Alterations of the Gut Microbiome in Chronic Hepatitis B Virus Infection Associated with Alanine Aminotransferase Level
Abstract
:1. Introduction
2. Materials and Methods
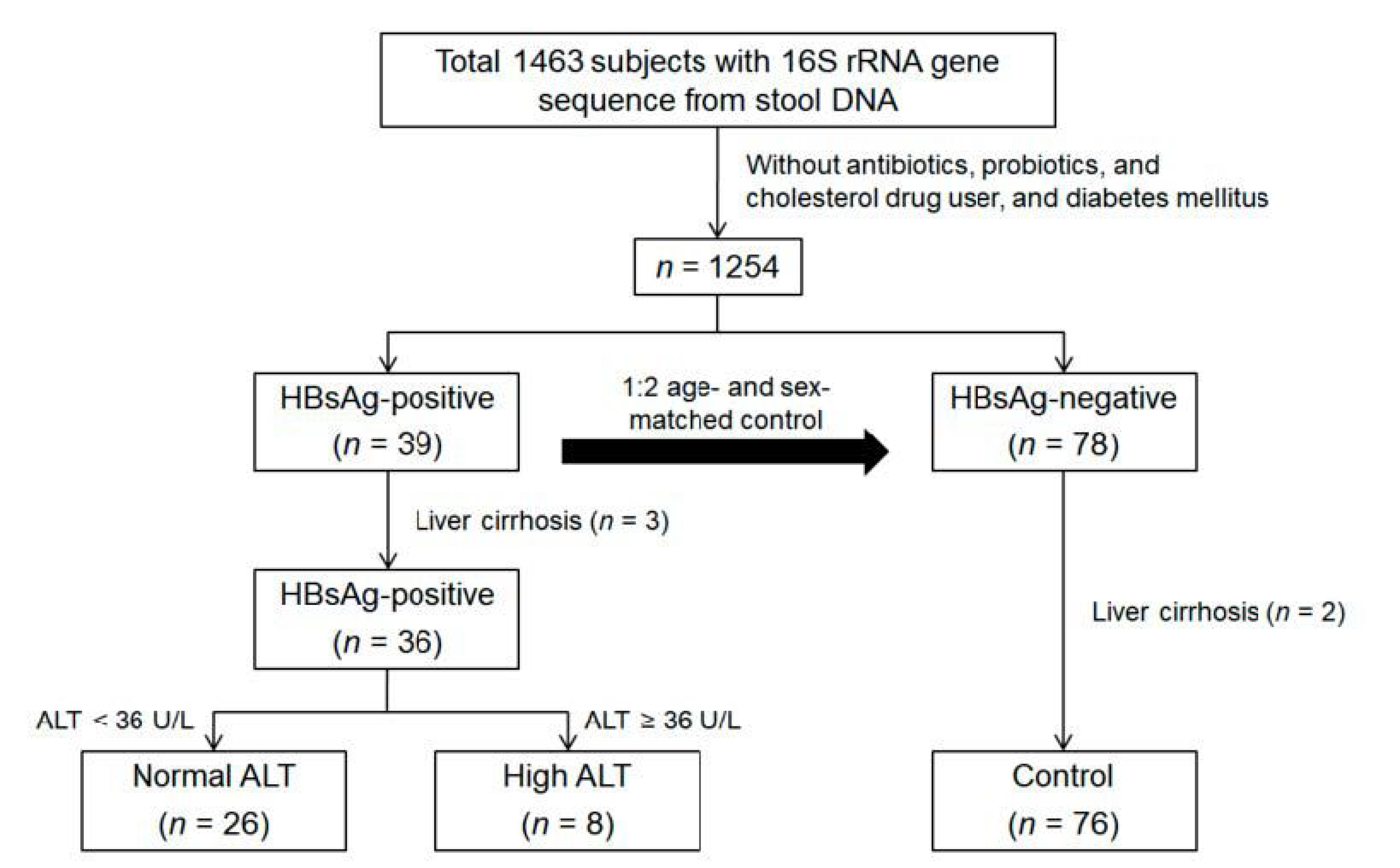
2.1. Study Subjects
2.2. Inclusion and Exclusion Criteria
2.3. Measurements
2.4. DNA Extraction and Sequence Data Generation
2.5. Sequence Analysis
2.6. Statistical Analysis
3. Results
3.1. Baseline Characteristics of the Study Population
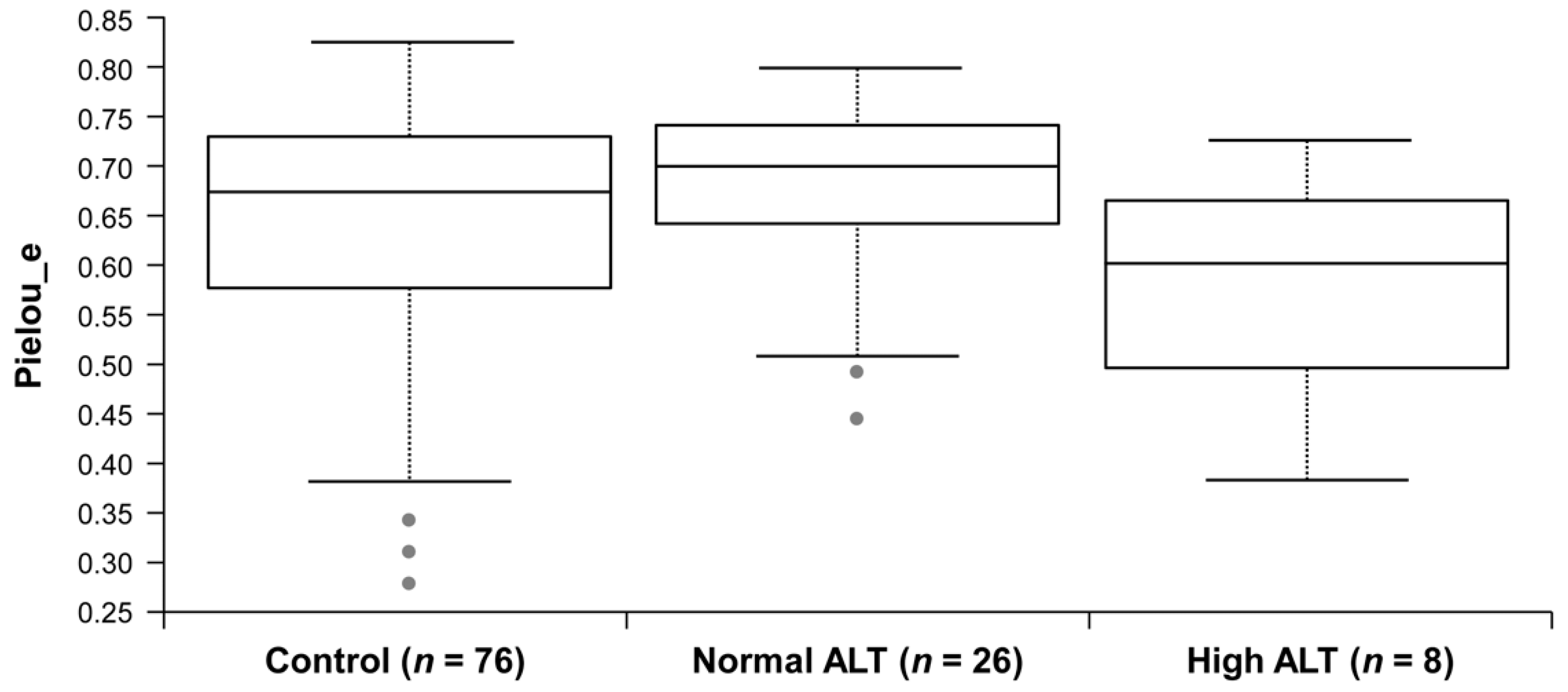
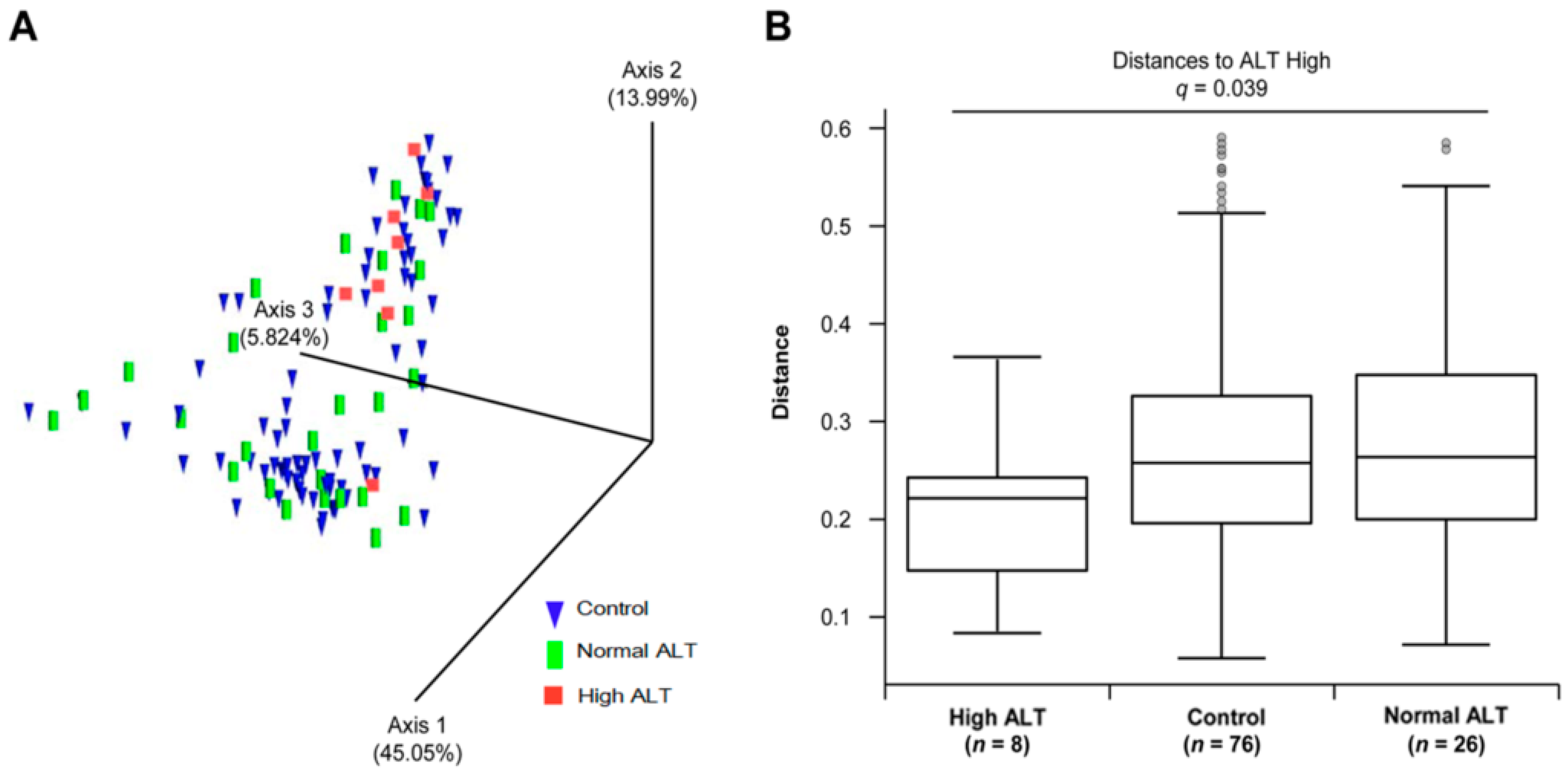
3.2. Microbial Diversity of the Gut Microbiota in Control, Normal ALT, and High ALT Groups
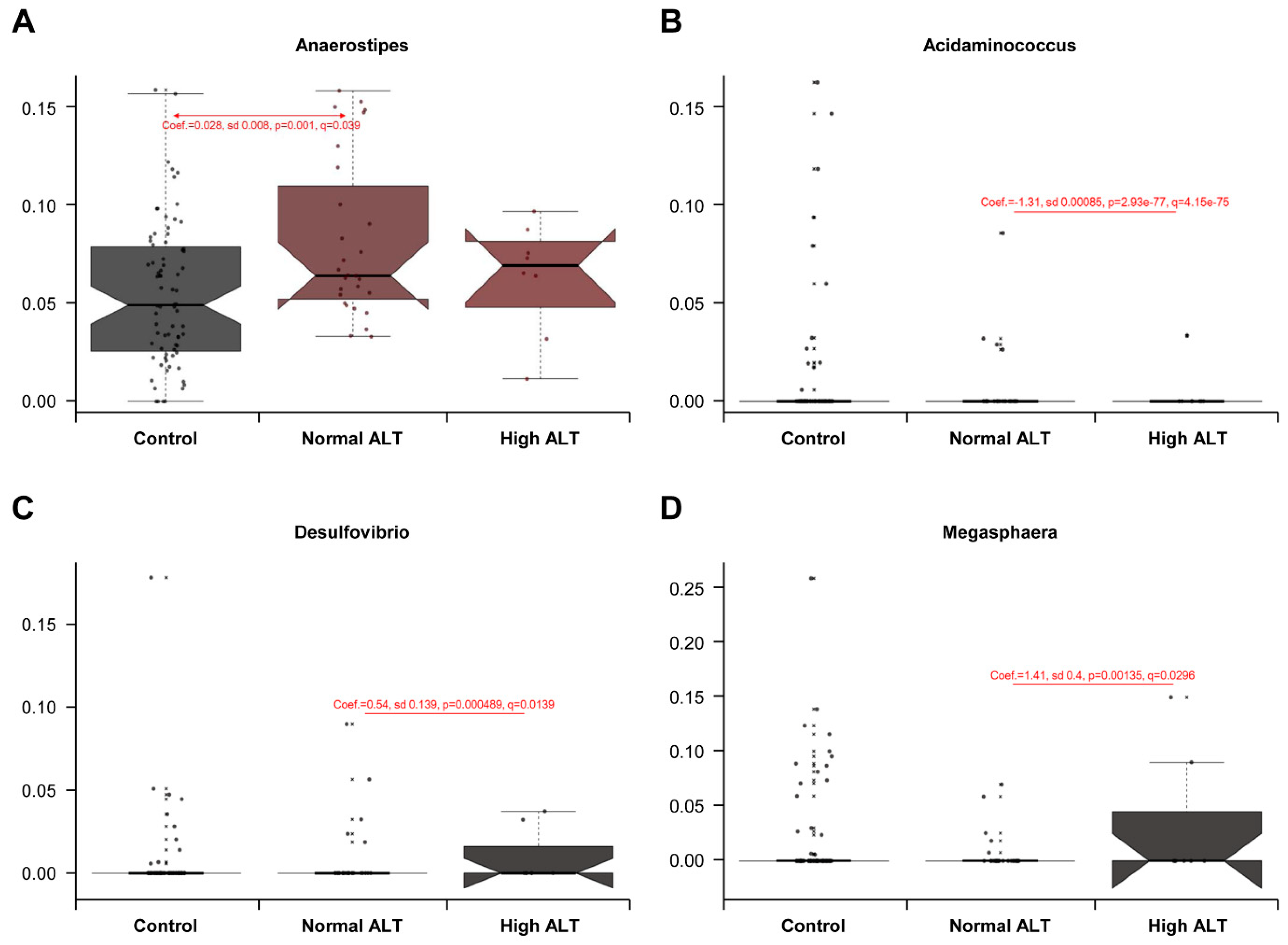
3.3. Taxonomic Comparison between Control, Normal ALT, and High ALT Groups
4. Discussion
Limitation
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Beasley, R.P. Rocks along the road to the control of HBV and HCC. Ann. Epidemiol. 2009, 19, 231–234. [Google Scholar] [CrossRef]
- Cho, E.J.; Kim, S.E.; Suk, K.T.; An, J.; Jeong, S.W.; Chung, W.J.; Kim, Y.J. Current status and strategies for hepatitis B control in Korea. Clin. Mol. Hepatol. 2017, 23, 205–211. [Google Scholar] [CrossRef]
- Chae, H.B.; Kim, J.H.; Kim, J.K.; Yim, H.J. Current status of liver diseases in Korea: Hepatitis B. Korean J. Hepatol. 2009, 15, S13–S24. [Google Scholar] [CrossRef] [PubMed]
- Song, I.H.; Kim, K.S. Current status of liver diseases in Korea: Hepatocellular carcinoma. Korean J. Hepatol. 2009, 15, S50–S59. [Google Scholar] [CrossRef] [PubMed]
- Chassaing, B.; Etienne-Mesmin, L.; Gewirtz, A.T. Microbiota-liver axis in hepatic disease. Hepatology 2014, 59, 328–339. [Google Scholar] [CrossRef]
- Qin, N.; Yang, F.; Li, A.; Prifti, E.; Chen, Y.; Shao, L.; Guo, J.; Le Chatelier, E.; Yao, J.; Wu, L.; et al. Alterations of the human gut microbiome in liver cirrhosis. Nature 2014, 513, 59–64. [Google Scholar] [CrossRef]
- Bajaj, J.S.; Betrapally, N.S.; Gillevet, P.M. Decompensated cirrhosis and microbiome interpretation. Nature 2015, 525, E1–E2. [Google Scholar] [CrossRef]
- Lu, H.; Wu, Z.; Xu, W.; Yang, J.; Chen, Y.; Li, L. Intestinal microbiota was assessed in cirrhotic patients with hepatitis B virus infection. Intestinal microbiota of HBV cirrhotic patients. Microb. Ecol. 2011, 61, 693–703. [Google Scholar] [CrossRef]
- Xu, M.; Wang, B.; Fu, Y.; Chen, Y.; Yang, F.; Lu, H.; Xu, J.; Li, L. Changes of fecal Bifidobacterium species in adult patients with hepatitis B virus-induced chronic liver disease. Microb. Ecol. 2012, 63, 304–313. [Google Scholar] [CrossRef] [PubMed]
- Sarin, S.K.; Kumar, M.; Lau, G.K.; Abbas, Z.; Chan, H.L.; Chen, C.J.; Chen, D.S.; Chen, H.L.; Chen, P.J.; Chien, R.N.; et al. Asian-Pacific clinical practice guidelines on the management of hepatitis B: A 2015 update. Hepatol. Int. 2016, 10, 1–98. [Google Scholar] [CrossRef] [PubMed]
- Xu, D.; Huang, Y.; Wang, J. Gut microbiota modulate the immune effect against hepatitis B virus infection. Eur. J. Clin. Microbiol. Infect. Dis. 2015, 34, 2139–2147. [Google Scholar] [CrossRef]
- Chang, Y.; Jung, H.-S.; Cho, J.; Zhang, Y.; Yun, K.E.; Lazo, M.; Pastor-Barriuso, R.; Ahn, J.; Kim, C.-W.; Rampal, S.; et al. Metabolically Healthy Obesity and the Development of Nonalcoholic Fatty Liver Disease. Am. J. Gastroenterol. 2016, 111, 1133–1140. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yun, Y.; Kim, H.N.; Kim, S.E.; Heo, S.G.; Chang, Y.; Ryu, S.; Shin, H.; Kim, H.L. Comparative analysis of gut microbiota associated with body mass index in a large Korean cohort. BMC Microbiol. 2017, 17, 151. [Google Scholar] [CrossRef] [Green Version]
- Chang, Y.; Kim, B.K.; Yun, K.E.; Cho, J.; Zhang, Y.; Rampal, S.; Zhao, D.; Jung, H.S.; Choi, Y.; Ahn, J.; et al. Metabolically-healthy obesity and coronary artery calcification. J. Am. Coll. Cardiol. 2014, 63, 2679–2686. [Google Scholar] [CrossRef] [PubMed]
- Fadrosh, D.W.; Ma, B.; Gajer, P.; Sengamalay, N.; Ott, S.; Brotman, R.M.; Ravel, J. An improved dual-indexing approach for multiplexed 16S rRNA gene sequencing on the Illumina MiSeq platform. Microbiome 2014, 2, 6. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C. UPARSE: Highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 2013, 10, 996–998. [Google Scholar] [CrossRef]
- Lozupone, C.; Knight, R. UniFrac: A New Phylogenetic Method for Comparing Microbial Communities. Appl. Environ. Microbiol. 2005, 71, 8228–8235. [Google Scholar] [CrossRef]
- Morgan, X.C.; Tickle, T.L.; Sokol, H.; Gevers, D.; Devaney, K.L.; Ward, D.V.; Reyes, J.A.; Shah, S.A.; LeLeiko, N.; Snapper, S.B.; et al. Dysfunction of the intestinal microbiome in inflammatory bowel disease and treatment. Genome Biol. 2012, 13, R79. [Google Scholar] [CrossRef]
- Wang, J.; Wang, Y.; Zhang, X.; Liu, J.; Zhang, Q.; Zhao, Y.; Peng, J.; Feng, Q.; Dai, J.; Sun, S.; et al. Gut Microbial Dysbiosis Is Associated with Altered Hepatic Functions and Serum Metabolites in Chronic Hepatitis B Patients. Front. Microbiol. 2017, 8, 2222. [Google Scholar] [CrossRef] [Green Version]
- Marounek, M.; Fliegrova, K.; Bartos, S. Metabolism and some characteristics of ruminal strains of Megasphaera elsdenii. Appl. Environ. Microbiol. 1989, 55, 1570–1573. [Google Scholar]
- Shetty, S.A.; Marathe, N.P.; Lanjekar, V.; Ranade, D.; Shouche, Y.S. Comparative genome analysis of Megasphaera sp. reveals niche specialization and its potential role in the human gut. PLoS ONE 2013, 8, e79353. [Google Scholar] [CrossRef]
- Oehler, N.; Volz, T.; Bhadra, O.D.; Kah, J.; Allweiss, L.; Giersch, K.; Bierwolf, J.; Riecken, K.; Pollok, J.M.; Lohse, A.W.; et al. Binding of hepatitis B virus to its cellular receptor alters the expression profile of genes of bile acid metabolism. Hepatology 2014, 60, 1483–1493. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Duncan, S.H.; Louis, P.; Flint, H.J. Lactate-utilizing bacteria, isolated from human feces, that produce butyrate as a major fermentation product. Appl. Environ. Microbiol. 2004, 70, 5810–5817. [Google Scholar] [CrossRef] [PubMed]
- Hamer, H.M.; Jonkers, D.; Venema, K.; Vanhoutvin, S.; Troost, F.J.; Brummer, R.J. Review article: The role of butyrate on colonic function. Aliment. Pharmacol. Ther. 2008, 27, 104–119. [Google Scholar] [CrossRef]
- Nobili, V.; Alkhouri, N.; Alisi, A.; Della Corte, C.; Fitzpatrick, E.; Raponi, M.; Dhawan, A. Nonalcoholic fatty liver disease: A challenge for pediatricians. JAMA Pediatr. 2015, 169, 170–176. [Google Scholar] [CrossRef] [PubMed]
- Joo, E.J.; Chang, Y.; Yeom, J.S.; Ryu, S. Hepatitis B virus infection and decreased risk of nonalcoholic fatty liver disease: A cohort study. Hepatology 2017, 65, 828–835. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.Y.; Lu, C.W.; Liu, Y.L.; Chiang, C.H.; Lee, L.T.; Huang, K.C. Relationship between chronic hepatitis B and metabolic syndrome: A structural equation modeling approach. Obesity 2016, 24, 483–489. [Google Scholar] [CrossRef]
- Jinjuvadia, R.; Liangpunsakul, S. Association between metabolic syndrome and its individual components with viral hepatitis B. Am. J. Med. Sci. 2014, 347, 23–27. [Google Scholar] [CrossRef]
- Wong, V.W.; Wong, G.L.; Chu, W.C.; Chim, A.M.; Ong, A.; Yeung, D.K.; Yiu, K.K.; Chu, S.H.; Chan, H.Y.; Woo, J.; et al. Hepatitis B virus infection and fatty liver in the general population. J. Hepatol. 2012, 56, 533–540. [Google Scholar] [CrossRef]
- Spadoni, I.; Zagato, E.; Bertocchi, A.; Paolinelli, R.; Hot, E.; Di Sabatino, A.; Caprioli, F.; Bottiglieri, L.; Oldani, A.; Viale, G.; et al. A gut-vascular barrier controls the systemic dissemination of bacteria. Science 2015, 350, 830–834. [Google Scholar] [CrossRef]
| Demographic Characteristics | HBsAg-Negative Control (n = 76) | HBsAg-Positive Normal ALT (n = 28) | HBsAg-Positive High ALT (n = 8) | p-Value a |
|---|---|---|---|---|
| Male (number) | 53 (70%) | 17 (37%) | 8 (100%) | 0.103 |
| Age (years) | 45.07 ± 6.74 | 45.82 ± 7.03 | 49.00 ±4.28 | 0.460 |
| BMI (kg/m2) | 23.44 ± 3.01 | 23.99 ± 2.72 | 26.89 ± 2.68 | 0.007 |
| Fat percent (%) | 24.41 ± 5.93 | 26.86 ± 6.43 | 26.24 ± 5.65 | 0.174 |
| Systolic BP (mmHg) | 110.39 ± 13.41 | 109.21 ± 13.46 | 117.50 ± 15.51 | 0.308 |
| Diastolic BP (mmHg) | 72.03 ± 10.61 | 71.36 ± 10.76 | 73.88 ± 10.52 | 0.839 |
| Glucose (mg/dL) | 94.99 ± 8.73 | 91.57 ± 7.14 | 94.13 ± 8.17 | 0.184 |
| Total-C (mg/dL) | 204.28 ± 36.69 | 191.54 ± 38.05 | 202.13 ± 40.83 | 0.306 |
| HDL-C (mg/dL) | 55.41 ± 15.00 | 55.18 ± 11.89 | 64.63 ± 12.86 | 0.207 |
| LDL cholesterol (mg/dL) | 128.25 ± 32.70 | 114.79 ± 33.01 | 121.63 ± 35.21 | 0.181 |
| Triglycerides (mg/dL) | 128.76 ± 75.03 | 115.46 ± 70.44 | 86.25 ± 28.97 | 0.241 |
| Total Bilirubin (mg/dL) | 0.83 ± 0.43 | 0.83 ± 0.36 | 0.86 ± 0.21 | 0.982 |
| AST (IU/L) | 23.39 ± 15.42 | 21.86 ± 7.19 | 39.25 ± 8.84 | 0.005 |
| ALT (IU/L) | 25.61 ± 26.21 | 20.36 ± 7.94 | 51.88 ± 17.07 | 0.003 |
| WBC (×103/mm3) | 5.93 ± 1.54 | 5.52 ± 1.19 | 5.11 ± 0.96 | 0.181 |
| Platelet (×103/mm3) | 250.09 ± 47.86 | 222.86 ± 45.37 | 182.38 ± 44.76 | <0.001 |
| HOMA-IR | 1.35 ± 0.80 | 1.28 ± 0.54 | 1.96 ± 0.67 | 0.066 |
| Fatty liver b (%) | 39 (30/76) | 18 (5/28) | 25 (2/8) | 0.102 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yun, Y.; Chang, Y.; Kim, H.-N.; Ryu, S.; Kwon, M.-J.; Cho, Y.K.; Kim, H.-L.; Cheong, H.S.; Joo, E.-J. Alterations of the Gut Microbiome in Chronic Hepatitis B Virus Infection Associated with Alanine Aminotransferase Level. J. Clin. Med. 2019, 8, 173. https://doi.org/10.3390/jcm8020173
Yun Y, Chang Y, Kim H-N, Ryu S, Kwon M-J, Cho YK, Kim H-L, Cheong HS, Joo E-J. Alterations of the Gut Microbiome in Chronic Hepatitis B Virus Infection Associated with Alanine Aminotransferase Level. Journal of Clinical Medicine. 2019; 8(2):173. https://doi.org/10.3390/jcm8020173
Chicago/Turabian StyleYun, Yeojun, Yoosoo Chang, Han-Na Kim, Seungho Ryu, Min-Jung Kwon, Yong Kyun Cho, Hyung-Lae Kim, Hae Suk Cheong, and Eun-Jeong Joo. 2019. "Alterations of the Gut Microbiome in Chronic Hepatitis B Virus Infection Associated with Alanine Aminotransferase Level" Journal of Clinical Medicine 8, no. 2: 173. https://doi.org/10.3390/jcm8020173
APA StyleYun, Y., Chang, Y., Kim, H.-N., Ryu, S., Kwon, M.-J., Cho, Y. K., Kim, H.-L., Cheong, H. S., & Joo, E.-J. (2019). Alterations of the Gut Microbiome in Chronic Hepatitis B Virus Infection Associated with Alanine Aminotransferase Level. Journal of Clinical Medicine, 8(2), 173. https://doi.org/10.3390/jcm8020173






