An Association of Gut Microbiota with Different Phenotypes in Chinese Patients with Rheumatoid Arthritis
Abstract
:1. Introduction
2. Experimental Section
2.1. Patients
2.2. Samples Collection and Bacterial DNA Extraction
2.3. 16S Ribosomal RNA (16S rRNA)-Based Deep Sequencing and Compositional Analysis
2.4. Determination of Serum Levels of Proinflammatory Cytokines, RF-IgM, and ACPA
2.5. Statistical Analysis
3. Results
3.1. Clinical Characteristics of 128 RA Patients and Demographic Data of HC
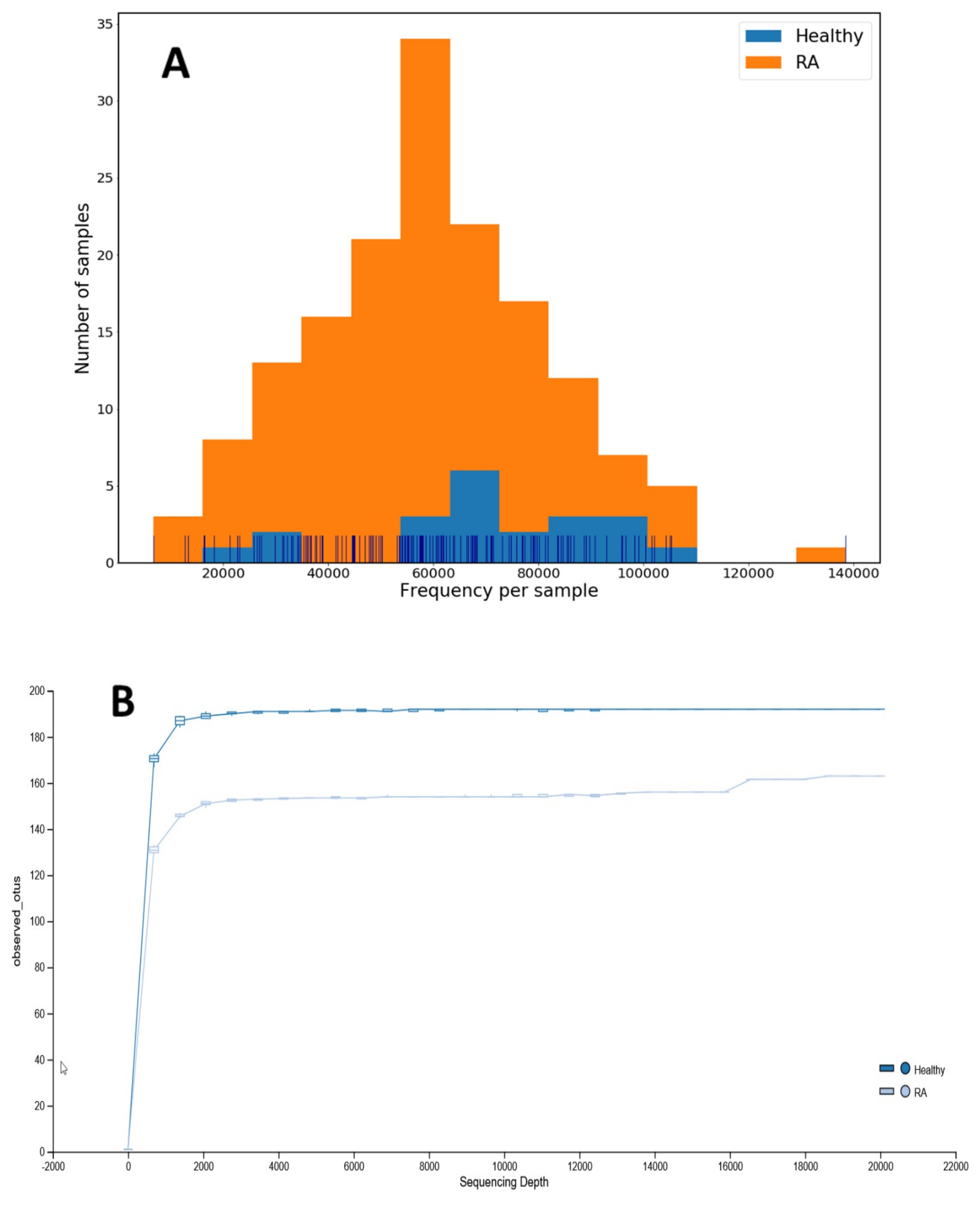
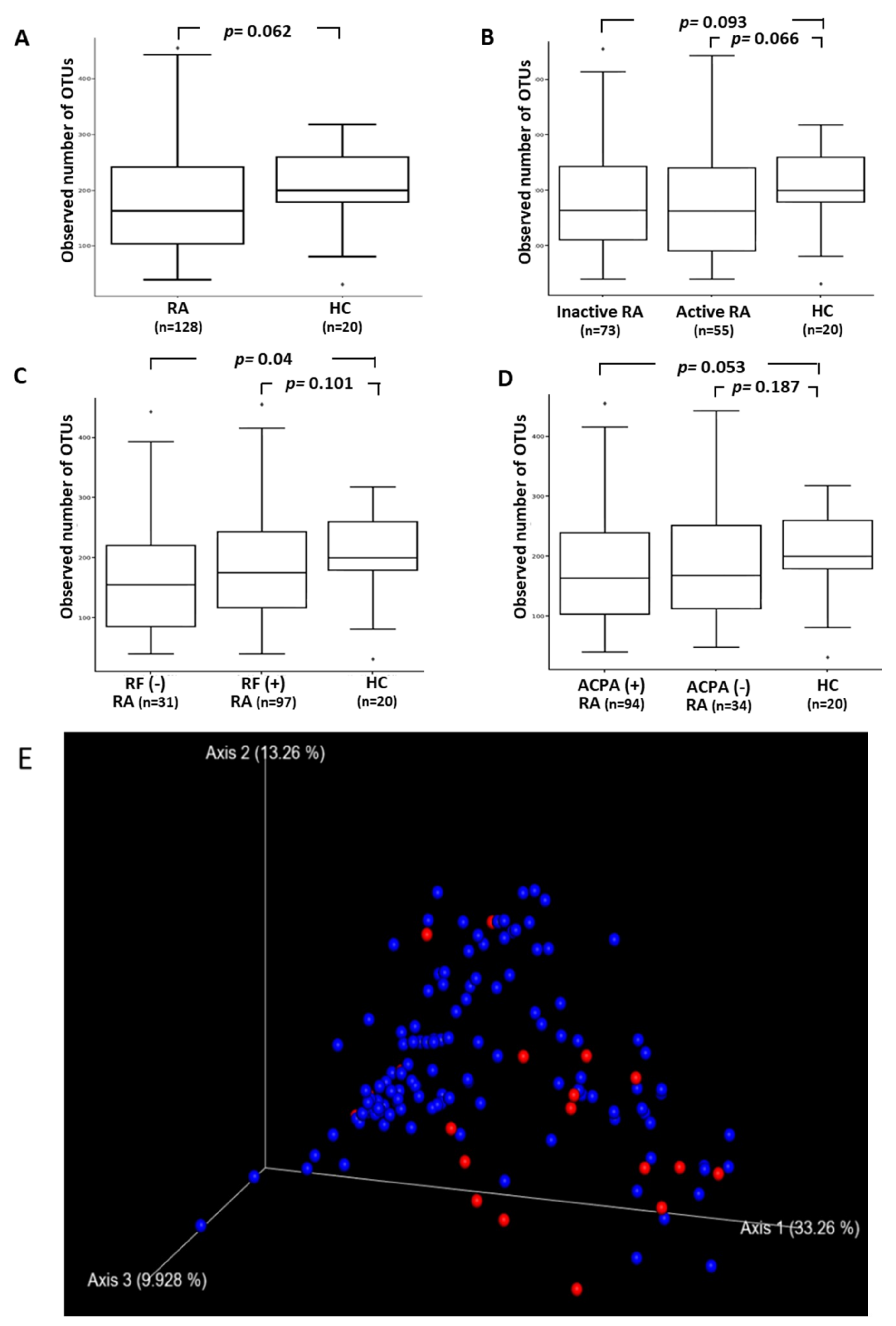
3.2. The Difference in the Diversity of Gut Microbiota within and among Groups
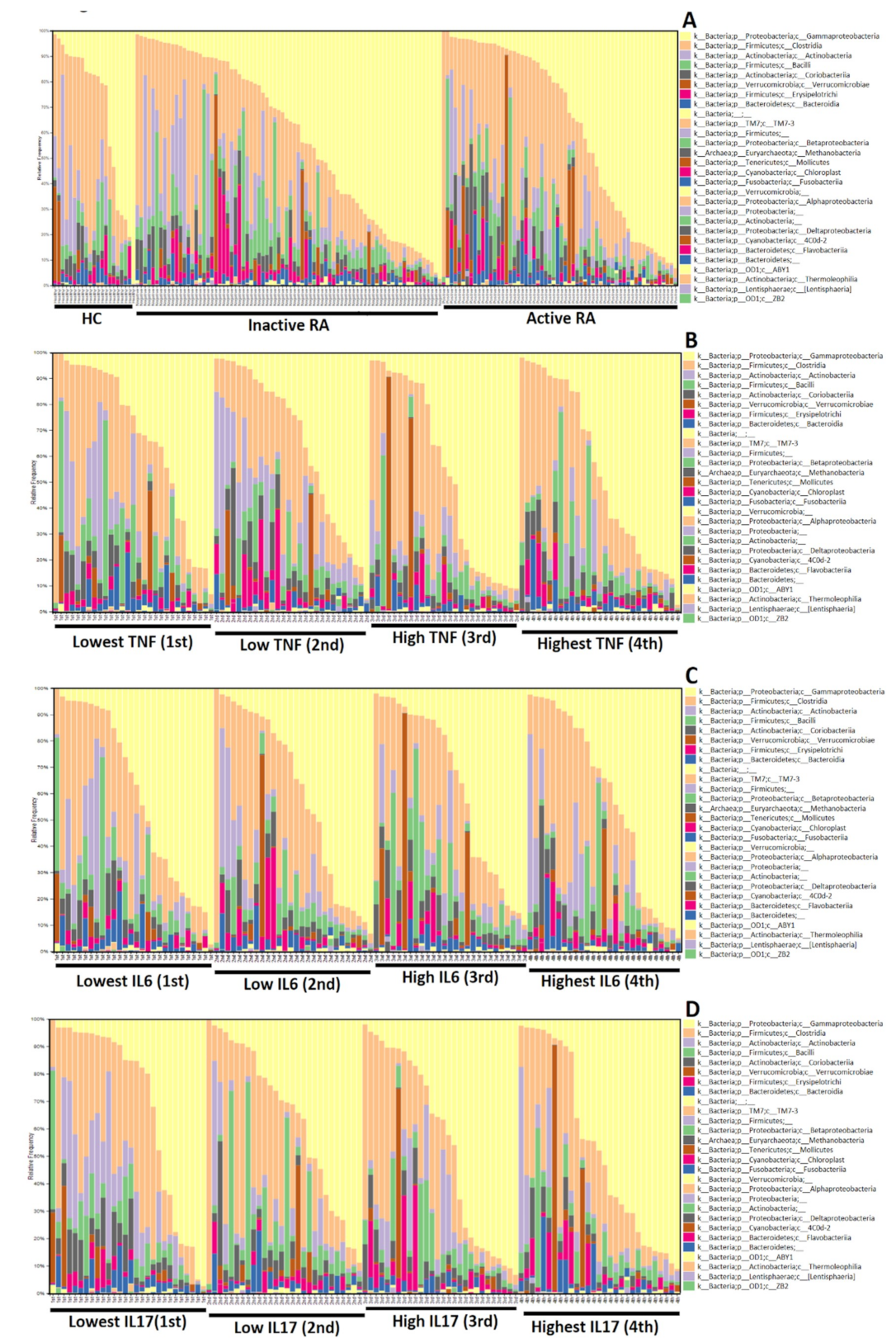
3.3. The Difference in Microbiota Composition
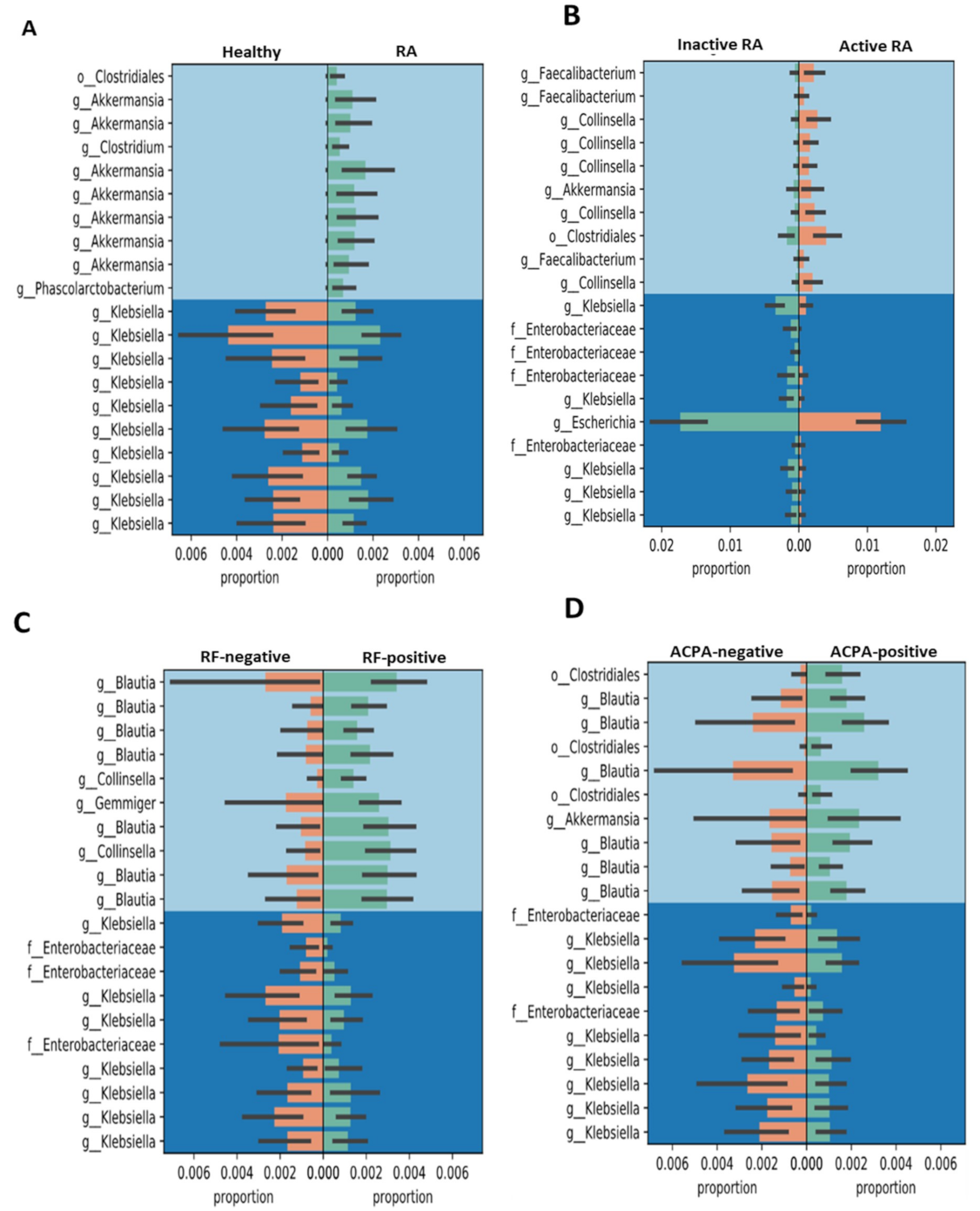
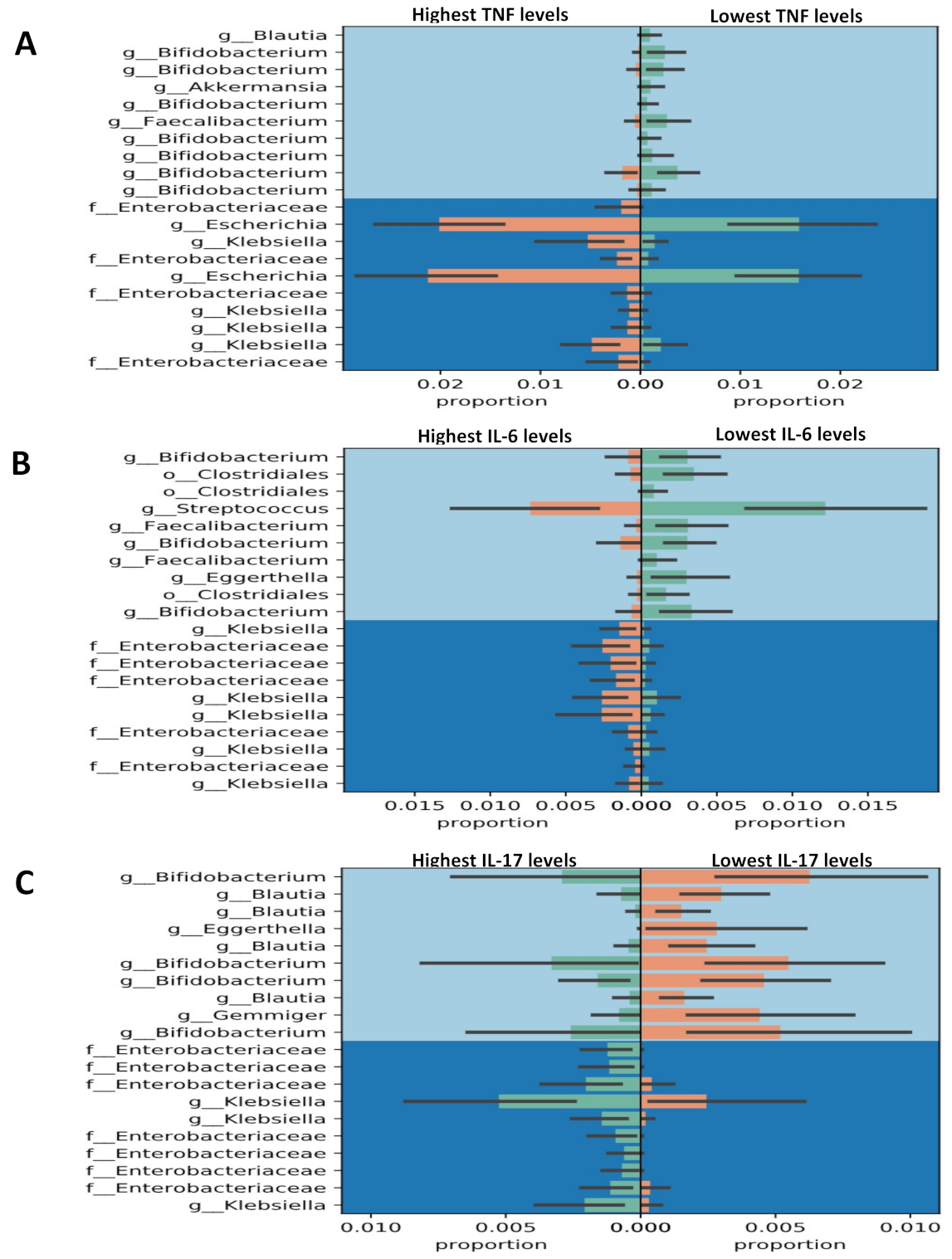
3.4. Gneiss Differential Abundance Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Arumugam, M.; Raes, J.; Pelletier, E.; Le Paslier, D.; Yamada, T.; Mende, D.R.; Fernandes, G.R.; Tap, J.; Bruls, T.; Batto, J.-M.; et al. Enterotypes of the human gut microbiome. Nature 2011, 473, 174–180. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.K.; Mazmanian, S.K. Has the Microbiota Played a Critical Role in the Evolution of the Adaptive Immune System? Science 2010, 330, 1768–1773. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Round, J.L.; O’Connell, R.M.; Mazmanian, S.K. Coordination of tolerogenic immune responses by the commensal microbiota. J. Autoimmun. 2010, 34, 220–225. [Google Scholar] [CrossRef] [PubMed]
- López, P.; De Paz, B.; Rodríguez-Carrio, J.; Hevia, A.; Sánchez, B.; Margolles, A.; Suárez, A. Th17 responses and natural IgM antibodies are related to gut microbiota composition in systemic lupus erythematosus patients. Sci. Rep. 2016, 6, 24072–24083. [Google Scholar] [CrossRef]
- Atarashi, K.; Tanoue, T.; Shima, T.; Imaoka, A.; Kuwahara, T.; Momose, Y.; Cheng, G.; Yamasaki, S.; Saito, T.; Ohba, Y. Induction of colonic regulatory T cells by indigenous Clostridium species. Science 2011, 331, 337–341. [Google Scholar] [CrossRef]
- Guerreiro, C.S.G.; Calado, Â.C.A.; Sousa, J.S.J.; Fonseca, J.E.F.J. Diet, microbiota and gut permeability—The unknown triad in rheumatoid arthritis. Front. Med. 2018, 5, 349–356. [Google Scholar] [CrossRef]
- Dobber, R.; Hertogh-Huijbregts, A.; Rozing, J.; Bottomly, K.; Nagelkerken, L. The Involvement of the Intestinal Microflora in the Expansion of CD4. Clin. Dev. Immunol. 1992, 2, 141–150. [Google Scholar] [CrossRef]
- Koboziev, I.; Webb, C.R.; Furr, K.L.; Grisham, M.B. Role of the enteric microbiota in intestinal homeostasis and inflammation. Free Radic. Biol. Med. 2014, 68, 122–133. [Google Scholar] [CrossRef]
- Horta-Baas, G.; Romero-Figueroa, M.d.S.; Montiel-Jarquín, A.J.; Pizano-Zárate, M.L.; García-Mena, J.; Ramírez-Durán, N. Intestinal dysbiosis and rheumatoid arthritis: A link between gut microbiota and the pathogenesis of rheumatoid arthritis. J. Immunol. Res. 2017, 2017, 1–13. [Google Scholar] [CrossRef]
- Chen, J.; Wright, K.; Davis, J.M.; Jeraldo, P.; Marietta, E.V.; Murray, J.; Nelson, H.; Matteson, E.L.; Taneja, V. An expansion of rare lineage intestinal microbes characterizes rheumatoid arthritis. Genome Med. 2016, 8, 43–56. [Google Scholar] [CrossRef]
- Zhang, X.; Zhang, D.; Jia, H.; Feng, Q.; Wang, D.; Liang, D.; Wu, X.; Li, J.; Tang, L.; Li, Y. The oral and gut microbiomes are perturbed in rheumatoid arthritis and partly normalized after treatment. Nat. Med. 2015, 21, 895–905. [Google Scholar] [CrossRef] [PubMed]
- Maeda, Y.; Kurakawa, T.; Umemoto, E.; Motooka, D.; Ito, Y.; Gotoh, K.; Hirota, K.; Matsushita, M.; Furuta, Y.; Narazaki, M. Dysbiosis contributes to arthritis development via activation of autoreactive T cells in the intestine. Arthr. Rheumatol. 2016, 68, 2646–2661. [Google Scholar] [CrossRef] [PubMed]
- Maeda, Y.; Takeda, K. Role of gut microbiota in rheumatoid arthritis. J. Clin. Med. 2017, 6, 60–66. [Google Scholar] [CrossRef] [PubMed]
- Furst, D.E.; Emery, P. Rheumatoid arthritis pathophysiology: Update on emerging cytokine and cytokine-associated cell targets. Rheumatology 2014, 53, 1560–1569. [Google Scholar] [CrossRef] [PubMed]
- Scher, J.U.; Abramson, S.B. The microbiome and rheumatoid arthritis. Nat. Rev. Rheumatol. 2011, 7, 569–578. [Google Scholar] [CrossRef]
- Picchianti-Diamanti, A.; Rosado, M.M.; D’Amelio, R. Infectious agents and inflammation: The role of microbiota in autoimmune arthritis. Front. Microbiol. 2018, 8, 2696–2704. [Google Scholar] [CrossRef]
- Caporaso, J.G.; Lauber, C.L.; Walters, W.A.; Berg-Lyons, D.; Lozupone, C.A.; Turnbaugh, P.J.; Fierer, N.; Knight, R. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc. Natl. Acad. Sci. USA 2011, 108, 4516–4522. [Google Scholar] [CrossRef]
- Nishimura, K.; Sugiyama, D.; Kogata, Y.; Tsuji, G.; Nakazawa, T.; Kawano, S.; Saigo, K.; Morinobu, A.; Koshiba, M.; Kuntz, K.M. Meta-analysis: Diagnostic accuracy of anti–cyclic citrullinated peptide antibody and rheumatoid factor for rheumatoid arthritis. Ann. Intern. Med. 2007, 146, 797–808. [Google Scholar] [CrossRef]
- Elshafie, A.I.; Elbagir, S.; Aledrissy, M.I.; Elagib, E.M.; Nur, M.A.; Rönnelid, J. Occurrence of anti-CCP2 and RF isotypes and their relation to age and disease severity among Sudanese patients with rheumatoid arthritis. Clin. Rheumatol. 2019, 38, 1545–1553. [Google Scholar] [CrossRef] [Green Version]
- Aletaha, D.; Neogi, T.; Silman, A.J.; Funovits, J.; Felson, D.T.; Bingham III, C.O.; Birnbaum, N.S.; Burmester, G.R.; Bykerk, V.P.; Cohen, M.D. 2010 rheumatoid arthritis classification criteria: An American College of Rheumatology/European League Against Rheumatism collaborative initiative. Arthritis Rheum. 2010, 62, 2569–2581. [Google Scholar] [CrossRef]
- Prevoo, M.; Van’T Hof, M.A.; Kuper, H.; Van Leeuwen, M.; Van De Putte, L.; Van Riel, P. Modified disease activity scores that include twenty-eight-joint counts development and validation in a prospective longitudinal study of patients with rheumatoid arthritis. Arthritis Rheum. Off. J. Am. Coll. Rheumatol. 1995, 38, 44–48. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Smolen, J.S.; Aletaha, D.; Bijlsma, J.W.; Breedveld, F.C.; Boumpas, D.; Burmester, G.; Combe, B.; Cutolo, M.; De Wit, M.; Dougados, M. Treating rheumatoid arthritis to target: Recommendations of an international task force. Ann. Rheum. Dis. 2010, 69, 631–637. [Google Scholar] [CrossRef] [PubMed]
- Nossa, C.W.; Oberdorf, W.E.; Yang, L.; Aas, J.A.; Paster, B.J.; DeSantis, T.Z.; Brodie, E.L.; Malamud, D.; Poles, M.A.; Pei, Z. Design of 16S rRNA gene primers for 454 pyrosequencing of the human foregut microbiome. World J. Gastroenterol. 2010, 16, 4135–4144. [Google Scholar] [CrossRef] [PubMed]
- Andrews, S. FastQC: A Quality Control Tool for High Throughput Sequence Data; Babraham Bioinformatics, Babraham Institute: Cambridge, UK, 2010. [Google Scholar]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Zhang, J.; Kobert, K.; Flouri, T.; Stamatakis, A. PEAR: A fast and accurate Illumina Paired-End reAd mergeR. Bioinformatics 2013, 30, 614–620. [Google Scholar] [CrossRef]
- Wang, Q.; Garrity, G.M.; Tiedje, J.M.; Cole, J.R. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 2007, 73, 5261–5267. [Google Scholar] [CrossRef]
- DeSantis, T.Z.; Hugenholtz, P.; Larsen, N.; Rojas, M.; Brodie, E.L.; Keller, K.; Huber, T.; Dalevi, D.; Hu, P.; Andersen, G.L. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl. Environ. Microbiol. 2006, 72, 5069–5072. [Google Scholar] [CrossRef]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Pena, A.G.; Goodrich, J.K.; Gordon, J.I. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef] [Green Version]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef] [Green Version]
- Pedregosa, F.; Varoquaux, G.; Gramfort, A.; Michel, V.; Thirion, B.; Grisel, O.; Blondel, M.; Prettenhofer, P.; Weiss, R.; Dubourg, V. Scikit-learn: Machine learning in Python. J. Mach. Learn. Res. 2011, 12, 2825–2830. [Google Scholar]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef] [PubMed]
- Morton, J.T.; Sanders, J.; Quinn, R.A.; McDonald, D.; Gonzalez, A.; Vázquez-Baeza, Y.; Navas-Molina, J.A.; Song, S.J.; Metcalf, J.L.; Hyde, E.R.; et al. Balance Trees Reveal Microbial Niche Differentiation. MSystems 2017, 2, 16–26. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef] [PubMed]
- Price, M.N.; Dehal, P.S.; Arkin, A.P. FastTree 2—Approximately maximum-likelihood trees for large alignments. PLoS ONE 2010, 5, e9490. [Google Scholar] [CrossRef] [PubMed]
- Lozupone, C.; Knight, R. UniFrac: A new phylogenetic method for comparing microbial communities. Appl. Environ. Microbiol. 2005, 71, 8228–8235. [Google Scholar] [CrossRef]
- Vázquez-Baeza, Y.; Pirrung, M.; Gonzalez, A.; Knight, R. EMPeror: A tool for visualizing high-throughput microbial community data. Gigascience 2013, 2, 16–19. [Google Scholar] [CrossRef]
- Chen, J.; Bittinger, K.; Charlson, E.S.; Hoffmann, C.; Lewis, J.; Wu, G.D.; Collman, R.G.; Bushman, F.D.; Li, H. Associating microbiome composition with environmental covariates using generalized UniFrac distances. Bioinformatics 2012, 28, 2106–2113. [Google Scholar] [CrossRef]
- Chang, Q.; Luan, Y.; Sun, F. Variance adjusted weighted UniFrac: A powerful beta diversity measure for comparing communities based on phylogeny. BMC Bioinform. 2011, 12, 118–131. [Google Scholar] [CrossRef]
- Anderson, M.J. A new method for non-parametric multivariate analysis of variance. Austral Ecol. 2001, 26, 32–46. [Google Scholar]
- Jeong, Y.; Kim, J.W.; You, H.J.; Park, S.J.; Lee, J.; Ju, J.H.; Park, M.S.; Jin, H.; Cho, M.L.; Kwon, B.; et al. Gut microbial composition and function are altered in patients with early rheumatoid arthritis. J. Clin. Med. 2019, 8, 693. [Google Scholar] [CrossRef]
- Eriksson, K.; Fei, G.; Lundmark, A.; Benchimol, D.; Lee, L.; Hu, Y.O.O.; Kats, A.; Saevarsdottir, S.; Catrina, A.I.; Klinge, B.; et al. Periodontal health and oral microbiota in patients with rheumatoid arthritis. J. Clin. Med. 2019, 8, 630. [Google Scholar] [CrossRef] [PubMed]
- Stoll, M.L.; Pierce, M.K.; Watkins, J.A.; Zhang, M.; Weiss, P.F.; Weiss, J.E.; Elson, C.O.; Cron, R.Q.; Kumar, R.; Morrow, C.D. Akkermansia muciniphila is permissive to arthritis in the K/BxN mouse model of arthritis. Genes Immun. 2018, 20, 158–166. [Google Scholar] [CrossRef] [PubMed]
- Derrien, M.; Van Baarlen, P.; Hooiveld, G.; Norin, E.; Muller, M.; de Vos, W. Modulation of mucosal immune response, tolerance, and proliferation in mice colonized by the mucin-degrader Akkermansia muciniphila. Front. Microbiol. 2011, 2, 166–179. [Google Scholar] [CrossRef] [PubMed]
- Ganesh, B.P.; Klopfleisch, R.; Loh, G.; Blaut, M. Commensal Akkermansia muciniphila exacerbates gut inflammation in Salmonella typhimurium-infected gnotobiotic mice. PLoS ONE 2013, 8, e74963. [Google Scholar] [CrossRef]
- Lucchino, B.; Spinelli, F.R.; Iannuccelli, C.; Guzzo, M.P.; Conti, F.; Franco, M.D. Mucosa-environment interactions in the pathogenesis of rheumatoid arthritis. Cells 2019, 8, 700. [Google Scholar] [CrossRef]
- Schirmer, M.; Smeekens, S.P.; Vlamakis, H.; Jaeger, M.; Oosting, M.; Franzosa, E.A.; Horst, R.; Jansen, T.; Jacobs, L.; Bonder, M.J.; et al. Linking the human gut microbiome to inflammatory cytokine production capacity. Cell 2016, 167, 1125–1136. [Google Scholar] [CrossRef]
- Liu, X.; Zeng, B.; Zhang, J.; Li, W.; Mou, F.; Wang, H.; Zou, Q.; Zhong, B.; Wu, L.; Wei, H.; et al. Role of the gut microbiome in modulating arthritis progression in mice. Sci. Rep. 2016, 6, 30594. [Google Scholar] [CrossRef]
- Picchianti-Diamanti, A.; Panebianco, C.; Salemi, S.; Sorgi, M.; Di Rosa, R.; Tropea, A.; Sgrulletti, M.; Salerno, G.; Terracciano, F.; D’amelio, R. Analysis of gut microbiota in rheumatoid arthritis patients: Disease-related Dysbiosis and modifications induced by Etanercept. Int. J. Mol. Sci. 2018, 19, 2938. [Google Scholar] [CrossRef]
- Ramos-Romero, S.; Hereu, M.; Atienza, L.; Casas, J.; Jáuregui, O.; Amézqueta, S.; Dasilva, G.; Medina, I.; Nogués, M.R.; Romeu, M. Mechanistically different effects of fat and sugar on insulin resistance, hypertension, and gut microbiota in rats. Am. J. Physiol. Endocrinol. Metab. 2018, 314, 552–563. [Google Scholar] [CrossRef]
- Pedersen, C.; Ijaz, U.Z.; Gallagher, E.; Horton, F.; Ellis, R.J.; Jaiyeola, E.; Duparc, T.; Russell-Jones, D.; Hinton, P.; Cani, P.D. Fecal Enterobacteriales enrichment is associated with increased in vivo intestinal permeability in humans. Physiol. Rep. 2018, 6, 13649–13665. [Google Scholar] [CrossRef]
- Vaahtovuo, J.; Munukka, E.; Korkeamäki, M.; Luukkainen, R.; Toivanen, P. Fecal microbiota in early rheumatoid arthritis. J. Rheumatol. 2008, 35, 1500–1505. [Google Scholar] [PubMed]
- Gul’neva, M.; Noskov, S. Colonic microbial biocenosis in rheumatoid arthritis. Klin. Med. 2011, 89, 45–48. [Google Scholar]
- Imaoka, A.; Shima, T.; Kato, K.; Mizuno, S.; Uehara, T.; Matsumoto, S.; Setoyama, H.; Hara, T.; Umesaki, Y. Anti-inflammatory activity of probiotic Bifidobacterium: Enhancement of IL-10 production in peripheral blood mononuclear cells from ulcerative colitis patients and inhibition of IL-8 secretion in HT-29 cells. World J. Gastroenterol. 2008, 14, 2511–2516. [Google Scholar] [CrossRef] [PubMed]
- Bjarnason, I.; So, A.; Levi, A.J.; Peters, T.; Williams, P.; Zanelli, G.; Gumpel, J.M.; Ansell, B. Intestinal permeability and inflammation in rheumatoid arthritis: Effects of non-steroidal anti-inflammatory drugs. Lancet 1984, 324, 1171–1174. [Google Scholar] [CrossRef]
- Muniz-Pedrogo, D.A.; Chen, J.; Hillmann, B.M.; Jeraldo, P.; Saffouri, G.; Al-Ghalith, G.A.; Friton, J.; Taneja, V.; Davis, J.M.; Knights, D. Mo1934-Gut Microbial Markers of Arthritis Including Inflammatory Bowel Disease Associated Arthropathy. Gastroenterology 2018, 154, S856. [Google Scholar] [CrossRef]
- De Filippo, C.; Cavalieri, D.; Di Paola, M.; Ramazzotti, M.; Poullet, J.B.; Massart, S.; Collini, S.; Pieraccini, G.; Lionetti, P. Impact of diet in shaping gut microbiota revealed by a comparative study in children from Europe and rural Africa. Proc. Natl. Acad. Sci. USA 2010, 107, 14691–14696. [Google Scholar] [CrossRef] [Green Version]
- Zamani, B.; Golkar, H.R.; Farshbaf, S.; Emadi-Baygi, M.; Tajabadi-Ebrahimi, M.; Jafari, P.; Akhavan, R.; Taghizadeh, M.; Memarzadeh, M.R.; Asemi, Z. Clinical and metabolic response to probiotic supplementation in patients with rheumatoid arthritis: A randomized, double-blind, placebo-controlled trial. Int. J. Rheum. Dis. 2016, 19, 869–879. [Google Scholar] [CrossRef]
- Sandhya, P.; Danda, D.; Sharma, D.; Scaria, V. Does the buck stop with the bugs?: An overview of microbial dysbiosis in rheumatoid arthritis. Int. J. Rheum. Dis. 2016, 19, 8–20. [Google Scholar] [CrossRef]
| RA Patients (n = 128) | Healthy Controls (n = 20) | |
|---|---|---|
| Mean age at entry of study, years | 55.5 ± 11.1 | 55.6 ± 9.8 |
| Female proportion | 102 (79.7%) | 16 (80.0%) |
| RF positivity at entry of study | 97 (75.8%) | NA |
| ACPA positivity at entry of study | 94 (73.4%) | NA |
| DAS-28 at entry of study | 3.38 ± 1.23 | NA |
| Concomitant drugs at entry of study | ||
| Corticosteroid, mg/day | 3.32 ± 2.31 | NA |
| Methotrexate | 94 (73.4%) | NA |
| Hydroxychloroquine | 40 (31.3%) | NA |
| Sulfasalazine | 38 (29.7%) | NA |
| Cyclosporine | 4 (3.1%) | NA |
| Biologics at entry of study | ||
| Etanercept | 17 (13.3%) | NA |
| Adalimumab | 34 (26.6%) | NA |
| Tocilizumab | 44 (34.3%) | NA |
| Rituximab | 11 (8.6%) | NA |
| Diabetes mellitus | 12 (9.4%) | 1 (5.0%) |
| Hypertension | 26 (20.4%) | 3 (15.0%) |
| Smoking | 11 (8.6%) | 2 (10.0%) |
| Alcohol consumption | 18 (14.1%) | 4 (20.0%) |
| Group Comparison | Pseudo-F | p-Value # | q-Value (FDR Adjusted p-Value) |
|---|---|---|---|
| HC vs. Total RA | 1.705 | 0.009 | 0.009 |
| HC vs. Inactive RA | 1.567 | 0.019 | 0.029 |
| HC vs. Active RA | 1.777 | 0.005 | 0.015 |
| Inactive RA vs. Active RA | 1.230 | 0.138 | 0.138 |
| HC vs. RF(−) RA | 1.794 | 0.005 | 0.015 |
| HC vs. RF(+) RA | 1.562 | 0.016 | 0.024 |
| RF(−) RA vs. RF(+) RA | 0.996 | 0.420 | 0.420 |
| HC vs. ACPA(−) RA | 1.665 | 0.010 | 0.030 |
| HC vs. ACPA(+) RA | 1.603 | 0.021 | 0.032 |
| ACPA(−) RA vs. ACPA(+) RA | 0.928 | 0.596 | 0.596 |
| AutoAb(−) RA vs. AutoAb(+) RA | 2.192 | 0.048 | 0.048 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chiang, H.-I.; Li, J.-R.; Liu, C.-C.; Liu, P.-Y.; Chen, H.-H.; Chen, Y.-M.; Lan, J.-L.; Chen, D.-Y. An Association of Gut Microbiota with Different Phenotypes in Chinese Patients with Rheumatoid Arthritis. J. Clin. Med. 2019, 8, 1770. https://doi.org/10.3390/jcm8111770
Chiang H-I, Li J-R, Liu C-C, Liu P-Y, Chen H-H, Chen Y-M, Lan J-L, Chen D-Y. An Association of Gut Microbiota with Different Phenotypes in Chinese Patients with Rheumatoid Arthritis. Journal of Clinical Medicine. 2019; 8(11):1770. https://doi.org/10.3390/jcm8111770
Chicago/Turabian StyleChiang, Hsin-I, Jian-Rong Li, Chun-Chi Liu, Po-Yu Liu, Hsin-Hua Chen, Yi-Ming Chen, Joung-Liang Lan, and Der-Yuan Chen. 2019. "An Association of Gut Microbiota with Different Phenotypes in Chinese Patients with Rheumatoid Arthritis" Journal of Clinical Medicine 8, no. 11: 1770. https://doi.org/10.3390/jcm8111770
APA StyleChiang, H.-I., Li, J.-R., Liu, C.-C., Liu, P.-Y., Chen, H.-H., Chen, Y.-M., Lan, J.-L., & Chen, D.-Y. (2019). An Association of Gut Microbiota with Different Phenotypes in Chinese Patients with Rheumatoid Arthritis. Journal of Clinical Medicine, 8(11), 1770. https://doi.org/10.3390/jcm8111770








