Gastrointestinal Microbiota and Type 1 Diabetes Mellitus: The State of Art
Abstract
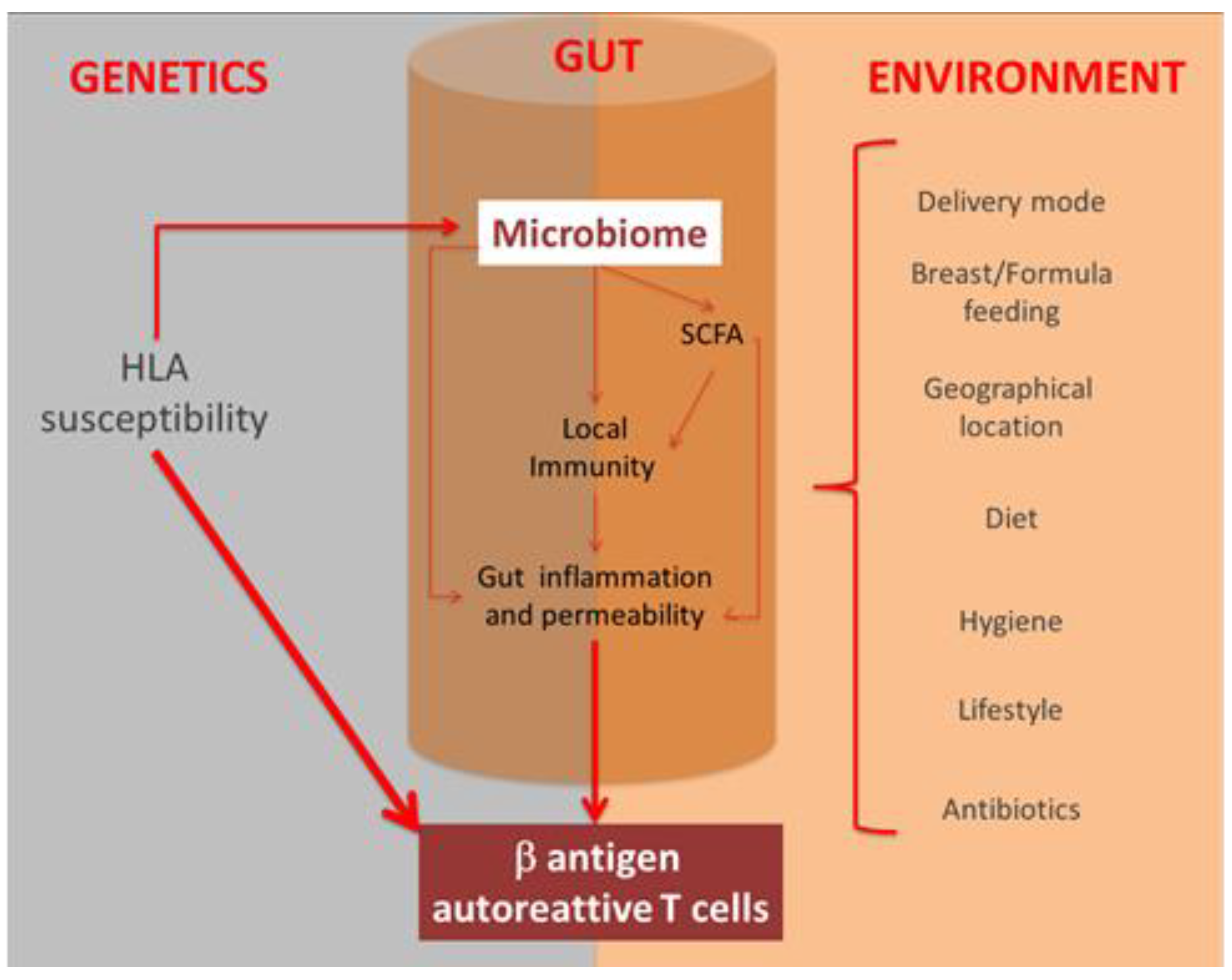
1. Introduction
2. Type 1 Diabetes Mellitus
3. The Microbiota
4. Importance of the Microbiota to the Host
5. Microbiota and Type 1 Diabetes Mellitus
5.1. The Microbiota in Experimental T1DM
5.2. Intervention Studies in Experimental Autoimmune Diabetes
5.3. The Microbiota in Human T1DM
6. Future perspective
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Gülden, E.; Wong, F.S.; Wen, L. The gut microbiota and type 1 diabetes. Clin. Immunol. 2015, 159, 143–153. [Google Scholar] [CrossRef] [PubMed]
- Paun, A.; Yau, C.; Danska, J.S. The influence of the microbiome on type 1 diabetes. J. Immunol. 2017, 198, 590–595. [Google Scholar] [CrossRef] [PubMed]
- Hu, C.; Wong, F.S.; Wen, L. Type 1 diabetes and gut microbiota: Friend or foe? Pharmacol. Res. 2015, 98, 9–15. [Google Scholar] [CrossRef] [PubMed]
- American Diabetes Association. Classification and diagnosis of diabetes: Standards of medical care in diabetes. Diabetes Care 2019, 4, S13–S28. [Google Scholar] [CrossRef]
- International Diabetes Federation. IDF Diabetes Atlas, 8th ed. 2017. Available online: http://www.diabetesatlas.org (accessed on 1 November 2019).
- Soltesz, G.; Patterson, C.C.; Dahlquist, G.; EURODIAB Study Group. Worldwide childhood type 1 diabetes incidence–what can we learn from epidemiology? Pediatric Diabetes 2007, 8, 6–14. [Google Scholar] [CrossRef]
- Patterson, C.C.; Gyürüs, E.; Rosenbauer, J.; Cinek, O.; Neu, A.; Schober, E.; Parslow, R.C.; Joner, G.; Svensson, J.; Castell, C. Trends in childhood type 1 diabetes incidence in Europe during 1989–2008: Evidence of non-uniformity over time in rates of increase. Diabetologia 2012, 55, 2142–2147. [Google Scholar] [CrossRef]
- Harjutsalo, V.; Sjöberg, L.; Tuomilehto, J. Time trends in the incidence of type 1 diabetes in Finnish children: A cohort study. Lancet 2008, 371, 1777–1782. [Google Scholar] [CrossRef]
- Knip, M.; Korhonen, S.; Kulmala, P.; Veijola, R.; Reunanen, A.; Raitakari, O.T.; Viikari, J.; Åkerblom, H.K. Prediction of type 1 diabetes in the general population. Diabetes Care 2010, 33, 1206–1212. [Google Scholar] [CrossRef]
- Regnell, S.E.; Lernmark, Å. Early prediction of autoimmune (type 1) diabetes. Diabetologia 2017, 60, 1370–1381. [Google Scholar] [CrossRef]
- Wolfsdorf, J.; Glaser, N.; Sperling, M.A. Diabetic ketoacidosis in infants, children, and adolescents: A consensus statement from the American Diabetes Association. Diabetes care 2006, 29, 1150–1159. [Google Scholar] [CrossRef]
- Dayan, C.M.; Korah, M.; Tatovic, D.; Bundy, B.N.; Herold, K.C. Changing the landscape for type 1 diabetes: The first step to prevention. Lancet 2019. [Google Scholar] [CrossRef]
- Pociot, F.; Lernmark, Å. Genetic risk factors for type 1 diabetes. Lancet 2016, 387, 2331–2339. [Google Scholar] [CrossRef]
- Sun, C.; Zhi, D.; Shen, S.; Luo, F.; Sanjeevi, C.B. SNPs in the exons of Toll-like receptors are associated with susceptibility to type 1 diabetes in Chinese population. Hum. Immunol. 2014, 75, 1084–1088. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Infante, M.; Ricordi, C.; Sanchez, J.; Clare-Salzler, M.J.; Padilla, N.; Fuenmayor, V.; Chavez, C.; Alvarez, A.; Baidal, D.; Alejandro, R.; et al. Influence of Vitamin D on Islet Autoimmunity and Beta-Cell Function in Type 1 Diabetes. Nutrients 2019, 11, 2185. [Google Scholar] [CrossRef] [PubMed]
- Nam, H.K.; Rhie, Y.J.; Lee, K.H. Vitamin D level and gene polymorphisms in Korean children with type 1 diabetes. Pediatric Diabetes 2019, 20, 750–758. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, A.E.A.; Sakhr, H.M.; Hassan, M.H.; El-Amir, M.I.; Ameen, H.H. Vitamin D receptor rs7975232, rs731236 and rs1544410 single nucleotide polymorphisms, and 25-hydroxyvitamin D levels in Egyptian children with type 1 diabetes mellitus: Effect of vitamin D co-therapy. Diabetes Metab. Syndr. Obes. 2019, 12, 703. [Google Scholar] [CrossRef] [PubMed]
- Rewers, M.; Ludvigsson, J. Environmental risk factors for type 1 diabetes. Lancet 2016, 387, 2340–2348. [Google Scholar] [CrossRef]
- Bäckhed, F.; Ley, R.E.; Sonnenburg, J.L.; Peterson, D.A.; Gordon, J.I. Host-bacterial mutualism in the human intestine. Science 2005, 307, 1915–1920. [Google Scholar] [CrossRef]
- Sender, R.; Fuchs, S.; Milo, R. Are we really vastly outnumbered? Revisiting the ratio of bacterial to host cells in humans. Cell 2016, 164, 337–340. [Google Scholar] [CrossRef]
- Fuks, G.; Elgart, M.; Amir, A.; Zeisel, A.; Turnbaugh, P.J.; Soen, Y.; Shental, N. Combining 16S rRNA gene variable regions enables high-resolution microbial community profiling. Microbiome 2018, 6, 17. [Google Scholar] [CrossRef]
- Poretsky, R.; Rodriguez-R, L.M.; Luo, C.; Tsementzi, D.; Konstantinidis, K.T. Strengths and limitations of 16S rRNA gene amplicon sequencing in revealing temporal microbial community dynamics. PLoS ONE 2014, 9, e93827. [Google Scholar] [CrossRef] [PubMed]
- Qin, J.; Li, R.; Raes, J.; Arumugam, M.; Burgdorf, K.S.; Manichanh, C.; Nielsen, T.; Pons, N.; Levenez, F.; Yamada, T.; et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature 2010, 464, 59. [Google Scholar] [CrossRef] [PubMed]
- Eckburg, P.B.; Bik, E.M.; Bernstein, C.N.; Purdom, E.; Dethlefsen, L.; Sargent, M.; Gill, S.R.; Nelson, K.E.; Relman, D.A. Diversity of the human intestinal microbial flora. Science 2005, 308, 1635–1638. [Google Scholar] [CrossRef] [PubMed]
- Yatsunenko, T.; Rey, F.E.; Manary, M.J.; Trehan, I.; Dominguez-Bello, M.G.; Contreras, M.; Magris, M.; Hidalgo, G.; Baldassano, R.N.; Anokhin, A.P.; et al. Human gut microbiome viewed across age and geography. Nature 2012, 486, 222. [Google Scholar] [CrossRef]
- Bäckhed, F.; Roswall, J.; Peng, Y.; Feng, Q.; Jia, H.; Kovatcheva-Datchary, P.; Li, Y.; Xia, Y.; Xie, H.; Zhong, H.; et al. Dynamics and stabilization of the human gut microbiome during the first year of life. Cell Host Microbe 2015, 17, 690–703. [Google Scholar] [CrossRef]
- Dominguez-Bello, M.G.; Costello, E.K.; Contreras, M.; Magris, M.; Hidalgo, G.; Fierer, N.; Knight, R. Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proc. Natl. Acad. Sci. USA 2010, 107, 11971–11975. [Google Scholar] [CrossRef]
- Ferretti, P.; Pasolli, E.; Tett, A.; Asnicar, F.; Gorfer, V.; Fedi, S.; Armanini, F.; Truong, D.T.; Manara, S.; Zolfo, M.; et al. Mother-to-infant microbial transmission from different body sites shapes the developing infant gut microbiome. Cell Host Microbe 2018, 24, 133–145. [Google Scholar] [CrossRef]
- Rinninella, E.; Raoul, P.; Cintoni, M.; Franceschi, F.; Miggiano, G.A.D.; Gasbarrini, A.; Mele, M.C. What is the healthy gut microbiota composition? a changing ecosystem across age, environment, diet, and diseases. Microorganisms 2019, 7, 14. [Google Scholar] [CrossRef]
- Praveen, P.; Jordan, F.; Priami, C.; Morine, M.J. The role of breast-feeding in infant immune system: A systems perspective on the intestinal microbiome. Microbiome 2015, 3, 41. [Google Scholar] [CrossRef]
- Singh, R.K.; Chang, H.W.; Yan, D.; Lee, K.M.; Ucmak, D.; Wong, K.; Abrouk, M.; Farahnik, B.; Nakamura, M.; Zhu, T.H.; et al. Influence of diet on the gut microbiome and implications for human health. J. Transl. Med. 2017, 15, 73. [Google Scholar] [CrossRef]
- De Filippo, C.; Cavalieri, D.; Di Paola, M.; Ramazzotti, M.; Poullet, J.B.; Massart, S.; Collini, S.; Pieraccini, G.; Lionetti, P. Impact of diet in shaping gut microbiota revealed by a comparative study in children from Europe and rural Africa. Proc. Natl. Acad. Sci. USA 2010, 107, 14691–14696. [Google Scholar] [CrossRef] [PubMed]
- Venkataraman, R.; Juwal, J.; Princy, J. The effect of probiotics on glycemic index. Panminerva Med. 2018, 60, 234–235. [Google Scholar] [CrossRef] [PubMed]
- Fjalstad, J.W.; Esaiassen, E.; Juvet, L.K.; van den Anker, J.N.; Klingenberg, C. Antibiotic therapy in neonates and impact on gut microbiota and antibiotic resistance development: A systematic review. J. Antimicrob. Chemother. 2017, 73, 569–580. [Google Scholar] [CrossRef] [PubMed]
- Antonopoulos, D.A.; Huse, S.M.; Morrison, H.G.; Schmidt, T.M.; Sogin, M.L.; Young, V.B. Reproducible community dynamics of the gastrointestinal microbiota following antibiotic perturbation. Infect. Immun. 2009, 77, 2367–2375. [Google Scholar] [CrossRef] [PubMed]
- Claesson, M.J.; Jeffery, I.B.; Conde, S.; Power, S.E.; O’connor, E.M.; Cusack, S.; Harris, H.M.; Coakley, M.; Lakshminarayanan, B.; O’Sullivan, O.; et al. Gut microbiota composition correlates with diet and health in the elderly. Nature 2012, 488, 178. [Google Scholar] [CrossRef] [PubMed]
- Krishnan, S.; Alden, N.; Lee, K. Pathways and functions of gut microbiota metabolism impacting host physiology. Curr. Opin. Biotechnol. 2015, 36, 137–145. [Google Scholar] [CrossRef]
- LeBlanc, J.G.; Chain, F.; Martín, R.; Bermúdez-Humarán, L.G.; Courau, S.; Langella, P. Beneficial effects on host energy metabolism of short-chain fatty acids and vitamins produced by commensal and probiotic bacteria. Microb. Cell Fact 2017, 16, 79. [Google Scholar] [CrossRef]
- Thaiss, C.A.; Zmora, N.; Levy, M.; Elinav, E. The microbiome and innate immunity. Nature 2016, 535, 65. [Google Scholar] [CrossRef]
- Luu, M.; Visekruna, A. Short-Chain fatty acids: Bacterial messengers modulating the immunometabolism of T cells. Eur. J. Immunol. 2019, 49, 842–848. [Google Scholar] [CrossRef]
- Lazar, V.; Ditu, L.; Pircalabioru, G.; Gheorghe, I.; Curutiu, C.; Holban, A.M.; Chifiriuc, C.M. Aspects of gut microbiota and immune system interactions in infectious diseases, immunopathology and cancer. Front. Immunol. 2018, 9, 1830. [Google Scholar] [CrossRef]
- Skonieczna-Żydecka, K.; Marlicz, W.; Misera, A.; Koulaouzidis, A.; Łoniewski, I. Microbiome—The Missing Link in the Gut-Brain Axis: Focus on Its Role in Gastrointestinal and Mental Health. J. Clin. Med. 2018, 7, 521. [Google Scholar] [CrossRef] [PubMed]
- Petersen, C.; Round, J.L. Defining dysbiosis and its influence on host immunity and disease. Cell Microbiol. 2014, 16, 1024–1033. [Google Scholar] [CrossRef] [PubMed]
- Vangay, P.; Ward, T.; Gerber, J.S.; Knights, D. Antibiotics, pediatric dysbiosis, and disease. Cell Host Microbe 2015, 17, 553–564. [Google Scholar] [CrossRef] [PubMed]
- Brüssow, H. Problems with the concept of gut microbiota dysbiosis. Microb. Biotechnol. 2019, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Brown, K.; Godovannyi, A.; Ma, C.; Zhang, Y.; Ahmadi-Vand, Z.; Dai, C.; Gorzelak, M.A.; Chan, Y.; Chan, J.M.; Lochner, A.; et al. Prolonged antibiotic treatment induces a diabetogenic intestinal microbiome that accelerates diabetes in NOD mice. ISME J. 2016, 10, 321. [Google Scholar] [CrossRef]
- Daft, J.G.; Ptacek, T.; Kumar, R.; Morrow, C.; Lorenz, R.G. Cross-fostering immediately after birth induces a permanent microbiota shift that is shaped by the nursing mother. Microbiome 2015, 3, 17. [Google Scholar] [CrossRef]
- Wen, L.; Ley, R.E.; Volchkov, P.Y.; Stranges, P.B.; Avanesyan, L.; Stonebraker, A.C.; Hu, C.; Wong, F.S.; Szot, G.L.; Bluestone, J.A.; et al. Innate immunity and intestinal microbiota in the development of Type 1 diabetes. Nature 2008, 455, 1109. [Google Scholar] [CrossRef]
- Miranda, M.C.G.; Oliveira, R.P.; Torres, L.; Aguiar, S.L.F.; Pinheiro Rosa, N.; Lemos, L.; Guimarães, M.A.; Reis, D.; Silveira, T.; Ferreira, Ê.; et al. Frontline Science: Abnormalities in the gut mucosa of non-obese diabetic mice precede the onset of type 1 diabetes. J. Leukoc. Biol. 2019, 106, 513–529. [Google Scholar] [CrossRef]
- Caviglia, G.P.; Rosso, C.; Ribaldone, D.G.; Dughera, F.; Fagoonee, S.; Astegiano, M.; Pellicano, R. Physiopathology of intestinal barrier and the role of zonulin. Minerva Biotecnol. 2019, 31, 83–92. [Google Scholar] [CrossRef]
- Neu, J.; Reverte, C.M.; Mackey, A.D.; Liboni, K.; Tuhacek-Tenace, L.M.; Hatch, M.; Li, N.; Caicedo, R.A.; Schatz, D.A.; Atkinson, M. Changes in intestinal morphology and permeability in the biobreeding rat before the onset of type 1 diabetes. J. Pediatric Gastroenterol. Nutr. 2005, 40, 589–595. [Google Scholar] [CrossRef]
- Vaarala, O.; Atkinson, M.A.; Neu, J. The “perfect storm” for type 1 diabetes: The complex interplay between intestinal microbiota, gut permeability, and mucosal immunity. Diabetes 2008, 57, 2555–2562. [Google Scholar] [CrossRef] [PubMed]
- Lee, A.S.; Gibson, D.L.; Zhang, Y.; Sham, H.P.; Vallance, B.A.; Dutz, J.P. Gut barrier disruption by an enteric bacterial pathogen accelerates insulitis in NOD mice. Diabetologia 2010, 53, 741–748. [Google Scholar] [CrossRef] [PubMed]
- Turley, S.J.; Lee, J.W.; Dutton-Swain, N.; Mathis, D.; Benoist, C. Endocrine self and gut non-self intersect in the pancreatic lymph nodes. Proc. Natl. Acad. Sci. USA 2005, 102, 17729–17733. [Google Scholar] [CrossRef] [PubMed]
- Sorini, C.; Cosorich, I.; Conte, M.L.; De Giorgi, L.; Facciotti, F.; Lucianò, R.; Rocchi, M.; Ferrarese, R.; Sanvito, F.; Canducci, F.; et al. Loss of gut barrier integrity triggers activation of islet-reactive T cells and autoimmune diabetes. Proc. Natl. Acad. Sci. USA 2019, 116, 15140–15149. [Google Scholar] [CrossRef] [PubMed]
- Calcinaro, F.; Dionisi, S.; Marinaro, M.; Candeloro, P.; Bonato, V.; Marzotti, S.; Corneli, R.B.; Ferretti, E.; Gulino, A.; Grasso, F.; et al. Oral probiotic administration induces interleukin-10 production and prevents spontaneous autoimmune diabetes in the non-obese diabetic mouse. Diabetologia 2005, 48, 1565–1575. [Google Scholar] [CrossRef] [PubMed]
- Dolpady, J.; Sorini, C.; Di Pietro, C.; Cosorich, I.; Ferrarese, R.; Saita, D.; Clementi, M.; Canducci, F.; Falcone, M. Oral probiotic VSL# 3 prevents autoimmune diabetes by modulating microbiota and promoting indoleamine 2, 3-dioxygenase-enriched tolerogenic intestinal environment. J. Diabetes Res. 2016, 2016, 7569431. [Google Scholar] [CrossRef] [PubMed]
- Valladares, R.; Sankar, D.; Li, N.; Williams, E.; Lai, K.K.; Abdelgeliel, A.S.; Gonzalez, C.F.; Wasserfall, C.H.; Larkin, J.; Schatz, D.; et al. Lactobacillus johnsonii N6. 2 mitigates the development of type 1 diabetes in BB-DP rats. PLoS ONE 2010, 5, e10507. [Google Scholar] [CrossRef]
- Chen, K.; Chen, H.; Faas, M.M.; de Haan, B.J.; Li, J.; Xiao, P.; Zhang, H.; Diana, J.; de Vos, P.; Sun, J. Specific inulin-type fructan fibers protect against autoimmune diabetes by modulating gut immunity, barrier function, and microbiota homeostasis. Mol. Nutr. Food Res. 2017, 61, 1601006. [Google Scholar] [CrossRef]
- Hansen, C.H.F.; Krych, L.; Nielsen, D.S.; Vogensen, F.K.; Hansen, L.H.; Sørensen, S.J.; Buschard, K.; Hansen, A.K. Early life treatment with vancomycin propagates Akkermansia muciniphila and reduces diabetes incidence in the NOD mouse. Diabetologia 2012, 55, 2285–2294. [Google Scholar] [CrossRef]
- Brugman, S.; Klatter, F.A.; Visser, J.T.J.; Wildeboer-Veloo, A.C.M.; Harmsen, H.J.M.; Rozing, J.; Bos, N.A. Antibiotic treatment partially protects against type 1 diabetes in the Bio-Breeding diabetes-prone rat. Is the gut flora involved in the development of type 1 diabetes? Diabetologia 2006, 49, 2105–2108. [Google Scholar] [CrossRef]
- Hu, Y.; Jin, P.; Peng, J.; Zhang, X.; Wong, F.S.; Wen, L. Different immunological responses to early-life antibiotic exposure affecting autoimmune diabetes development in NOD mice. J. Autoimmun. 2016, 72, 47–56. [Google Scholar] [CrossRef] [PubMed]
- Livanos, A.E.; Greiner, T.U.; Vangay, P.; Pathmasiri, W.; Stewart, D.; McRitchie, S.; Li, H.; Chung, J.; Sohn, J.; Kim, S.; et al. Antibiotic-Mediated gut microbiome perturbation accelerates development of type 1 diabetes in mice. Nat. Microbiol. 2016, 1, 16140. [Google Scholar] [CrossRef] [PubMed]
- Hara, N.; Alkanani, A.K.; Ir, D.; Robertson, C.E.; Wagner, B.D.; Frank, D.N.; Zipris, D. Prevention of virus-induced type 1 diabetes with antibiotic therapy. J. Immunol. 2012, 189, 3805–3814. [Google Scholar] [CrossRef] [PubMed]
- Mariño, E.; Richards, J.L.; McLeod, K.H.; Stanley, D.; Yap, Y.A.; Knight, J.; McKenzie, C.; Kranich, J.; Oliveira, A.C.; Rossello, F.J.; et al. Gut microbial metabolites limit the frequency of autoimmune T cells and protect against type 1 diabetes. Nat. Immunol. 2017, 18, 552. [Google Scholar] [CrossRef] [PubMed]
- Needell, J.C.; Ir, D.; Robertson, C.E.; Kroehl, M.E.; Frank, D.N.; Zipris, D. Maternal treatment with short-chain fatty acids modulates the intestinal microbiota and immunity and ameliorates type 1 diabetes in the offspring. PLoS ONE 2017, 12, e0183786. [Google Scholar] [CrossRef] [PubMed]
- Candon, S.; Perez-Arroyo, A.; Marquet, C.; Valette, F.; Foray, A.P.; Pelletier, B.; Milani, C.; Ventura, M.; Bach, J.F.; Chatenoud, L. Antibiotics in early life alter the gut microbiome and increase disease incidence in a spontaneous mouse model of autoimmune insulin-dependent diabetes. PLoS ONE 2015, 10, e0125448. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.S.; Li, J.; Krautkramer, K.A.; Badri, M.; Battaglia, T.; Borbet, T.C.; Koh, H.; Ng, S.; Sibley, R.A.; Li, Y.; et al. Antibiotic-induced acceleration of type 1 diabetes alters maturation of innate intestinal immunity. Elife 2018, 7, e37816. [Google Scholar] [CrossRef]
- Wu, C.; Pan, L.L.; Niu, W.; Fang, X.; Liang, W.; Li, J.; Li, H.; Pan, X.; Chen, W.; Zhang, H.; et al. Modulation of gut microbiota by low methoxyl pectin attenuates type 1 diabetes in non-obese diabetic mice. Front. Immunol. 2019, 10, 1733. [Google Scholar] [CrossRef]
- Jia, L.; Shan, K.; Pan, L.L.; Feng, N.; Lv, Z.; Sun, Y.; Li, J.; Wu, C.; Zhang, H.; Chen, W.; et al. Clostridium butyricum CGMCC0313.1 protects against autoimmune diabetes by modulating intestinal immune homeostasis and inducing pancreatic regulatory T cells. Front. Immunol. 2017, 8, 1345. [Google Scholar] [CrossRef]
- Kostic, A.D.; Gevers, D.; Siljander, H.; Vatanen, T.; Hyötyläinen, T.; Hämäläinen, A.M.; Peet, A.; Tillmann, V.; Pöhö, P.; Mattila, I.; et al. The dynamics of the human infant gut microbiome in development and in progression toward type 1 diabetes. Cell Host Microbe 2015, 17, 260–273. [Google Scholar] [CrossRef]
- De Goffau, M.C.; Luopajärvi, K.; Knip, M.; Ilonen, J.; Ruohtula, T.; Härkönen, T.; Orivuori, L.; Hakala, S.; Welling, G.W.; Harmsen, H.J.; et al. Fecal microbiota composition differs between children with β-cell autoimmunity and those without. Diabetes 2012, 62, 1238–1244. [Google Scholar] [CrossRef] [PubMed]
- Giongo, A.; Gano, K.A.; Crabb, D.B.; Mukherjee, N.; Novelo, L.L.; Casella, G.; Drew, J.C.; Ilonen, J.; Knip, M.; Hyöty, H.; et al. Toward defining the autoimmune microbiome for type 1 diabetes. ISME J. 2011, 5, 82. [Google Scholar] [CrossRef] [PubMed]
- De Goffau, M.C.; Fuentes, S.; van den Bogert, B.; Honkanen, H.; de Vos, W.M.; Welling, G.W.; Hyöty, H.; Harmsen, H.J. Aberrant gut microbiota composition at the onset of type 1 diabetes in young children. Diabetologia 2014, 57, 1569–1577. [Google Scholar] [CrossRef] [PubMed]
- Murri, M.; Leiva, I.; Gomez-Zumaquero, J.M.; Tinahones, F.J.; Cardona, F.; Soriguer, F.; Queipo-Ortuño, M.I. Gut microbiota in children with type 1 diabetes differs from that in healthy children: A case-control study. BMC Med. 2013, 11, 46. [Google Scholar] [CrossRef] [PubMed]
- Davis-Richardson, A.G.; Ardissone, A.N.; Dias, R.; Simell, V.; Leonard, M.T.; Kemppainen, K.M.; Drew, J.C.; Schatz, D.; Atkinson, M.A.; Kolaczkowski, B.; et al. Bacteroides dorei dominates gut microbiome prior to autoimmunity in Finnish children at high risk for type 1 diabetes. Front. Microbiol. 2014, 5, 678. [Google Scholar] [CrossRef]
- Vatanen, T.; Franzosa, E.A.; Schwager, R.; Tripathi, S.; Arthur, T.D.; Vehik, K.; Lernmark, Å.; Hagopian, W.A.; Rewers, M.J.; She, J.X.; et al. The human gut microbiome in early-onset type 1 diabetes from the TEDDY study. Nature 2018, 562, 589. [Google Scholar] [CrossRef]
- Endesfelder, D.; Engel, M.; Davis-Richardson, A.G.; Ardissone, A.N.; Achenbach, P.; Hummel, S.; Winkler, C.; Atkinson, M.; Schatz, D.; Triplett, E.; et al. Towards a functional hypothesis relating anti-islet cell autoimmunity to the dietary impact on microbial communities and butyrate production. Microbiome 2016, 4, 17. [Google Scholar] [CrossRef]
- Gavin, P.G.; Mullaney, J.A.; Loo, D.; Lê Cao, K.A.; Gottlieb, P.A.; Hill, M.M.; Zipris, D.; Hamilton-Williams, E.E. Intestinal metaproteomics reveals host-microbiota interactions in subjects at risk for type 1 diabetes. Diabetes Care 2018, 41, 2178–2186. [Google Scholar] [CrossRef]
- Russell, J.T.; Roesch, L.F.; Ördberg, M.; Ilonen, J.; Atkinson, M.A.; Schatz, D.A.; Triplett, E.W.; Ludvigsson, J. Genetic risk for autoimmunity is associated with distinct changes in the human gut microbiome. Nat. Commun. 2019, 10, 1–12. [Google Scholar] [CrossRef]
- Ho, J.; Nicolucci, A.C.; Virtanen, H.; Schick, A.; Meddings, J.; Reimer, R.A.; Huang, C. Effect of prebiotic on microbiota, intestinal permeability and glycemic control in children with type 1 diabetes. J. Clin. Endocrinol. Metab. 2019, 104, 4427–4440. [Google Scholar] [CrossRef]
- Uusitalo, U.; Liu, X.; Yang, J.; Aronsson, C.A.; Hummel, S.; Butterworth, M.; Lernmark, Å.; Rewers, M.; Hagopian, W.; She, J.X.; et al. Association of early exposure of probiotics and islet autoimmunity in the TEDDY study. JAMA Pediatric 2016, 170, 20–28. [Google Scholar] [CrossRef] [PubMed]
- Canani, R.B.; Di Costanzo, M.; Leone, L.; Pedata, M.; Meli, R.; Calignano, A. Potential beneficial effects of butyrate in intestinal and extraintestinal diseases. World J. Gastroenterol. 2011, 17, 1519. [Google Scholar] [CrossRef] [PubMed]
- Vatanen, T.; Kostic, A.D.; d’Hennezel, E.; Siljander, H.; Franzosa, E.A.; Yassour, M.; Kolde, R.; Vlamakis, H.; Arthur, T.D.; Hämäläinen, A.M.; et al. Variation in microbiome LPS immunogenicity contributes to autoimmunity in humans. Cell 2016, 165, 842–853. [Google Scholar] [CrossRef] [PubMed]
- Maffeis, C.; Martina, A.; Corradi, M.; Quarella, S.; Nori, N.; Torriani, S.; Plebani, M.; Contreas, G.; Felis, G.E. Association between intestinal permeability and faecal microbiota composition in Italian children with beta cell autoimmunity at risk for type 1 diabetes. Diabetes Metab. Res. Rev. 2016, 32, 700–709. [Google Scholar] [CrossRef] [PubMed]
- Harbison, J.E.; Roth-Schulze, A.J.; Giles, L.C.; Tran, C.D.; Ngui, K.M.; Penno, M.A.; Thomson, R.L.; Wentworth, J.M.; Colman, P.G.; Craig, M.E.; et al. Gut microbiome dysbiosis and increased intestinal permeability in children with islet autoimmunity and type 1 diabetes: A prospective cohort study. Pediatric Diabetes 2019, 20, 574–583. [Google Scholar] [CrossRef]
- Dominguez-Bello, M.G.; De Jesus-Laboy, K.M.; Shen, N.; Cox, L.M.; Amir, A.; Gonzalez, A.; Bokulich, N.A.; Song, S.J.; Hoashi, M.; Rivera-Vinas, J.I.; et al. Partial restoration of the microbiota of cesarean-born infants via vaginal microbial transfer. Nat. Med. 2016, 22, 250. [Google Scholar] [CrossRef]
- Mishra, S.; Wang, S.; Nagpal, R.; Miller, B.; Singh, R.; Taraphder, S.; Yadav, H. Probiotics and prebiotics for the amelioration of type 1 diabetes: Present and future perspectives. Microorganisms 2019, 7, 67. [Google Scholar] [CrossRef]
- Groele, L.; Szajewska, H.; Szypowska, A. Effects of Lactobacillus rhamnosus GG and Bifidobacterium lactis Bb12 on beta-cell function in children with newly diagnosed type 1 diabetes: Protocol of a randomised controlled trial. BMJ Open 2017, 7, e017178. [Google Scholar] [CrossRef]
| Study Design | Study Sample | Main Findings | Ref. |
|---|---|---|---|
| Prospective | In total, 33 genetically predisposed infants | Microbial diversity in T1DM progressors in the time window between seroconversion and T1DM onset and inflammation-favoring organisms | [71] |
| Case Control | In total, 18 Ab (+) vs. 18 Ab (-) children matched for HLA | Ab-positive children: microbial diversity, Bacteroidetes (Bacteroides and Prevotella), Bifidobacterium species, short-chain fatty acid (SCFA)-producing bacteria | [72] |
| Case Control (longitudinal data) | In total, four HLA-matched case (Ab+) –control (Ab-) pairs (three time points) | Bacteroidetes (Bacteroidales) and Firmicutes (Clostriales) in cases vs. controls at all time points. Children progressing T1DM: microbial diversity | [73] |
| Case Control | In total, 28 newly diagnosed type 1 diabetes (T1DM) (average duration 4.8 years) vs. 27 age-matched control children | In children < 3 years: Bacteroidetes in cases and Clostridium clusters IV and XIVa in controls | [74] |
| Case Control | In total, 16 T1DM vs. 16 healthy children | Cases: Bacteroidetes, Firmicutes and Actinobacteria | [75] |
| Case Control (longitudinal data) | In total, 29 Ab (+) cases vs. 47 Ab (-) healthy controls | B. dorei and B. vulgatus in cases prior to seroconversion | [76] |
| Prospective | In total, 783 genetically predisposed children | SCFA-producing bacteria genes in children who seroconverted or developed T1DM | [77] |
| Prospective | In total, 19 Ab (+) and 21 Ab (-) children | Bacteroides Akkermansia SCFA-producing bacteria genes. A functional association between diet (early introduction of non-milk diet), gut microbiome (Bacteroides), metagenomic changes (genes for the production of butyrate) and development of islet Ab | [78] |
| Case Control | In total, 33 recent-onset T1DM, 17 Ab (+), 29 Ab (-), and 22 healthy subjects | T1DM: microbial taxa associated with host proteins involved in maintaining the function of the mucous barrier, microvilli adhesion, and exocrine pancreas | [79] |
| Cohort Study | In total, 403 children (age = 1 year) | Genetic risk for developing T1DM autoimmunity is associated with distinct changes in the gut microbiome | [80] |
| RCT | T1DM children (8–17 years) randomized to prebiotic oligofructose-enriched inulin (n = 17) or placebo (n = 21) for 12 weeks | At 3 months, C-peptide was significantly higher in the group that received prebiotics | [81] |
| Prospective Cohort | In total, 7473 children (4–10 years) | Early probiotic supplementation (at the age of 0–27 days) associated with a decreased risk of islet autoimmunity in children with the HLADR3/4 genotype | [82] |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Durazzo, M.; Ferro, A.; Gruden, G. Gastrointestinal Microbiota and Type 1 Diabetes Mellitus: The State of Art. J. Clin. Med. 2019, 8, 1843. https://doi.org/10.3390/jcm8111843
Durazzo M, Ferro A, Gruden G. Gastrointestinal Microbiota and Type 1 Diabetes Mellitus: The State of Art. Journal of Clinical Medicine. 2019; 8(11):1843. https://doi.org/10.3390/jcm8111843
Chicago/Turabian StyleDurazzo, Marilena, Arianna Ferro, and Gabriella Gruden. 2019. "Gastrointestinal Microbiota and Type 1 Diabetes Mellitus: The State of Art" Journal of Clinical Medicine 8, no. 11: 1843. https://doi.org/10.3390/jcm8111843
APA StyleDurazzo, M., Ferro, A., & Gruden, G. (2019). Gastrointestinal Microbiota and Type 1 Diabetes Mellitus: The State of Art. Journal of Clinical Medicine, 8(11), 1843. https://doi.org/10.3390/jcm8111843





