The Role of MicroRNAs in the Regulation of Gastric Cancer Stem Cells: A Meta-Analysis of the Current Status
Abstract
1. Introduction
2. MicroRNAs and Cancer Stem Cells
3. Gastric Cancer Stem Cells
4. MicroRNAs and Gastric Cancer Stem Cells
5. Meta-Analysis of Up/Down miRNAs in GCSCs Features: A Focus on Stemness-Related Pathways
6. Conclusions
Supplementary Materials
Author Contributions
Conflicts of Interest
References
- McLean, M.H.; El-Omar, E.M. Genetics of gastric cancer. Nat. Rev. Gastroenterol. Hepatol. 2014, 11, 664–674. [Google Scholar] [CrossRef]
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics, 2016. CA Cancer J. Clin. 2016, 66, 7–30. [Google Scholar] [CrossRef] [PubMed]
- Bernards, N.; Creemers, G.J.; Nieuwenhuijzen, G.A.P.; Bosscha, K.; Pruijt, J.F.M.; Lemmens, V.E.P.P. No improvement in median survival for patients with metastatic gastric cancer despite increased use of chemotherapy. Ann. Oncol. 2013, 24, 3056–3060. [Google Scholar] [CrossRef]
- Ferlay, J.; Soerjomataram, I.; Dikshit, R.; Eser, S.; Mathers, C.; Rebelo, M.; Parkin, D.M.; Forman, D.; Bray, F. Cancer incidence and mortality worldwide: Sources, methods and major patterns in GLOBOCAN 2012. Int. J. Cancer 2015, 136, 359–386. [Google Scholar] [CrossRef] [PubMed]
- Torre, L.A.; Bray, F.; Siegel, R.L.; Ferlay, J.; Lortet-Tieulent, J.; Jemal, A. Global cancer statistics, 2012. CA Cancer J. Clin. 2015, 65, 87–108. [Google Scholar] [CrossRef]
- Gullo, I.; Carneiro, F.; Oliveira, C.; Almeida, G.M. Heterogeneity in Gastric Cancer: From Pure Morphology to Molecular Classifications. Pathobiology 2018, 85, 50–63. [Google Scholar] [CrossRef]
- Gao, J.P.; Xu, W.; Liu, W.T.; Yan, M.; Zhu, Z.G. Tumor heterogeneity of gastric cancer: From the perspective of tumor-initiating cell. World J. Gastroenterol. 2018, 24, 2567–2581. [Google Scholar] [CrossRef] [PubMed]
- McGranahan, N.; Swanton, C. Clonal Heterogeneity and Tumor Evolution: Past, Present, and the Future. Cell 2017, 168, 613–628. [Google Scholar] [CrossRef]
- Baccelli, I.; Trumpp, A. The evolving concept of cancer and metastasis stem cells. J. Cell Biol. 2012, 198, 281–293. [Google Scholar] [CrossRef] [PubMed]
- Kreso, A.; Dick, J.E. Evolution of the cancer stem cell model. Cell Stem Cell 2014, 14, 275–291. [Google Scholar] [CrossRef]
- Iseghohi, S.O. Cancer stem cells may contribute to the difficulty in treating cancer. Genes Dis. 2016, 3, 7–10. [Google Scholar] [CrossRef]
- Clarke, M.F.; Dick, J.E.; Dirks, P.B.; Eaves, C.J.; Jamieson, C.H.M.; Jones, D.L.; Visvader, J.; Weissman, I.L.; Wahl, G.M. Cancer stem cells-perspective on current status and future directions: AACR workshop on cancer stem cells. Cancer Res. 2006, 66, 9339–9344. [Google Scholar] [CrossRef]
- Lapidot, T.; Sirard, C.; Vormoor, J.; Murdoch, B.; Hoang, T.; Caceres-Cortes, J.; Minden, M.; Paterson, B.; Caligiuri, M.A.; Dick, J.E. A cell initiating human acute myeloid leukaemia after transplantation into SCID mice. Nature 1994, 367, 645–648. [Google Scholar] [CrossRef]
- Al-Hajj, M.; Wicha, M.S.; Benito-Hernandez, A.; Morrison, S.J.; Clarke, M.F. Prospective identification of tumorigenic breast cancer cells. Proc. Natl. Acad. Sci. USA 2003, 100, 3983–3988. [Google Scholar] [CrossRef]
- Singh, S.K.; Clarke, I.D.; Terasaki, M.; Bonn, V.E.; Hawkins, C.; Squire, J.; Dirks, P.B. Identification of a cancer stem cell in human brain tumors. Cancer Res. 2003, 63, 5821–5828. [Google Scholar]
- Collins, A.T.; Berry, P.A.; Hyde, C.; Stower, M.J.; Maitland, N.J. Prospective identification of tumorigenic prostate cancer stem cells. Cancer Res. 2005, 65, 10946–10951. [Google Scholar] [CrossRef]
- Fang, D.; Nguyen, T.K.; Leishear, K.; Finko, R.; Kulp, A.N.; Hotz, S.; Van Belle, P.A.; Xu, X.; Elder, D.E.; Herlyn, M. A tumorigenic subpopulation with stem cell properties in melanomas. Cancer Res. 2005, 65, 9328–9337. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Heidt, D.G.; Dalerba, P.; Burant, C.F.; Zhang, L.; Adsay, V.; Wicha, M.; Clarke, M.F.; Simeone, D.M. Identification of pancreatic cancer stem cells. Cancer Res. 2007, 67, 1030–1037. [Google Scholar] [CrossRef] [PubMed]
- Yashiro, M. Gastric Cancer Stem Cells and Resistance to Cancer Therapy. Chemotherapy 2014, 3, 135. [Google Scholar]
- Takaishi, S.; Okumura, T.; Tu, S.; Wang, S.S.; Shibata, W.; Vigneshwaran, R.; Gordon, S.A.; Shimada, Y.; Wang, T.C. Identification of gastric cancer stem cells using the cell surface marker CD44. Stem Cells 2009, 27, 1006–1020. [Google Scholar] [CrossRef] [PubMed]
- Riquelme, I.; Letelier, P.; Riffo-Campos, A.L.; Brebi, P.; Roa, J.C. Emerging Role of miRNAs in the Drug Resistance of Gastric Cancer. Int. J. Mol. Sci. 2016, 17, 424. [Google Scholar] [CrossRef]
- Ali Hosseini Rad, S.; Bavarsad, M.S.; Arefian, E.; Jaseb, K.; Shahjahani, M.; Saki, N. The Role of microRNAs in Stemness of Cancer Stem Cells. Oncol. Rev. 2013, 7, 8. [Google Scholar] [CrossRef][Green Version]
- Iorio, M.V.; Croce, C.M. MicroRNA dysregulation in cancer: Diagnostics, monitoring and therapeutics. A comprehensive review. EMBO Mol. Med. 2012, 4, 143–159. [Google Scholar] [CrossRef] [PubMed]
- Croce, C.M.; Calin, G.A. miRNAs, cancer, and stem cell division. Cell 2005, 122, 6–7. [Google Scholar] [CrossRef]
- Farazi, T.A.; Hoell, J.I.; Morozov, P.; Tuschl, T. MicroRNAs in human cancer. Adv. Exp. Med. Biol. 2013, 774, 1–20. [Google Scholar]
- Lee, R.C.; Feinbaum, R.L.; Ambros, V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 1993, 75, 843–854. [Google Scholar] [CrossRef]
- Pasquinelli, A.E.; Reinhart, B.J.; Slack, F.; Martindale, M.Q.; Kuroda, M.I.; Maller, B.; Hayward, D.C.; Ball, E.E.; Degnan, B.; Müller, P.; et al. Conservation of the sequence and temporal expression of let-7 heterochronic regulatory RNA. Nature 2000, 408, 86–89. [Google Scholar] [CrossRef]
- Yu, F.; Yao, H.; Zhu, P.; Zhang, X.; Pan, Q.; Gong, C.; Huang, Y.; Hu, X.; Su, F.; Lieberman, J.; Song, E. Let-7 regulates self-renewal and tumorigenicity of breast cancer cells. Cell 2007, 131, 1109–1123. [Google Scholar] [CrossRef] [PubMed]
- Shimono, Y.; Zabala, M.; Cho, R.W.; Lobo, N.; Dalerba, P.; Qian, D.; Diehn, M.; Liu, H.; Panula, S.P.; Chiao, E.; et al. Downregulation of miRNA-200c links breast cancer stem cells with normal stem cells. Cell 2009, 138, 592–603. [Google Scholar] [CrossRef] [PubMed]
- Iliopoulos, D.; Lindahl-Allen, M. Loss of miR-200 inhibition of Suz12 leads to polycomb-mediated repression required for the formation and maintenance of cancer stem cells. Mol. Cell 2010, 39, 761–772. [Google Scholar] [CrossRef] [PubMed]
- Lim, Y.; Wright, J.; Attema, J.; Gregory, P.; Bert, A.; Smith, E.; Thomas, D.; Drew, P.; Khew-Goodall, Y.; Goodall, G. Epigenetic modulation of the miR-200 family is associated with transition to a breast cancer stem cell-like state. J. Cell Sci. 2013, 126, 2256–2266. [Google Scholar] [CrossRef] [PubMed]
- Yu, F.; Deng, H.; Yao, H.; Liu, Q.; Su, F.; Song, E. MiR-30 reduction maintains self-renewal and inhibits apoptosis in breast tumor-initiating cells. Oncogene 2010, 29, 4194–4204. [Google Scholar] [CrossRef] [PubMed]
- Ouzounova, M.; Vuong, T.; Ancey, P.B.; Ferrand, M.; Durand, G.; Le-Calvez Kelm, F.; Croce, C.; Matar, C.; Herceg, Z.; Hernandez-Vargas, H. MicroRNA miR-30 family regulates non-attachment growth of breast cancer cells. BMC Genom. 2013, 14, 139. [Google Scholar] [CrossRef]
- Song, S.J.; Ito, K.; Ala, U.; Kats, L.; Webster, K.; Sun, S.M.; Jongen-Lavrencic, M.; Manova-Todorova, K.; Teruya-Feldstein, J.; Avigan, D.E.; et al. The oncogenic microRNA miR-22 targets the TET2 tumor suppressor to promote hematopoietic stem cell self-renewal and transformation. Cell Stem Cell 2013, 13, 87–101. [Google Scholar] [CrossRef]
- Khalaj, M.; Woolthuis, C.M.; Hu, W.; Durham, B.H.; Chu, S.H.; Qamar, S.; Armstrong, S.A.; Park, C.Y. miR-99 regulates normal and malignant hematopoietic stem cell self-renewal. J. Exp. Med. 2017, 214, 2453–2470. [Google Scholar] [CrossRef] [PubMed]
- Lechman, E.R.; Gentner, B.; Ng, S.W.; Schoof, E.M.; van Galen, P.; Kennedy, J.A.; Nucera, S.; Ciceri, F.; Kaufmann, K.B.; Takayama, N.; et al. miR-126 Regulates Distinct Self-Renewal Outcomes in Normal and Malignant Hematopoietic Stem Cells. Cancer Cell 2016, 29, 214–228. [Google Scholar] [CrossRef] [PubMed]
- Xu, D.D.; Zhou, P.J.; Wang, Y.; Zhang, Y.; Zhang, R.; Zhang, L.; Chen, S.H.; Fu, W.Y.; Ruan, B.B.; Xu, H.P.; et al. MiR-150 suppresses the proliferation and tumorigenicity of leukemia stem cells by targeting the nanog signaling pathway. Front. Pharm. 2016, 7, 439. [Google Scholar] [CrossRef]
- Kleinová, R.; Slabý, O.; Šána, J. Te Relevance of MicroRNAs in Glioblastoma Stem Cells. Klin. Onkol. Cas Ceské Slov. Onkol. Spol. 2015, 28, 338–344. [Google Scholar]
- Godlewski, J.; Nowicki, M.O.; Bronisz, A.; Williams, S.; Otsuki, A.; Nuovo, G.; Raychaudhury, A.; Newton, H.B.; Chiocca, E.A.; Lawler, S. Targeting of the Bmi-1 oncogene/stem cell renewal factor by microRNA-128 inhibits glioma proliferation and self-renewal. Cancer Res. 2008, 68, 9125–9130. [Google Scholar] [CrossRef]
- Gal, H.; Pandi, G.; Kanner, A.A.; Ram, Z.; Lithwick-Yanai, G.; Amariglio, N.; Rechavi, G.; Givol, D. MIR-451 and Imatinib mesylate inhibit tumor growth of Glioblastoma stem cells. Biochem. Biophys. Res. Commun. 2008, 376, 86–90. [Google Scholar] [CrossRef]
- Nan, Y.; Han, L.; Zhang, A.; Wang, G.; Jia, Z.; Yang, Y.; Yue, X.; Pu, P.; Zhong, Y.; Kang, C. MiRNA-451 plays a role as tumor suppressor in human glioma cells. Brain Res. 2010, 1359, 14–21. [Google Scholar] [CrossRef] [PubMed]
- Guessous, F.; Zhang, Y.; Kofman, A.; Catania, A.; Li, Y.; Schiff, D.; Purow, B.; Abounader, R. microRNA-34a is tumor suppressive in brain tumors and glioma stem cells. Cell Cycle 2010, 9, 1031–1036. [Google Scholar] [CrossRef]
- Wu, N.; Xiao, L.; Zhao, X.; Zhao, J.; Wang, J.; Wang, F.; Cao, S.; Lin, X. miR-125b regulates the proliferation of glioblastoma stem cells by targeting E2F2. FEBS Lett. 2012, 586, 3831–3839. [Google Scholar] [CrossRef] [PubMed]
- Chung, H.J.; Choi, Y.E.; Kim, E.S.; Han, Y.H.; Park, M.J.; Bae, I.H. miR-29b attenuates tumorigenicity and stemness maintenance in human glioblastoma multiforme by directly targeting BCL2L2. Oncotarget 2015, 6, 18429–18444. [Google Scholar] [CrossRef] [PubMed]
- Ma, S.; Chan, K.W.; Hu, L.; Lee, T.K.; Wo, J.Y.; Ng, I.O.; Zheng, B.J.; Guan, X.Y. Identification and characterization of tumorigenic liver cancer stem/progenitor cells. Gastroenterology 2007, 132, 2542–2556. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Yang, X.; Ruan, B.; Dai, B.; Gao, Y.; Duan, J.; Qu, S.; Tao, K.; Dou, K.; Li, H. Overexpression of miR-200a suppresses epithelial-mesenchymal transition of liver cancer stem cells. Tumour Biol. 2015, 36, 2447–2456. [Google Scholar] [CrossRef] [PubMed]
- Lou, W.; Liu, J.; Gao, Y.; Zhong, G.; Ding, B.; Xu, L.; Fan, W. MicroRNA regulation of liver cancer stem cells. Am. J. Cancer Res. 2018, 8, 1126–1141. [Google Scholar]
- Cai, H.; Chen, Y.; Yang, X.; Ma, S.; Wang, Q.; Zhang, Y.; Niu, X.; Ding, G.; Yuan, Y. Let7b modulates the Wnt/beta-catenin pathway in liver cancer cells via downregulated Frizzled4. Tumour Biol. 2017, 39. [Google Scholar] [CrossRef]
- Jin, B.; Wang, W.; Meng, X.X.; Du, G.; Li, J.; Zhang, S.Z.; Zhou, B.H.; Fu, Z.H. Let-7 inhibits self-renewal of hepatocellular cancer stem-like cells through regulating the epithelial-mesenchymal transition and the Wnt signaling pathway. BMC Cancer 2016, 16, 863. [Google Scholar] [CrossRef]
- Jiang, C.; Yu, M.; Xie, X.; Huang, G.; Peng, Y.; Ren, D.; Lin, M.; Liu, B.; Liu, M.; Wang, W.; Kuang, M. miR-217 targeting DKK1 promotes cancer stem cell properties via activation of the Wnt signaling pathway in hepatocellular carcinoma. Oncol. Rep. 2017, 38, 2351–2359. [Google Scholar] [CrossRef]
- Zheng, Z.; Liu, J.; Yang, Z.; Wu, L.; Xie, H.; Jiang, C.; Lin, B.; Chen, T.; Xing, C.; Liu, Z.; et al. MicroRNA-452 promotes stem-like cells of hepatocellular carcinoma by inhibiting Sox7 involving Wnt/beta-catenin signaling pathway. Oncotarget 2016, 7, 28000–28012. [Google Scholar] [CrossRef] [PubMed]
- Mukohyama, J.; Shimono, Y.; Minami, H.; Kakeji, Y.; Suzuki, A. Roles of microRNAs and RNA-Binding Proteins in the Regulation of Colorectal Cancer Stem Cells. Cancers 2017, 9, 143. [Google Scholar] [CrossRef] [PubMed]
- Yan, T.T.; Ren, L.L.; Shen, C.Q.; Wang, Z.H.; Yu, Y.N.; Liang, Q.; Tang, J.Y.; Chen, Y.X.; Sun, D.F.; Zgodziński, W.; et al. MicroRNA-508 defines the stem-like/mesenchymal subtype in colorectal cancer. Cancer Res. 2018, 78, 1751–1765. [Google Scholar] [CrossRef]
- Fang, Y.X.; Chang, Y.L.; Gao, W.Q. MicroRNAs targeting prostate cancer stem cells. Exp. Biol. Med. 2015, 240, 1071–1078. [Google Scholar] [CrossRef]
- Liu, C.; Liu, R.; Zhang, D.; Deng, Q.; Liu, B.; Chao, H.P.; Rycaj, K.; Takata, Y.; Lin, K.; Lu, Y. MicroRNA-141 suppresses prostate cancer stem cells and metastasis by targeting a cohort of pro-metastasis genes. Nat. Commun. 2017, 8, 14270. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Ye, J.; Yan, H.; Tang, Z.; Shen, J.; Zhang, J.; Yang, L. MiR-491 attenuates cancer stem cells-like properties of hepatocellular carcinoma by inhibition of GIT-1/NF-kappaB-mediated EMT. Tumour Biol. 2016, 37, 201–209. [Google Scholar] [CrossRef]
- Hsieh, I.S.; Chang, K.C.; Tsai, Y.T.; Ke, J.Y.; Lu, P.J.; Lee, K.H.; Yeh, S.D.; Hong, T.M.; Chen, Y.L. MicroRNA-320 suppresses the stem cell-like characteristics of prostate cancer cells by downregulating the Wnt/beta-catenin signaling pathway. Carcinogenesis 2013, 34, 530–538. [Google Scholar] [CrossRef]
- Fu, Y.; Li, H.; Hao, X. The self-renewal signaling pathways utilized by gastric cancer stem cells. Tumour Biol. 2017, 39. [Google Scholar] [CrossRef]
- Huang, Q.; Zou, X. Clinicopathology of Early Gastric Carcinoma: An Update for Pathologists and Gastroenterologists. Gastrointest. Tumors 2017, 3, 115–124. [Google Scholar] [CrossRef]
- Houghton, J.; Stoicov, C.; Nomura, S.; Rogers, A.B.; Carlson, J.; Li, H.; Cai, X.; Fox, J.G.; Goldering, J.R.; Wang, T.C. Gastric cancer originating from bone marrow-derived cells. Science 2004, 306, 1568–1571. [Google Scholar] [CrossRef]
- Varon, C.; Dubus, P.; Mazurier, F.; Asencio, C.; Chambonnier, L.; Ferrand, J.; Giese, A.; Senant-Dugot, N.; Carlotti, M.; Mégraud, F. Helicobacter pylori infection recruits bone marrow-derived cells that participate in gastric preneoplasia in mice. Gastroenterology 2012, 142, 281–291. [Google Scholar] [CrossRef]
- Zhang, S.; Kim, W.; Pham, T.T.; Rogers, A.B.; Houghton, J.M.; Moss, S.F. Native and bone marrow-derived cell mosaicism in gastric carcinoma in H. pylori-infected p27-deficient mice. Oncotarget 2016, 7, 69136–69148. [Google Scholar] [CrossRef][Green Version]
- Telford, W.G. Stem cell side population analysis and sorting using DyeCycle violet. Curr. Protoc. Cytom. 2010, 9, 30. [Google Scholar]
- Greve, B.; Kelsch, R.; Spaniol, K.; Eich, H.T.; Gotte, M. Flow Cytometry in Cancer Stem Cell Analysis and Separation. Cytom. A 2012, 81, 284–293. [Google Scholar] [CrossRef]
- Shimoda, M.; Ota, M.; Okada, Y. Isolation of cancer stem cells by side population method. Methods Mol. Biol. 2018, 1692, 49–59. [Google Scholar]
- Pastrana, E.; Silva-Vargas, V.; Doetsch, F. Eyes wide open: A critical review of sphere-formation as an assay for stem cells. Cell Stem Cell 2011, 8, 486–498. [Google Scholar] [CrossRef] [PubMed]
- Tang, D.G. Understanding cancer stem cell heterogeneity and plasticity. Cell Res. 2012, 22, 457–472. [Google Scholar] [CrossRef] [PubMed]
- Sun, M.; Zhou, W.; Zhang, Y.Y.; Wang, D.L.; Wu, X.L. CD44+ gastric cancer cells with stemness properties are chemoradioresistant and highly invasive. Oncol. Lett. 2013, 5, 1793–1798. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Li, C.; He, F.; Cai, Y.; Yang, H. Identification of CD44+CD24+ gastric cancer stem cells. J. Cancer Res. Clin. Oncol. 2011, 137, 1679–1686. [Google Scholar] [CrossRef] [PubMed]
- Chen, T.; Yang, K.; Yu, J.; Meng, W.; Yuan, D.; Bi, F.; Liu, F.; Liu, J.; Dai, B.; Chen, X.; et al. Identification and expansion of cancer stem cells in tumor tissues and peripheral blood derived from gastric adenocarcinoma patients. Cell Res. 2012, 2, 248–258. [Google Scholar] [CrossRef]
- Nishikawa, S.; Konno, M.; Hamabe, A.; Hasegawa, S.; Kano, Y.; Fukusumi, T.; Satoh, T.; Takiguchi, S.; Mori, M.; Doki, Y.; et al. Surgically resected human tumors reveal the biological significance of the gastric cancer stem cell markers CD44 and CD26. Oncol. Lett. 2015, 9, 2361–2367. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Han, M.E.; Jeon, T.Y.; Hwang, S.H.; Lee, Y.S.; Kim, H.J.; Shim, H.E.; Yoon, S.; Baek, S.Y.; Kim, B.S.; Kang, C.D.; et al. Cancer spheres from gastric cancer patients provide an ideal model system for cancer stem cell research. Cell Mol. Life Sci. 2011, 68, 3589–3605. [Google Scholar] [CrossRef]
- Lau, W.M.; Teng, E.; Chong, H.S.; Lopez, K.A.; Tay, A.Y.; Salto-Tellez, M.; Shabbir, A.; So, J.B.; Chan, S.L. CD44v8-10 is a cancer-specific marker for gastric cancer stem cells. Cancer Res. 2014, 74, 2630–2641. [Google Scholar] [CrossRef]
- Ohkuma, M.; Haraguchi, N.; Ishii, H.; Mimori, K.; Tanaka, F.; Kim, H.M.; Shimomura, M.; Hirose, H.; Yanaga, K.; Mori, M. Absence of CD71 transferrin receptor characterizes human gastric adenosquamous carcinoma stem cells. Ann. Surg. Oncol. 2012, 19, 1357–1364. [Google Scholar] [CrossRef] [PubMed]
- Jiang, J.; Zhang, Y.; Chuai, S.; Wang, Z.; Zheng, D.; Xu, F.; Zhang, Y.; Li, C.; Liang, Y.; Chen, Z. Trastuzumab (Herceptin) targets gastric cancer stem cells characterized by CD90 phenotype. Oncogene 2012, 31, 671–682. [Google Scholar] [CrossRef]
- Chen, D.H.; Lu, R.Q.; Ni, X.C.; Wu, J.G.; Wang, S.L.; Jiang, B.J.; Yu, J.W. Influence of CD133+ expression on patients’ survival and resistance of CD133+ cells to anti-tumor reagents in gastric cancer. Asian Pac. J. Trop. Biomed. 2015, 5, 996–1004. [Google Scholar] [CrossRef]
- Katsuno, Y.; Ehata, S.; Yashiro, M.; Yanagihara, K.; Hirakawa, K.; Miyazono, K. Coordinated expression of REG4 and aldehyde dehydrogenase 1 regulating tumourigenic capacity of diffuse-type gastric carcinoma initiating cells is inhibited by TGF-β. J. Pathol. 2012, 228, 391–404. [Google Scholar] [CrossRef]
- Zhang, L.; Guo, X.; Zhang, D.; Fan, Y.; Qin, L.; Dong, S.; Zhang, L. Upregulated miR-132 in Lgr5+ gastric cancer stem cell-like cells contributes to cisplatin-resistance via SIRT1/CREB/ABCG2 signaling pathway. Mol. Carcinog. 2017, 56, 2022–2034. [Google Scholar] [CrossRef]
- Golestaneh, A.F.; Atashi, A.; Langroudi, L.; Shafiee, A.; Ghaemi, N.; Soleimani, M. miRNAs expressed differently in cancer stem cells and cancer cells of human gastric cancer cell line MKN-45. Cell Biochem. Funct. 2012, 30, 411–508. [Google Scholar] [CrossRef]
- Zeng, J.F.; Ma, X.Q.; Wang, L.P.; Wang, W. MicroRNA-145 exerts tumor-suppressive and chemo-resistance lowering effects by targeting CD44 in gastric cancer. World J. Gastroenterol. 2017, 23, 2337–2345. [Google Scholar] [CrossRef]
- Xiao, W.; Li, D.; Tang, Y.; Chen, Y.; Deng, W.B.; Chen, J.; Zhou, W.W.; Liao, A. Inhibition of epithelialmesenchymal transition in gastric cancer cells by miR-711-mediated downregulation of CD44 expression. Oncol. Rep. 2018, 40, 2844–2853. [Google Scholar]
- Lee, S.D.; Yu, D.; Lee, D.Y.; Shin, H.S.; Jo, J.H.; Lee, Y.C. Upregulated miR-193a-3p is responsible for cisplatin resistance in CD44(+) gastric cancer cells. Cancer Sci. 2019, 110, 662–673. [Google Scholar] [CrossRef]
- Pan, Y.; Shu, X.; Sun, L.; Yu, L.; Sun, L.; Yang, Z.; Ran, Y. miR196a5p modulates gastric cancer stem cell characteristics by targeting Smad4. Int. J. Oncol. 2017, 50, 1965–1976. [Google Scholar] [CrossRef]
- Shao, Q.; Xu, J.; Guan, X.; Zhou, B.; Wei, W.; Deng, R.; Li, D.; Xu, X.; Zhu, H. In vitro and in vivo effects of miRNA-19b/20a/92a on gastric cancer stem cells and the related mechanism. Int. J. Med. Sci. 2018, 15, 86–94. [Google Scholar] [CrossRef] [PubMed]
- Beachy, P.A.; Karhadkar, S.S.; Berman, D.M. Tissue repair and stem cell renewal in carcinogenesis. Nature 2004, 432, 324–331. [Google Scholar] [CrossRef]
- Xiao, H.J.; Ji, Q.; Yang, L.; Li, R.T.; Zhang, C.; Hou, J.M. In vivo and in vitro effects of microRNA-124 on human gastric cancer by targeting JAG1 through the Notch signaling pathway. J. Cell. Biochem. 2017, 119, 2520–2534. [Google Scholar] [CrossRef] [PubMed]
- Wu, Q.; Yang, Z.; Wang, F.; Hu, S.; Yang, L.; Shi, Y.; Fan, D. MiR-19b/20a/92a regulates the self-renewal and proliferation of gastric cancer stem cells. J. Cell Sci. 2013, 126, 4220–4229. [Google Scholar] [CrossRef] [PubMed]
- Fan, D.; Ren, B.; Yang, X.; Liu, J.; Zhang, Z. Upregulation of miR-501-5p activates the wnt/β-catenin signaling pathway and enhances stem cell-like phenotype in gastric cancer. J. Exp. Clin. Cancer Res. 2016, 35, 177. [Google Scholar] [CrossRef] [PubMed]
- Peng, Y.; Zhang, X.; Ma, Q.; Yan, R.; Qin, Y.; Zhao, Y.; Cheng, Y.; Yang, M.; Wang, Q.; Feng, X.; et al. MiRNA-194 activates the Wnt/β-catenin signaling pathway in gastric cancer by targeting the negative Wnt regulator, SUFU. Cancer Lett. 2017, 385, 117–127. [Google Scholar] [CrossRef]
- Song, B.; Lin, H.X.; Dong, L.L.; Ma, J.J.; Jiang, Z.G. MicroR- NA-338 inhibits proliferation, migration, and invasion of gastric cancer cells by the Wnt/β-catenin signaling pathway. Eur. Rev. Med. Pharm. Sci. 2018, 22, 1290–1296. [Google Scholar]
- Yu, D.; Shin, H.S.; Lee, Y.S.; Lee, Y.C. miR-106b modulates cancer stem cell characteristics through TGF-beta/Smad signaling in CD44-positive gastric cancer cells. Lab. Investig. 2014, 94, 1370–1381. [Google Scholar] [CrossRef]
- Li, L.; Zhao, J.; Huang, S.; Wang, Y.; Zhu, L.; Cao, Y.; Xiong, J.; Deng, J. MiR-93-5p promotes gastric cancer-cell progression via inactivation of the Hippo signaling pathway. Gene 2018, 641, 240–247. [Google Scholar] [CrossRef]
- Kang, W.; Huang, T.; Zhou, Y.; Zhang, J.; Lung, R.W.M.; Tong, J.H.M.; Chan, A.W.H.; Zhang, B.; Wong, C.C.; Wu, F.; et al. miR-375 is involved in Hippo pathway by targeting YAP1/TEAD4-CTGF axis in gastric carcinogenesis. Cell Death Dis. 2018, 9, 92. [Google Scholar] [CrossRef]
- Kim, T.M.; Ko, Y.H.; Ha, S.J.; Lee, H.H. Impact of in vitro driven expression signatures of CD133 stem cell marker and tumor stroma on clinical outcomes in gastric cancers. BMC Cancer 2019, 19, 119. [Google Scholar] [CrossRef]
- Schmuck, R.; Warneke, V.; Behrens, H.M.; Simon, E.; Weichert, W.; Röcken, C. Genotypic and phenotypic characterization of side population of gastric cancer cell lines. Am. J. Pathol. 2011, 178, 1792–1804. [Google Scholar] [CrossRef]
- Ritchie, M.E.; Phipson, B.; Wu, D.; Hu, Y.; Law, C.W.; Shi, W.; Smyth, G.K. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015, 43, e47. [Google Scholar] [CrossRef]
- Yu, G.; Wang, L.G.; Han, Y.; He, Q.Y. clusterProfiler: An R package for comparing biological themes among gene clusters. OMICS 2012, 16, 284–287. [Google Scholar] [CrossRef]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef]
- Liberzon, A.; Birger, C.; Thorvaldsdóttir, H.; Ghandi, M.; Mesirov, J.P.; Tamayo, P. The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst. 2015, 1, 417–425. [Google Scholar] [CrossRef]
- Khalili, M.; Sadeghizadeh, M.; Ghorbanian, K.; Malekzadeh, R.; Vasei, M.; Mowla, S.J. Down-regulation of miR-302b, an ESC-specific microRNA, in Gastric Adenocarcinoma. Cell J. 2012, 13, 251–258. [Google Scholar]
- Ma, G.; Li, Q.; Dai, W.; Yang, X.; Sang, A. Prognostic Implications of miR-302a/b/c/d in Human Gastric Cancer. Pathol. Oncol. Res. 2017, 23, 899–905. [Google Scholar] [CrossRef]
- Chen, L.; Min, L.; Wang, X.; Zhao, J.; Chen, H.; Qin, J.; Chen, W.; Shen, Z.; Tang, Z.; Gan, Q.; Ruan, Y.; et al. Loss of RACK1 Promotes Metastasis of Gastric Cancer by Inducing a miR-302c/IL8 Signaling Loop. Cancer Res. 2015, 75, 3832–3841. [Google Scholar] [CrossRef]
- Li, S.; Zhang, H.; Ning, T.; Wang, X.; Liu, R.; Yang, H.; Han, Y.; Deng, T.; Zhou, L.; Zhang, L.; et al. MiR-520b/e Regulates Proliferation and Migration by Simultaneously Targeting EGFR in Gastric Cancer. Cell Physiol. Biochem. 2016, 40, 1303–1315. [Google Scholar] [CrossRef] [PubMed]
- Fan, Q.-H.; Yu, R.; Huang, W.-X.; Cui, X.-X.; Luo, B.-H.; Zhang, L.-Y. The has-miR-526b binding-site rs8506G>a polymorphism in the lincRNA-NR_024015 exon identified by GWASs predispose to non-cardia gastric cancer risk. PLoS ONE 2014, 9, e90008. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Wang, Y.; Zheng, X.; Zhang, Z.; Zhou, J.; Zhao, G.; Yang, J.; Xia, L.; Wang, R.; Cai, X.; Hu, H.; et al. MicroRNA-149 inhibits proliferation and cell cycle progression through the targeting of ZBTB2 in human gastric cancer. PLoS ONE 2012, 7, e41693. [Google Scholar] [CrossRef]
- Zheng, L.; Qi, T.; Yang, D.; Qi, M.; Li, D.; Xiang, X.; Huang, K.; Tong, Q. microRNA-9 suppresses the proliferation, invasion and metastasis of gastric cancer cells through targeting cyclin D1 and Ets1. PLoS ONE 2013, 8, e55719. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Dong, J.; Han, Z.; Zhang, K. MicroRNA-219-5p Represses the Proliferation, Migration, and Invasion of Gastric Cancer Cells by Targeting the LRH-1/Wnt/β-Catenin Signaling Pathway. Oncol. Res. 2017, 25, 617–627. [Google Scholar] [CrossRef]
- Jian, B.; Li, Z.; Xiao, D.; He, G.; Bai, L.; Yang, Q. Downregulation of microRNA-193-3p inhibits tumor proliferation migration and chemoresistance in human gastric cancer by regulating PTEN gene. Tumour Biol. 2016, 37, 8941–8949. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Yan, Z.; Zhang, J.; Gong, L.; Li, W.; Cui, J.; Liu, Y.; Gao, Z.; Li, J.; Shen, L.; Lu, Y. Combination of hsa-miR-375 and hsa-miR-142-5p as a predictor for recurrence risk in gastric cancer patients following surgical resection. Ann. Oncol. 2011, 22, 2257–2266. [Google Scholar] [CrossRef]
- Chiang, Y.; Zhou, X.; Wang, Z.; Song, Y.; Liu, Z.; Zhao, F.; Zhu, J.; Xu, H. Expression levels of microRNA-192 and -215 in gastric carcinoma. Pathol. Oncol. Res. 2012, 18, 585–591. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Ma, L.; Xu, J.; Liu, C.; Zhang, J.; Liu, J.; Chen, R.; Zhou, Y. Spheroid body-forming cells in the human gastric cancer cell line MKN-45 possess cancer stem cell properties. Int. J. Oncol. 2013, 42, 453–459. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Qian, Y.; Li, F.; Bei, S.; Li, M.; Feng, L. microRNA-9 selectively targets LMX1A to promote gastric cancer cell progression. Biochem. Biophys. Res. Commun. 2018, 505, 405–412. [Google Scholar] [CrossRef] [PubMed]
- Vlachos, I.S.; Zagganas, K.; Paraskevopoulou, M.D.; Georgakilas, G.; Karagkouni, D.; Vergoulis, T.; Dalamagas, T.; Hatzigeorgiou, A.G. DIANA-miRPath v3.0: Deciphering microRNA function with experimental support. Nucleic Acids Res. 2015, 43, 460–466. [Google Scholar] [CrossRef]
- R Development Core Team. R: A Language and Environment for Statistical Computing. Available online: http://www.R-project.org (accessed on 13 April 2019).
- Reimers, M.; Carey, V.J. Bioconductor: an open source framework for bioinformatics and computational biology. Methods Enzymol. 2006, 411, 119–134. [Google Scholar] [PubMed]
- Mishra, L.; Derynck, R.; Mishra, B. Transforming Growth Factor-ß Signaling in Stem Cells and Cancer. Science 2005, 310, 68–71. [Google Scholar] [CrossRef]
- Han, B.; Cai, H.; Chen, Y.; Hu, B.; Luo, H.; Wu, Y.; Wu, J. The role of TGFBI (βig-H3) in gastrointestinal tract tumorigenesis. Mol. Cancer 2015, 24, 14–64. [Google Scholar] [CrossRef]
- Choi, Y.J.; Kim, N.; Chang, H. Helicobacter pylori-induced epithelial-mesenchymal transition, a potential role of gastric cancer initiation and an emergence of stem cells. Carcinogenesis 2015, 36, 553–563. [Google Scholar] [CrossRef]
- Nishimura, K.; Semba, S.; Aoyagi, K.; Sasaki, H.; Yokozaki, H. Mesenchymal stem cells provide an advantageous tumor microenvironment for the restoration of cancer stem cells. Pathobiology 2012, 79, 290–306. [Google Scholar] [CrossRef]
- Donnelly, J.M.; Engevik, A.C.; Engevik, M. Gastritis promotes an activated bone marrow-derived mesenchymal stem cell with a phenotype reminiscent of a cancer-promoting cell. Dig. Dis. Sci. 2014, 59, 569–582. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Peng, X.; Liu, G.; Peng, H.; Chen, A.; Zha, L.; Wang, Z. SOX4 contributes to TGF-β-induced epithelial–mesenchymal transition and stem cell characteristics of gastric cancer cells. Genes Dis. 2018, 5, 49–61. [Google Scholar] [CrossRef]
- Zhou, G.X.; Li, X.Y.; Zhang, Q.; Zhao, K.; Zhang, C.P.; Xue, C.H.; Yang, K.; Tian, Z.B. Effects of the hippo signaling pathway in human gastric cancer. Asian Pac. J. Cancer Prev. 2013, 14, 5199–5205. [Google Scholar] [CrossRef] [PubMed]
- Qiao, Y.; Li, T.; Zheng, S.; Wang, H. The Hippo pathway as a drug target in gastric cancer. Cancer Lett. 2018, 420, 14–25. [Google Scholar] [CrossRef]
- Luo, W.; Brouwer, C. Pathview: An R/Bioconductor package for pathway-based data integration and visualization. Bioinformatics 2013, 29, 1830–1831. [Google Scholar] [CrossRef]
- Abe, Y.; Minegishi, T.; Leung, P.C. Activin receptor signaling. Growth Factors 2004, 22, 105–110. [Google Scholar] [CrossRef]
- Shi, L.; Resaul, J.; Owen, S.; Ye, L.; Jiang, W.G. Clinical and Therapeutic Implications of Follistatin in Solid Tumours. Cancer Genom. Proteom. 2016, 13, 425–435. [Google Scholar] [CrossRef]
- Mo, J.S.; Park, H.W.; Guan, K.L. The Hippo signaling pathway in stem cell biology and cancer. EMBO Rep. 2014, 15, 642–656. [Google Scholar] [CrossRef]
- Park, J.H.; Shin, J.E.; Park, H.W. The Role of Hippo Pathway in Cancer Stem Cell Biology. Mol. Cells 2018, 41, 83–92. [Google Scholar]
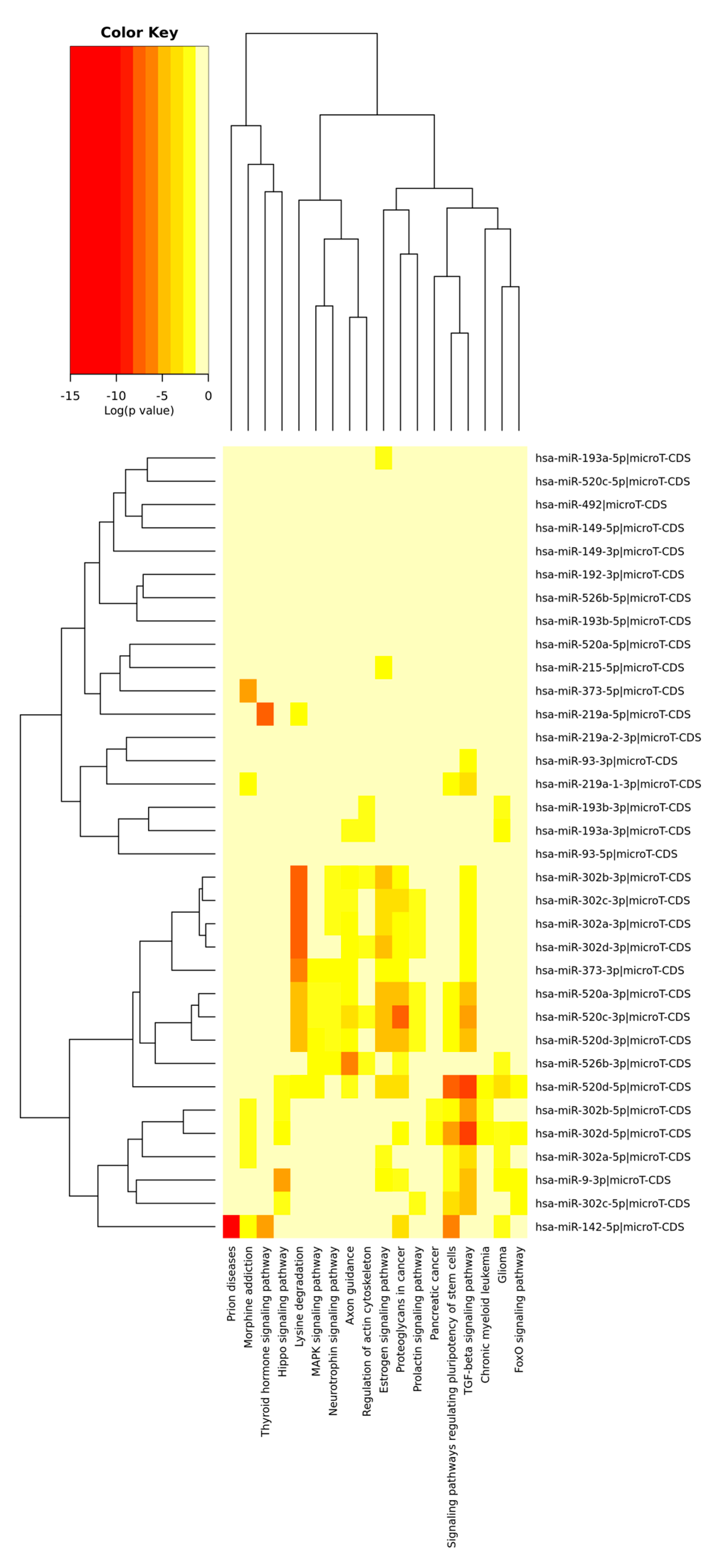
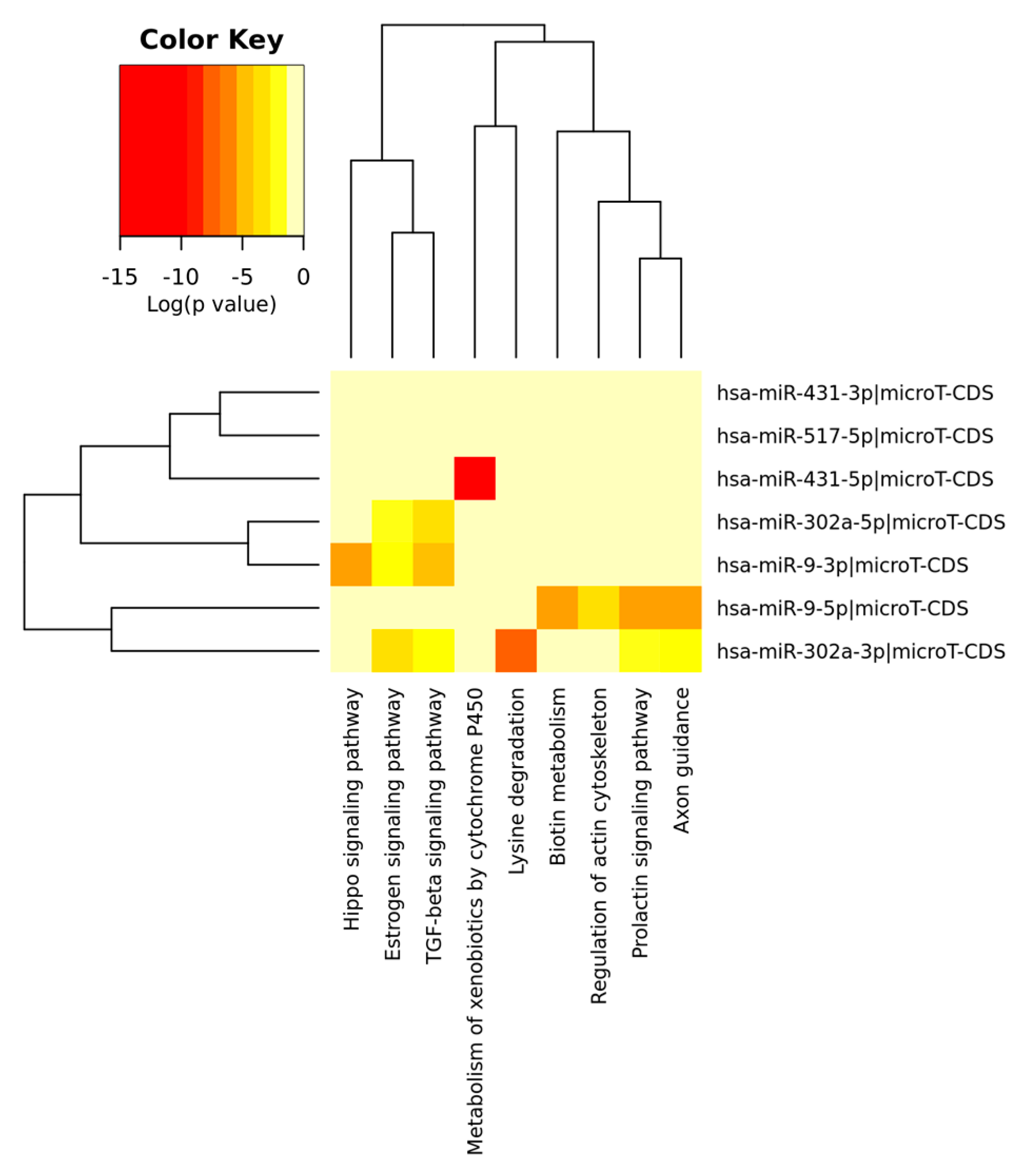
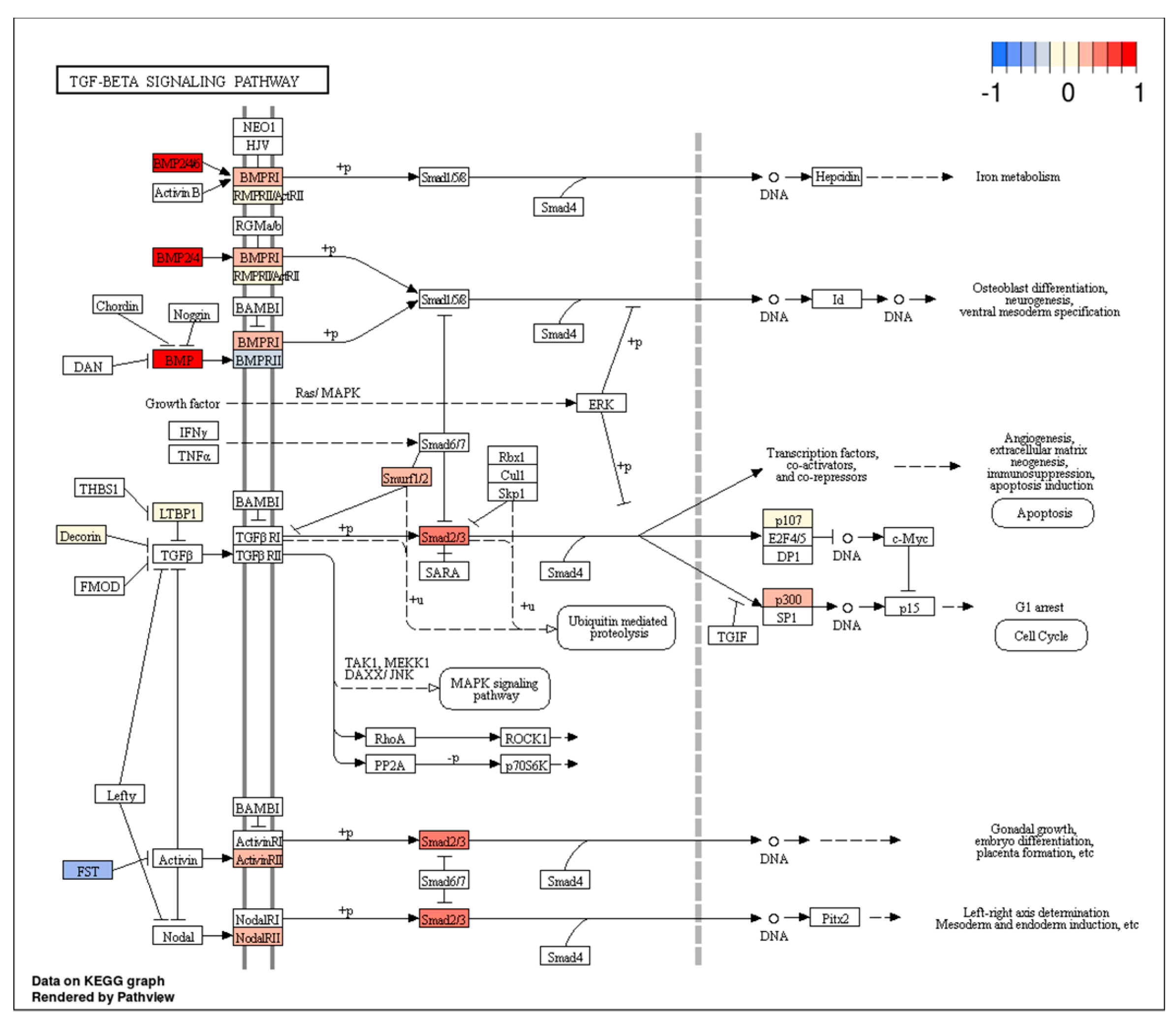
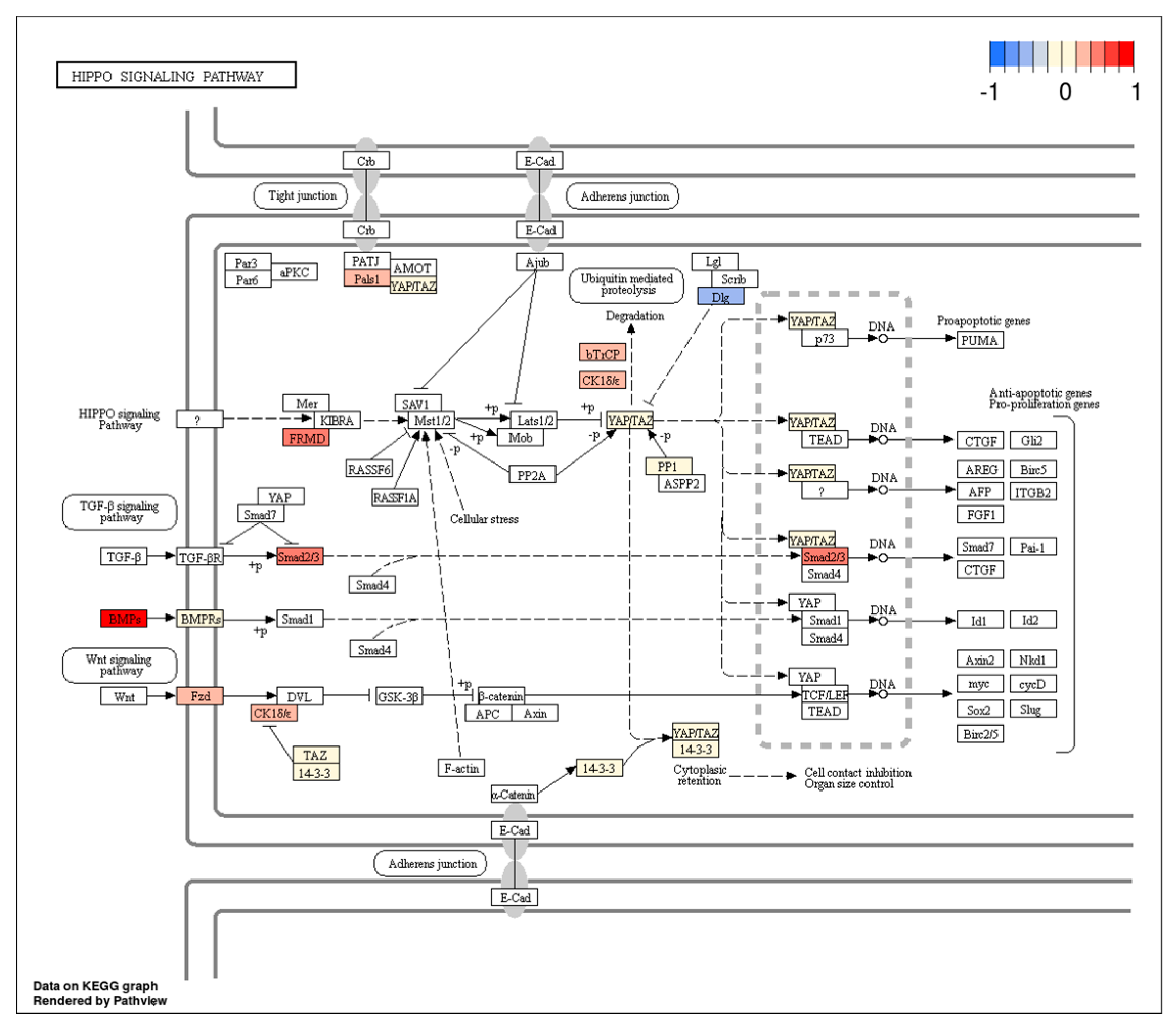
| MiRNA(s) | Target Gene(s) | Function | Cancer Type | Up/Downregulated | Reference(s) |
|---|---|---|---|---|---|
| Let-7 | RAS, HMGA2 | Regulation of self-renewal and multipotent differentiation capabilities | Breast | Down-regulated | Yu et al. [28] |
| MiR-200 family | BMI1 | Regulator of stem cell self-renewal | Breast | Down-regulated | Shimono et al. [29] |
| Suz12 | Inhibition of mammosphere, in combination with chemotherapy suppression of tumor growth | Breast | Down-regulated | Iliopoulos and Lindahl-Allen [30] | |
| ZEB1, ZEB2, TWIST1 | Transition to a breast cancer stem cell-like state | Breast | Down-regulated | Lim et al. [31] | |
| MiR-30 family | Ubc9, ITGB3, AVEN | Involvement in to apoptosis, proliferation and in tumor initiating BCSCs | Breast | Down-regulated | Yu et al. [32] Ouzounova et al. [33] |
| MiR-22 | TET2 | Promotion of self- renewal and transformation | Hematological malignancies | Up-regulated | Song et al. [34] |
| MiR-99 | Hoxa1 | Regulation of self-renewal and multipotent differentiation capabilities | Hematological malignancies | Up-regulated | Khalaj et al. [35] |
| MiR-126 | PI3K/AKT/mTOR pathway | Controller of cell cycle progression | Hematological malignancies | Up-regulated | Lechman et al. [36] |
| MiR-150 | Nanog | Suppression of proliferation and tumorigenicity of LSC | Hematological malignancies | Down-regulated | Xu et al. [37] |
| MiR-128 | Bmi-1 | Regulator of stem cell self-renewal | Glioblastoma | Down-regulated | Godlewski et al. [39] |
| MiR-451 | PI3K/AKT | Regulation of cell proliferation, invasion and apoptosis | Glioblastoma | Down-regulated | Gal et al. [40] Nan et al. [41] |
| MiR-34a | c-Met and Notch-1/2 | Tumor suppressor | Glioblastoma | Down-regulated | Guessous et al. [42] |
| MiR-125b | E2F2 | Regulation of cancer stem cell self-renewal and differentiation | Glioblastoma | Down-regulated | Wu et al. [43] |
| MiR-29b | BCL2L2 | Attenuates tumorigenicity and stemness maintenance | Glioblastoma | Down-regulated | Chung et al [44]. |
| MiR-200a | ZEB2 | Inhibition of EMT | Liver | Down-regulated | Wang et al [46] |
| MiR-491 | GIT-1/NF-kb | Inhibition of EMT | Liver | Down-regulated | Yang et al. [56] |
| let-7 b | Frizzled 4 | Reduces the proportion of cancer stem cells | Liver | Down-regulated | Cai et al. [48] |
| let-7 a | Wnt pathway | Inhibition of EMT | Liver | Down-regulated | Jin et al. [49] |
| MiR-217 | DKK1 | Regulation of the CSC-like phenotypes | Liver | Up-regulated | Jiang et al. [50] |
| MiR-452 | SOX 7 | Promotion of stem-like cells | Liver | Down-regulated | Zheng et al. [51] |
| MiR-200b/c, MiR-203 | BMI1, ZEB1 | Coordinately function for the suppression of the stem cell properties of CSCs | Colorectal | Down-regulated | Mukohyama et al. [52] |
| MiR-34a | Notch | Maintenance of CSCs | Colorectal | Down-regulated | Mukohyama et al. [52] |
| MiR-137 | MSI1 DCLK1 FMNL1 CDC42 | Enhances the stem cell properties of CSCs | Colorectal | Up-regulated | Mukohyama et al. [52] |
| MiR-221 | PTEN p27, p57 | ||||
| MiR-508 | ZEB1, BMI1, and SALL4, | Inhibits the CRC EMT and stemness process. | Colorectal | Down-regulated | Yan et al. [53] |
| MiR-320 | Wnt/beta-catenin signaling pathway | Promotes cancer stem cell-like properties | Prostate | Down-regulated | Hsieh et al. [57] |
| MiR-141 | CD44, EZH2 and Rho GTPases | Suppresses tumor growth and metastasis | Prostate | Down-regulated | Liu et al. [55] |
| Marker(s) Expression | In Vitro Assay | Efficiency (%) a | In Vivo Assay | Efficiency (%) b | Reference(s) |
|---|---|---|---|---|---|
| CD44+ | Spheroid colony formation | 10 cells/well NCI-N87, MKN-74, MKN-45 1 20 cells/well Human gastric cancer tissues 2 | Tumorigenicity (SCID mice) 1 (Nude mice) 2 | 20,000–30,000 cells injected 1 Skin NCI-N87, MKN-74, MKN-45 (100% efficiency) Stomach MKN-45 (100% efficiency) MKN-74 (75% efficiency) NCI-N87 (50% efficiency) 10,000 cells injected 2 Human gastric cancer tissues (80% efficiency) | 1. Takaishi et al. [20] 2. Sun et al. [68] |
| CD44+/CD24+ | Tumoroid sphere formation | 100 cell/well AGS | Tumorigenicity (NOD/SCID mice) | 200 cells injected AGS (50% efficiency) | Zhang et al. [69] |
| CD44+/CD54+ | Spheroid formation | 1 cell/well Human gastric cancer tissues 10,000 cells/dish Blood samples | Tumorigenicity (SCID/Nude mice) | 1000 cells injected Human gastric cancer tissues (100% efficiency) 9000 cells from spheres injected gastric cancer cells in circulation | Chen et al. [70] |
| CD44+/CD26+ | Spheroid formation | ≤5 × 106 cells/dish MKN7, MKN28, MKN45, AZ521 | Tumorigenicity (NOD/SCID mice) | 100 cells injected Human gastric cancer tissues (100% efficiency) | Nishikawa et al. [71] |
| CD44+/EpCAM+ | Spheroid formation | 1 cell/well Human gastric cancer tissues | Tumorigenicity (Nude mice) | 500 cells injected Human gastric cancer tissues (50% efficiency) | Han et al. [72] |
| CD44v8–10+ | Spheroid formation | 100 cells/dish (35-mm) GC45, GC84 xenograft tumors | Tumorigenicity (NOD/SCID mice) | 200 cells injected (75% efficiency) | Lau et al. [73] |
| CD71− | Colony formation | 500–1000 cells/dish (35-mm) MKN-1 | Tumorigenicity (NOD/SCID mice) | 100 cells injected (80% efficiency) | Ohkuma M. et al. [74] |
| CD90+ | Spheroid formation | 5000 cells/mL Gastric primary tumor model | Tumorigenicity (Nude mice) | 100 cells injected High Tumorigenicity group (100% efficiency) | Jiang et al. [75] |
| CD133+ | Colony formation | 1 cell/well KATO-III | Tumorigenicity (Nude mice) | 10,000 cells injected (100% efficiency) | Chen et al. [76] |
| ALDH1+ | Colony formation | 20,000 cells/well OCUM2-LMN | Tumorigenicity (Nude mice) | 100 cells injected (100% efficiency) | Katsuno et al. [77] |
| LGR5+ | Spheroid formation | 10,000 cells/well MKN-45, MKN-28 | Tumorigenicity (Nude mice) | 10,000 cells injected MKN-45, MKN-28 (100% efficiency) | Zhang et al. [78] |
| ID | Motif | Adjusted p-Value | Functional Relevance in GC |
|---|---|---|---|
| MiR-93 | AGCACTT | 0.017 | OncomiR [92] |
| MiR-302A_MiR-302B_MiR-302C | Tumor suppressor [100,101,102] | ||
| MiR-302D | No data | ||
| MiR-372_MiR-373_ MiR-520C | Tumor suppressor [79] | ||
| MiR-520E_MiR-520A_ MiR-520B _MiR-520D | Tumor suppressor [103] | ||
| MiR-526B | Tumor suppressor [104] | ||
| MiR-149 | GAGCCAG | 0.030 | Tumor suppressor [105] |
| MiR-9 | ACCAAAG | 0.030 | Tumor suppressor [106] |
| MiR-219 | GACAATC | 0.031 | Tumor suppressor [107] |
| MiR-193A_MiR-193B | GGCCAGT | 0.031 | OncomiR [108] |
| MiR-492 | CAGGTCC | 0.031 | No data |
| MiR-142_5P | ACTTTAT | 0.031 | Tumor suppressor [109] |
| MiR-192_MiR-215 | TAGGTCA | 0.039 | Tumor suppressor [110] |
| ID | Motif | p-Value | Functional Relevance in GC |
|---|---|---|---|
| MiR-9 | TAGCTTT | 0.007 | OncomiR [112] |
| MiR-431 | GCAAGAC | 0.014 | No data |
| MiR-302A | CACGTTT | 0.025 | OncomiR [79] |
| MiR-517 | TCTAGAG | 0.040 | No data |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ruggieri, V.; Russi, S.; Zoppoli, P.; La Rocca, F.; Angrisano, T.; Falco, G.; Calice, G.; Laurino, S. The Role of MicroRNAs in the Regulation of Gastric Cancer Stem Cells: A Meta-Analysis of the Current Status. J. Clin. Med. 2019, 8, 639. https://doi.org/10.3390/jcm8050639
Ruggieri V, Russi S, Zoppoli P, La Rocca F, Angrisano T, Falco G, Calice G, Laurino S. The Role of MicroRNAs in the Regulation of Gastric Cancer Stem Cells: A Meta-Analysis of the Current Status. Journal of Clinical Medicine. 2019; 8(5):639. https://doi.org/10.3390/jcm8050639
Chicago/Turabian StyleRuggieri, Vitalba, Sabino Russi, Pietro Zoppoli, Francesco La Rocca, Tiziana Angrisano, Geppino Falco, Giovanni Calice, and Simona Laurino. 2019. "The Role of MicroRNAs in the Regulation of Gastric Cancer Stem Cells: A Meta-Analysis of the Current Status" Journal of Clinical Medicine 8, no. 5: 639. https://doi.org/10.3390/jcm8050639
APA StyleRuggieri, V., Russi, S., Zoppoli, P., La Rocca, F., Angrisano, T., Falco, G., Calice, G., & Laurino, S. (2019). The Role of MicroRNAs in the Regulation of Gastric Cancer Stem Cells: A Meta-Analysis of the Current Status. Journal of Clinical Medicine, 8(5), 639. https://doi.org/10.3390/jcm8050639





