicobacter pylori. Proc. Natl. Acad. Sci. USA 2000, 97, 7533–7538. [Google Scholar] [CrossRef] [PubMed]
- Tabassam, F.H.; Graham, D.Y.; Yamaoka, Y. OipA plays a role in Helicobacter pylori-induced focal adhesion kinase activation and cytoskeletal re-organization. Cell. Microbiol. 2008, 10, 1008–1020. [Google Scholar] [CrossRef]
- Ando, T.; Peek, R.; Pride, D.; Levine, S.; Takata, T.; Lee, Y.C.; Kusugami, K.; Van der Ende, A.; Kuipers, E.; Kusters, J. Polymorphisms of Helicobacter pylori HP0638 reflect geographic origin and correlate with cagA status. J. Clin. Microbiol. 2002, 40, 239–246. [Google Scholar] [CrossRef]
- Belogolova, E.; Bauer, B.; Pompaiah, M.; Asakura, H.; Brinkman, V.; Ertl, C.; Bartfeld, S.; Nechitaylo, T.Y.; Haas, R.; Machuy, N. Helicobacter pylori outer membrane protein HopQ identified as a novel T4SS-associated virulence factor. Cell. Microbiol. 2013, 15, 1896–1912. [Google Scholar]
- Yu, J.; Leung, W.; Go, M.; Chan, M.; To, K.; Ng, E.; Chan, F.; Ling, T.; Chung, S.; Sung, J. Relationship between Helicobacter pylori babA2 status with gastric epithelial cell turnover and premalignant gastric lesions. Gut 2002, 51, 480–484. [Google Scholar] [CrossRef] [PubMed]
- Ito, Y.; Azuma, T.; Ito, S.; Miyaji, H.; Hirai, M.; Yamazaki, Y.; Sato, F.; Kato, T.; Kohli, Y.; Kuriyama, M. Analysis and typing of the vacA gene from cagA-positive strains of Helicobacter pylori isolated in Japan. J. Clin. Microbiol. 1997, 35, 1710–1714. [Google Scholar] [PubMed]
- Lai, C.H.; Kuo, C.H.; Chen, Y.C.; Chao, F.Y.; Poon, S.K.; Chang, C.S.; Wang, W.C. High prevalence of cagA-and babA2-positive Helicobacter pylori clinical isolates in Taiwan. J. Clin. Microbiol. 2002, 40, 3860–3862. [Google Scholar] [CrossRef] [PubMed]
- Fischbach, W.; Malfertheiner, P.; Jansen, P.L.; Bolten, W.; Bornschein, J.; Buderus, S.; Glocker, E.; Hoffmann, J.; Koletzko, S.; Labenz, J. S2k-Leitlinie Helicobacter pylori und gastroduodenale Ulkuskrankheit. Zeitschrift für Gastroenterologie 2016, 54, 327–363. [Google Scholar] [PubMed]


| Age (years) | Mean | 45 ± 15 |
| Range | 17–81 | |
| Sex (Count) | Female | 25 |
| Male | 16 | |
| Ethnicity (Count) | African | 2 |
| Caucasian | 36 | |
| Chinese | 1 | |
| Hispanic | 2 | |
| Gastritis (Count) | Mild | 3 |
| Moderate | 27 | |
| Marked | 11 | |
| Helicobacter pylori in gastric biopsy (Count) | Few/Some (+) | 9 |
| Abundant (++) | 18 | |
| Highly Abundant (+++) | 14 | |
| Drug-resistant Helicobacter pylori (Count) | Amoxicillin | 0 |
| Clarithromycin | 35 | |
| Metronidazole | 30 | |
| Levofloxacin | 12 | |
| Rifampicin | 1 | |
| Tetracycline | 0 |
| Gene | cagA-positive H. pylori | cagA-negative H. pylori | Difference between cagA-positive and cagA-negative H. pylori strains | ||
|---|---|---|---|---|---|
| In-frame (“status-on”) | Out-of-frame (“status-off”) | In-frame (“status-on”) | Out-of-frame (“status-off”) | ||
| hopZ | 9 | 10 | 13 | 9 | P > 0.05 |
| oipA | 14 | 5 | 6 | 16 | P < 0.001 |
| sabA | 10 | 9 | 6 | 16 | P > 0.05 |
| sabB | 10 | 9 | 8 | 14 | P > 0.05 |
| Patient | Age | Sex | Ethnicity | cagA | vacA | iceA | babA | babB | dupA | hopZ | oipA | sabA | sabB | hopQ |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 31 | F | Caucasian | + | s1a/m1 | iceA1 | babA2 | babB2 | - | off | on | on | off | allele 1 |
| 2 | 50 | M | Caucasian | + | s1a/m2 | iceA2 | babA2 | babB2 | - | on | on | off | off | allele 1 |
| 3 | 28 | M | Chinese | + | s1a/m2 | iceA1 | babA2 | - | - | off | on | on | on | allele 1 |
| 4 | 26 | M | Caucasian | + | s1a/m1 | iceA1 | babA2 | babB3 | - | off | off | on | on | allele 2 |
| 5 | 54 | M | Caucasian | + | s1a/m1 | iceA1 | babA2 | - | - | on | on | on | on | allele 1 |
| 6 | 54 | M | Caucasian | + | s1b/m1 | iceA2 | babA2 | babB1 | off | on | off | on | allele 2 | |
| 7 | 50 | F | Caucasian | + | s1b/m1 | iceA2 | babA2 | - | + | off | off | off | off | allele 2 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Imkamp, F.; Lauener, F.N.; Pohl, D.; Lehours, P.; Vale, F.F.; Jehanne, Q.; Zbinden, R.; Keller, P.M.; Wagner, K. Rapid Characterization of Virulence Determinants in Helicobacter pylori Isolated from Non-Atrophic Gastritis Patients by Next-Generation Sequencing. J. Clin. Med. 2019, 8, 1030. https://doi.org/10.3390/jcm8071030
Imkamp F, Lauener FN, Pohl D, Lehours P, Vale FF, Jehanne Q, Zbinden R, Keller PM, Wagner K. Rapid Characterization of Virulence Determinants in Helicobacter pylori Isolated from Non-Atrophic Gastritis Patients by Next-Generation Sequencing. Journal of Clinical Medicine. 2019; 8(7):1030. https://doi.org/10.3390/jcm8071030
Chicago/Turabian StyleImkamp, Frank, Francis N. Lauener, Daniel Pohl, Philippe Lehours, Filipa F. Vale, Quentin Jehanne, Reinhard Zbinden, Peter M. Keller, and Karoline Wagner. 2019. "Rapid Characterization of Virulence Determinants in Helicobacter pylori Isolated from Non-Atrophic Gastritis Patients by Next-Generation Sequencing" Journal of Clinical Medicine 8, no. 7: 1030. https://doi.org/10.3390/jcm8071030
APA StyleImkamp, F., Lauener, F. N., Pohl, D., Lehours, P., Vale, F. F., Jehanne, Q., Zbinden, R., Keller, P. M., & Wagner, K. (2019). Rapid Characterization of Virulence Determinants in Helicobacter pylori Isolated from Non-Atrophic Gastritis Patients by Next-Generation Sequencing. Journal of Clinical Medicine, 8(7), 1030. https://doi.org/10.3390/jcm8071030






