Quantification of HPV16 E6/E7 mRNA Spliced Isoforms Viral Load as a Novel Diagnostic Tool for Improving Cervical Cancer Screening
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Lines
2.2. Patients
2.3. Uterine Cervical Samples Analysis and Criteria of Selection
2.4. HR-HPV DNA Genotyping
2.5. HR-HPV E6/E7 mRNA Detection
2.6. Nucleic Acid Extractions
2.7. HPV16 DNA VL Quantification Assay
2.8. HPV16 E6/E7 mRNA VLs Quantification Assay
2.9. Statistical Analysis
3. Results
3.1. Validation of HPV16 DNA and E6/E7 mRNA VLs Quantification Assays
3.2. Quantification of HPV16 DNA and E6/E7 mRNA VLs in UCS
3.3. HPV16 E6/E7 mRNA VLs Are Increased in High Grade Cervical Lesions
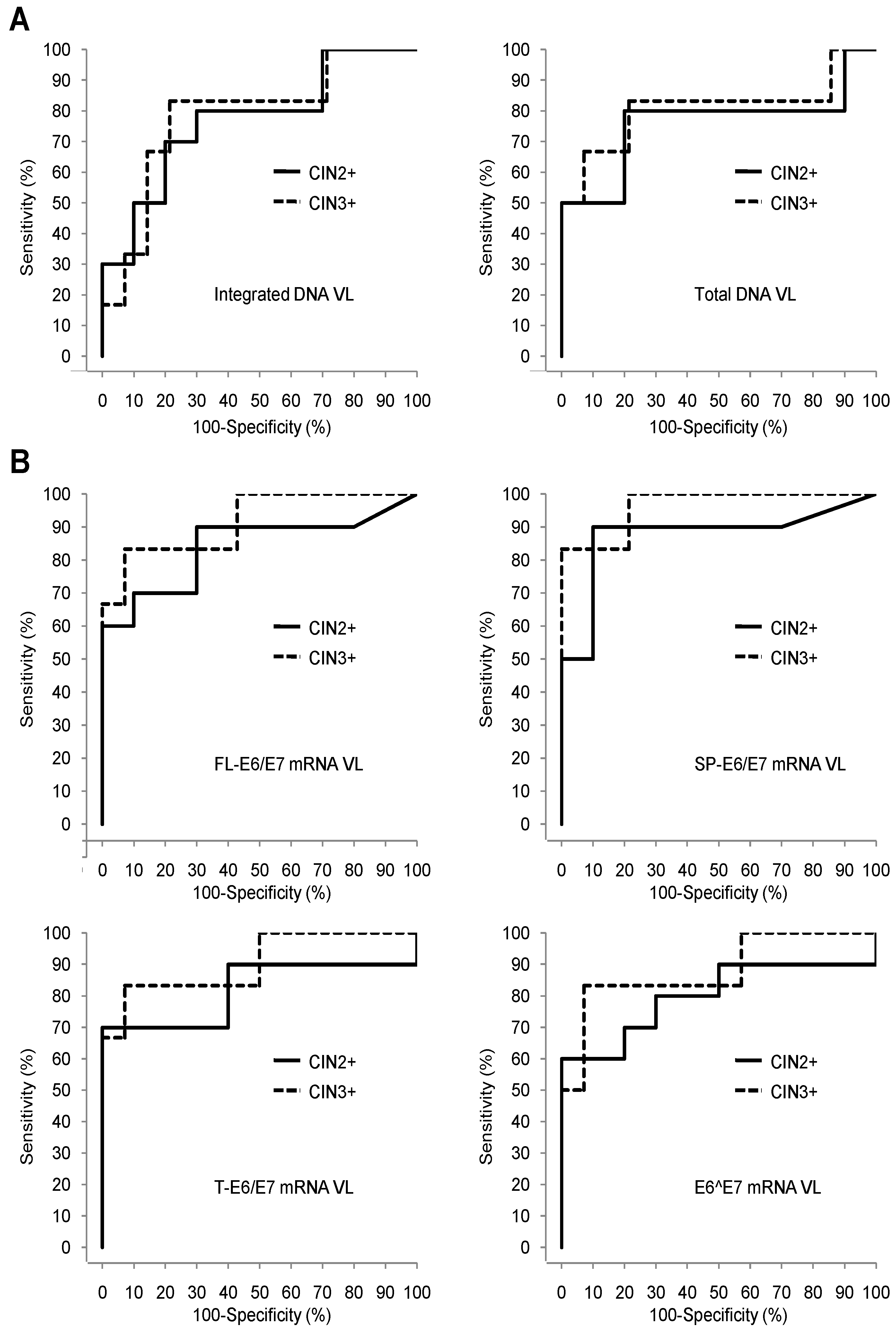
3.4. Comparison of the Pap Test, HPV16 DNA VLs, and HPV16 E6/E7 mRNA VLs Sets Diagnostic Performances for Detection of High Grade Cervical Lesions
4. Discussion
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| HPVs | human papillomaviruses |
| FL | full length E6/E7 mRNA |
| SP | + spliced E6/E7 mRNA containing intact E7 ORF |
| T | total E6/E7 mRNA corresponding to SP + E6^E7 mRNA |
| VL | viral loads |
| E6^E7 | E6/E7 mRNA containing disrupted E6 and E7 ORFs calculated by the following subtraction T-SP |
| ASC-US | atypical squamous cells of unknown significance |
| LSIL | low-grade squamous intraepithelial lesion |
| CC | cervical cancer |
| CIN | cervical intraepithelial neoplasia |
| CIN2+ | CIN of grade 2 or more: CIN2, CIN3, cancer |
| CIN3+ | CIN of grade 3 or more: CIN3, cancer |
| UCS | uterine cervical smears |
| ASC-H | atypical squamous cells- cannot exclude high grade |
| HSIL | high grade squamous intraepithelial lesion |
| ROC | receiver operating curves |
| AUC | area under roc curve |
| NPV | negative predictive value |
| PPV | positive predictive value |
References
- Ferlay, J.; Soerjomataram, I.; Dikshit, R.; Eser, S.; Mathers, C.; Rebelo, M.; Parkin, D.M.; Forman, D.; Bray, F. Cancer incidence and mortality worldwide: Sources, methods and major patterns in GLOBOCAN 2012. Int. J. Cancer 2015, 136, 359–386. [Google Scholar] [CrossRef] [PubMed]
- Walboomers, J.M.; Jacobs, M.V.; Manos, M.M.; Bosch, F.X.; Kummer, J.A.; Shah, K.V.; Snijders, P.J.; Peto, J.; Meijer, C.J.; Munoz, N. Human papillomavirus is a necessary cause of invasive cervical cancer worldwide. J. Pathol. 1999, 189, 12–19. [Google Scholar] [CrossRef]
- Munoz, N. Human papillomavirus and cancer: The epidemiological evidence. J. Clin. Virol. 2000, 19, 1–5. [Google Scholar] [CrossRef]
- Doorbar, J.; Egawa, N.; Griffin, H.; Kranjec, C.; Murakami, I. Human papillomavirus molecular biology and disease association. Rev. Med. Virol. 2015, 25, 2–23. [Google Scholar] [CrossRef]
- Bouvard, V.; Baan, R.; Straif, K.; Grosse, Y.; Secretan, B.; El Ghissassi, F.; Benbrahim-Tallaa, L.; Guha, N.; Freeman, C.; Galichet, L.; et al. A review of human carcinogens—Part b: Biological agents. Lancet Oncol. 2009, 10, 321–322. [Google Scholar] [CrossRef]
- De Villiers, E.M.; Fauquet, C.; Broker, T.R.; Bernard, H.U.; zur Hausen, H. Classification of papillomaviruses. Virology 2004, 324, 17–27. [Google Scholar] [CrossRef]
- Munoz, N.; Bosch, F.X.; de Sanjose, S.; Herrero, R.; Castellsague, X.; Shah, K.V.; Snijders, P.J.; Meijer, C.J. Epidemiologic classification of human papillomavirus types associated with cervical cancer. N. Engl. J. Med. 2003, 348, 518–527. [Google Scholar] [CrossRef]
- Clifford, G.M.; Smith, J.S.; Plummer, M.; Munoz, N.; Franceschi, S. Human papillomavirus types in invasive cervical cancer worldwide: A meta-analysis. Br. J. Cancer 2003, 88, 63–73. [Google Scholar] [CrossRef]
- de Sanjose, S.; Quint, W.G.; Alemany, L.; Geraets, D.T.; Klaustermeier, J.E.; Lloveras, B.; Tous, S.; Felix, A.; Bravo, L.E.; Shin, H.R.; et al. Human papillomavirus genotype attribution in invasive cervical cancer: A retrospective cross-sectional worldwide study. Lancet Oncol. 2010, 11, 1048–1056. [Google Scholar] [CrossRef]
- IARC Working Group on the Evaluation of Carcinogenic Risks to Humans. Human papillomaviruses. IARC Monogr. Eval. Carcinog Risks Hum. 2007, 90, 1–636. [Google Scholar]
- French, D.; Lorenzon, L. HPV infections: Basis of neoplastic transformation and related molecular tests. Curr. Pharm. Des. 2013, 19, 1371–1378. [Google Scholar] [PubMed]
- Schlecht, N.F.; Kulaga, S.; Robitaille, J.; Ferreira, S.; Santos, M.; Miyamura, R.A.; Duarte-Franco, E.; Rohan, T.E.; Ferenczy, A.; Villa, L.L.; et al. Persistent human papillomavirus infection as a predictor of cervical intraepithelial neoplasia. JAMA 2001, 286, 3106–3114. [Google Scholar] [CrossRef]
- Comparetto, C.; Borruto, F. Cervical cancer screening: A never-ending developing program. World J. Clin. Cases 2015, 3, 614–624. [Google Scholar] [CrossRef] [PubMed]
- Grosclaude, P. Contributions of the descriptive epidemiology, the registers and the troops. French situation seen by the registers of population. Med. Sci. (Paris) 2007, 23, 22–25. [Google Scholar] [CrossRef] [PubMed]
- SEER Cancer Statistics Review, 1975–2006. Available online: https://seer.cancer.gov/archive/csr/1975_2006/ (accessed on 28 November 2018).
- Cuzick, J.; Arbyn, M.; Sankaranarayanan, R.; Tsu, V.; Ronco, G.; Mayrand, M.H.; Dillner, J.; Meijer, C.J. Overview of human papillomavirus-based and other novel options for cervical cancer screening in developed and developing countries. Vaccine 2008, 26, 29–41. [Google Scholar] [CrossRef]
- Boulanger, J.C.; Fauvet, R.; Urrutiaguer, S.; Drean, Y.; Sevestre, H.; Ganry, O.; Bergeron, C.; Gondry, J. Cytological history of cases of invasive cervical cancer diagnosed in france in 2006. Gynecol. Obstet. Fertil. 2007, 35, 764–771. [Google Scholar] [CrossRef] [PubMed]
- Lorin, L.; Bertaut, A.; Hudry, D.; Beltjens, F.; Roignot, P.; Bone-Lepinoy, M.C.; Douvier, S.; Arveux, P. About invasive cervical cancer: A french population based study between 1998 and 2010. Eur. J. Obstet. Gynecol. Reprod. Biol. 2015, 191, 1–6. [Google Scholar] [CrossRef]
- Wright, T.C., Jr.; Schiffman, M.; Solomon, D.; Cox, J.T.; Garcia, F.; Goldie, S.; Hatch, K.; Noller, K.L.; Roach, N.; Runowicz, C.; et al. Interim guidance for the use of human papillomavirus DNA testing as an adjunct to cervical cytology for screening. Obstet. Gynecol. 2004, 103, 304–309. [Google Scholar] [CrossRef]
- Agorastos, T.; Chatzistamatiou, K.; Katsamagkas, T.; Koliopoulos, G.; Daponte, A.; Constantinidis, T.; Constantinidis, T.C. Primary screening for cervical cancer based on high-risk human papillomavirus (HPV) detection and HPV 16 and HPV 18 genotyping, in comparison to cytology. PLoS ONE 2015, 10, e0119755. [Google Scholar] [CrossRef]
- Ramzan, M.; Noor ul, A.; Ilyas, S.; Umer, M.; Bano, S.; Sarwar, S.; Shahzad, N.; Shakoori, A.R. A cornucopia of screening and diagnostic techniques for human papillomavirus associated cervical carcinomas. J. Virol. Methods 2015, 222, 192–201. [Google Scholar] [CrossRef]
- Tornesello, M.L.; Buonaguro, L.; Giorgi-Rossi, P.; Buonaguro, F.M. Viral and cellular biomarkers in the diagnosis of cervical intraepithelial neoplasia and cancer. Biomed. Res. Int. 2013, 519619. [Google Scholar] [CrossRef] [PubMed]
- Dutta, S.; Chakraborty, C.; Dutta, A.K.; Mandal, R.K.; Roychoudhury, S.; Basu, P.; Panda, C.K. Physical and methylation status of human papillomavirus 16 in asymptomatic cervical infections changes with malignant transformation. J. Clin. Pathol. 2015, 68, 206–211. [Google Scholar] [CrossRef] [PubMed]
- Tsakogiannis, D.; Kyriakopoulou, Z.; Ruether, I.G.; Amoutzias, G.D.; Dimitriou, T.G.; Diamantidou, V.; Kotsovassilis, C.; Markoulatos, P. Determination of human papillomavirus 16 physical status through E1/E6 and E2/E6 ratio analysis. J. Med. Microbiol. 2014, 63, 1716–1723. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.; He, Y.F.; Chen, M.; Chen, C.M.; Zhu, Q.J.; Lu, H.; Wei, Z.H.; Li, F.; Zhang, X.X.; Xu, C.J.; et al. Diagnosis of 25 genotypes of human papillomaviruses for their physical statuses in cervical precancerous/cancerous lesions: A comparison of E2/E6E7 ratio-based vs. Multiple E1-L1/E6E7 ratio-based detection techniques. J. Transl. Med. 2014, 12, 282. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.; Qin, Y.; Yu, L.; Lin, C.; Wang, H.; Cui, J.; Liu, B.; Liao, Y.; Warren, D.; Zhang, X.; et al. Association between human papillomavirus (HPV) 16, HPV18, and other HR-HPV viral load and the histological classification of cervical lesions: Results from a large-scale cross-sectional study. J. Med. Virol. 2017, 89, 535–541. [Google Scholar] [CrossRef] [PubMed]
- Marongiu, L.; Godi, A.; Parry, J.V.; Beddows, S. Human papillomavirus 16, 18, 31 and 45 viral load, integration and methylation status stratified by cervical disease stage. BMC Cancer 2014, 14, 384. [Google Scholar] [CrossRef]
- Origoni, M.; Cristoforoni, P.; Carminati, G.; Stefani, C.; Costa, S.; Sandri, M.T.; Mariani, L.; Preti, M. E6/E7 mrna testing for human papilloma virus-induced high-grade cervical intraepithelial disease (CIN2/CIN3): A promising perspective. Ecancermedicalscience 2015, 9, 533. [Google Scholar] [CrossRef]
- Ajiro, M.; Jia, R.; Zhang, L.; Liu, X.; Zheng, Z.M. Intron definition and a branch site adenosine at nt 385 control RNA splicing of HPV16 E6*I and E7 expression. PLoS ONE 2012, 7, e46412. [Google Scholar] [CrossRef]
- Shirasawa, H.; Jin, M.H.; Shimizu, K.; Akutsu, N.; Shino, Y.; Simizu, B. Transcription-modulatory activity of full-length E6 and E6*I proteins of human papillomavirus type 16. Virology 1994, 203, 36–42. [Google Scholar] [CrossRef]
- Ajiro, M.; Zheng, Z.M. E6^E7, a novel splice isoform protein of human papillomavirus 16, stabilizes viral E6 and E7 oncoproteins via HSP90 and GRP78. Am. Soc. Microbiol. 2015, 6. [Google Scholar] [CrossRef]
- Chen, J. Signaling pathways in HPV-associated cancers and therapeutic implications. Rev. Med. Virol. 2015, 25, 24–53. [Google Scholar] [CrossRef] [PubMed]
- Wang-Johanning, F.; Lu, D.W.; Wang, Y.; Johnson, M.R.; Johanning, G.L. Quantitation of human papillomavirus 16 E6 and E7 DNA and rna in residual material from thinprep papanicolaou tests using real-time polymerase chain reaction analysis. Cancer 2002, 94, 2199–2210. [Google Scholar] [CrossRef] [PubMed]
- Ho, C.M.; Lee, B.H.; Chang, S.F.; Chien, T.Y.; Huang, S.H.; Yan, C.C.; Cheng, W.F. Type-specific human papillomavirus oncogene messenger RNA levels correlate with the severity of cervical neoplasia. Int. J. Cancer 2010, 127, 622–632. [Google Scholar] [CrossRef] [PubMed]
- Baker, C.C.; Phelps, W.C.; Lindgren, V.; Braun, M.J.; Gonda, M.A.; Howley, P.M. Structural and transcriptional analysis of human papillomavirus type 16 sequences in cervical carcinoma cell lines. J. Virol. 1987, 61, 962–971. [Google Scholar] [PubMed]
- Solomon, D.; Davey, D.; Kurman, R.; Moriarty, A.; O’Connor, D.; Prey, M.; Raab, S.; Sherman, M.; Wilbur, D.; Wright, T., Jr.; et al. The 2001 bethesda system: Terminology for reporting results of cervical cytology. JAMA 2002, 287, 2114–2119. [Google Scholar] [CrossRef] [PubMed]
- Peitsaro, P.; Johansson, B.; Syrjanen, S. Integrated human papillomavirus type 16 is frequently found in cervical cancer precursors as demonstrated by a novel quantitative real-time PCR technique. J. Clin. Microbiol. 2002, 40, 886–891. [Google Scholar] [CrossRef]
- Steinau, M.; Rajeevan, M.S.; Unger, E.R. DNA and RNA references for qRT-PCR assays in exfoliated cervical cells. J. Mol. Diagn. 2006, 8, 113–118. [Google Scholar] [CrossRef]
- Ginsburg, O.; Bray, F.; Coleman, M.P.; Vanderpuye, V.; Eniu, A.; Kotha, S.R.; Sarker, M.; Huong, T.T.; Allemani, C.; Dvaladze, A.; et al. The global burden of women’s cancers: A grand challenge in global health. Lancet 2017, 389, 847–860. [Google Scholar] [CrossRef]
- Mustafa, R.A.; Santesso, N.; Khatib, R.; Mustafa, A.A.; Wiercioch, W.; Kehar, R.; Gandhi, S.; Chen, Y.; Cheung, A.; Hopkins, J.; et al. Systematic reviews and meta-analyses of the accuracy of HPV tests, visual inspection with acetic acid, cytology, and colposcopy. Int. J. Gynaecol. Obstet. 2016, 132, 259–265. [Google Scholar] [CrossRef]
- Burger, E.A.; Kornor, H.; Klemp, M.; Lauvrak, V.; Kristiansen, I.S. HPV mRNA tests for the detection of cervical intraepithelial neoplasia: A systematic review. Gynecol. Oncol. 2011, 120, 430–438. [Google Scholar] [CrossRef]
- Woodman, C.B.; Collins, S.I.; Young, L.S. The natural history of cervical hpv infection: Unresolved issues. Nat. Rev. Cancer 2007, 7, 11–22. [Google Scholar] [CrossRef]
- Ruttkay-Nedecky, B.; Jimenez Jimenez, A.M.; Nejdl, L.; Chudobova, D.; Gumulec, J.; Masarik, M.; Adam, V.; Kizek, R. Relevance of infection with human papillomavirus: The role of the p53 tumor suppressor protein and E6/E7 zinc finger proteins (Review). Int. J. Oncol. 2013, 43, 1754–1762. [Google Scholar] [CrossRef] [PubMed]
- Cricca, M.; Venturoli, S.; Leo, E.; Costa, S.; Musiani, M.; Zerbini, M. Molecular analysis of HPV 16 E6I/E6II spliced mRNAs and correlation with the viral physical state and the grade of the cervical lesion. J. Med. Virol. 2009, 81, 1276–1282. [Google Scholar] [CrossRef] [PubMed]
- Schmitt, M.; Dalstein, V.; Waterboer, T.; Clavel, C.; Gissmann, L.; Pawlita, M. The HPV16 transcriptome in cervical lesions of different grades. Mol. Cell. Probes 2011, 25, 260–265. [Google Scholar] [CrossRef] [PubMed]
- Tang, S.; Tao, M.; McCoy, J.P., Jr.; Zheng, Z.M. The E7 oncoprotein is translated from spliced E6*I transcripts in high-risk human papillomavirus type 16- or type 18-positive cervical cancer cell lines via translation reinitiation. J. Virol. 2006, 80, 4249–4263. [Google Scholar] [CrossRef] [PubMed]
- Pim, D.; Massimi, P.; Banks, L. Alternatively spliced Hpv-18 E6* protein inhibits E6 mediated degradation of p53 and suppresses transformed cell growth. Oncogene 1997, 15, 257–264. [Google Scholar] [CrossRef]

| Primer/Probe Set | Sequences | |
|---|---|---|
| HPV16 E6 | Forward | 5′-GAGAACTGCAATGTTTCAGGACC-3′ |
| Reverse | 5′- TGTATAGTTGTTTGCAGCTCTGTGC-3′ | |
| Probe | JOE-5′-CAGGAGCGACCCAGAAAGTTACCACAGTT-3′-TAMRA | |
| HPV16 E2 | Forward | 5′-AACGAAGTATCCTCTCCTGAAATTATTAG-3′ |
| Reverse | 5′-CCAAGGCGACGGCTTTG-3′ | |
| Probe | FAM-5′-CACCCCGCCGCGACCCATA-3′-TAMRA | |
| β-globin | Forward | 5′-TGCATCTGACTCCTGAGGAGAA-3′ |
| Reverse | 5′-GGGCCTCACCACCAACTTC-3′ | |
| Probe | TET-5′-CTGCCGTTACTGCCCT-3′-TAMRA |
| Primer/Probe Set | Sequences | |
|---|---|---|
| FL-E6/E7 | Forward | 5′-GTGTACTGCAAGCAACAGTTA-3′ |
| Reverse | 5′-CCCATCTCTATATACTATGCATAAATCC-3′ | |
| Probe | FAM-5′-CTGCGACGTGAGGTATATGACTTTGCT-3′-TAMRA | |
| SP-E6/E7 | Forward | 5′-GATTTGCAACCAGAGACAACTG-3′ |
| Reverse | 5′-GCTGGACCATCTATTTCATCCT-3′ | |
| Probe | FAM-5′-TGAGCAATTAAATGACAGCTCAGAGGAGG-3′-TAMRA | |
| T-E6/E7 | Forward | 5′-GACTCTACGCTTCGGTTGTG-3′ |
| Reverse | 5′-TGTGCCCATTAACAGGTCTT-3′ | |
| Probe | FAM-5′-CGTACAAAGCACACACGTAGACATTCG-3′-TAMRA | |
| β-actin | Forward | 5′-GACCCAGATCATGTTTGAGACC-3′ |
| Reverse | 5′-CCAGAGGCGTACAGGGATA-3′ | |
| Probe | FAM-5′-TGTACGTTGCTATCCAGGCTGTGC-3′-TAMRA |
| Viral Load Type | DNA VL (Copies/Cell ± SD (Range)) | E6/E7 mRNA VL (log10 (Copies/106 β-actin mRNA Copies) ±SD (Range)) | |
|---|---|---|---|
| DNA | Total | 0.93 ± 0.2 (0.7–1.06) | |
| Episomal | 0 | ||
| Integrated | 0.93 ± 0.2 (0.7–1.06) | ||
| mRNA | FL-E6/E7 | 4.47 ± 0.18 (4.27–4.59) | |
| SP-E6/E7 | 4.86 ± 0.21 (4.63–5.00) | ||
| T-E6/E7 | 4.93 ± 0.23 (4.68-5.12) | ||
| E6^E7 | 4.09 ± 0.37 (3.73-4.48) |
| Patient ID | Cytology Grade | Histology Grade | HPV16 DNA VL (log10 (Copies/106 Cells)) | HPV16 mRNA VL (log10 (Copies/106 β-actin mRNA Copies)) | |||||
|---|---|---|---|---|---|---|---|---|---|
| Total | Episomal | Integrated | FL-E6/E7 | SP-E6/E7 | T-E6/E7 | E6^E7 | |||
| 31 | Normal | Unlesional | 5.92 | 5.62 | 5.61 | 2.02 | 2.34 | 2.86 | 2.71 |
| 44 | Normal | Unlesional | 6.23 | 5.77 | 6.04 | 0.00 | 0.00 | 2.90 | 2.90 |
| 15 | LSIL | CIN1 | 6.80 | 6.01 | 6.73 | 2.15 | 2.73 | 3.99 | 3.97 |
| 18 | LSIL | CIN1 | 7.56 | 6.86 | 7.46 | 1.78 | 2.15 | 3.50 | 3.48 |
| 34 | Normal | CIN1 | 6.08 | 6.06 | 4.81 | 3.45 | 4.06 | 4.09 | 2.94 |
| 42 | Normal | CIN1 | 5.35 | 5.06 | 5.03 | 1.76 | 0.00 | 3.31 | 3.31 |
| 43 | Normal | CIN1 | 6.59 | 6.35 | 6.22 | 0.00 | 1.59 | 2.95 | 2.93 |
| 45 | ASC-US | CIN1 | 7.61 | 6.76 | 7.55 | 1.80 | 2.49 | 4.34 | 4.34 |
| 51 | LSIL | CIN1 | 6.95 | 6.36 | 6.83 | 2.97 | 0.00 | 4.30 | 4.30 |
| 55 | ASC-US | CIN1 | 5.89 | 5.79 | 5.19 | 2.91 | 3.04 | 3.27 | 2.87 |
| 2 | ASC-H | CIN2 | 6.97 | 6.17 | 6.89 | 2.80 | 3.16 | 3.88 | 3.79 |
| 4 | ASC-US | CIN2 | 5.37 | 4.71 | 5.27 | 0.00 | 0.00 | 1.91 | 1.91 |
| 9 | LSIL | CIN2 | 7.83 | 7.09 | 7.75 | 3.05 | 3.79 | 5.01 | 4.99 |
| 47 | ASC-H | CIN2 | 6.96 | 6.47 | 6.79 | 3.53 | 3.94 | 4.45 | 4.29 |
| 1 | HSIL | CIN3 | 8.09 | 7.48 | 7.97 | 3.52 | 4.19 | 4.72 | 4.56 |
| 5 | ASC-H | CIN3 | 7.19 | 6.55 | 7.08 | 4.25 | 4.81 | 5.10 | 4.79 |
| 41 | HSIL | CIN3 | 5.51 | 5.12 | 5.28 | 2.54 | 3.17 | 3.51 | 3.23 |
| 50 | ASC-H | CIN3 | 7.63 | 6.94 | 7.52 | 4.12 | 5.16 | 5.62 | 5.44 |
| 3 | HSIL | Invasive Cancer | 7.88 | 7.67 | 7.47 | 3.91 | 4.49 | 5.67 | 5.64 |
| 6 | HSIL | Invasive Cancer | 7.97 | 7.62 | 7.72 | 4.11 | 4.81 | 5.26 | 5.07 |
| HPV16 Viral Load Type | CIN2+ Histology Threshold | CIN3+ Histology Threshold | ||||||
|---|---|---|---|---|---|---|---|---|
| Histology <CIN2 | Histology CIN2+ | p-Value (Wilcoxon) | Histology <CIN3 | Histology CIN3+ | p-Value (Wilcoxon) | |||
| DNA VL (log10 (copies/106 cells)) | Total | Mean ± SD | 6.5 ± 0.7 | 7.1 ± 1.0 | 0.0690 | 6.6±0.8 | 7.4 ± 1.0 | 0.0490 |
| Integrated | Mean ± SD | 6.1 ± 1 | 7.0 ± 1.0 | 0.0596 | 6.3±1.0 | 7.2 ± 1.0 | 0.0676 | |
| E6/E7 mRNA VL (log10 (copies/106 β-actin mRNA copies)) | FL-E6/E7 | Mean ± SD | 1.9 ± 1.2 | 3.2 ± 1.3 | 0.0201 | 2.0±1.2 | 3.7 ± 0.6 | 0.0102 |
| SP-E6/E7 | Mean ± SD | 1.8 ± 1.4 | 3.8 ± 1.5 | 0.0112 | 2.1±1.5 | 4.4 ± 0.7 | 0.0048 | |
| T-E6/E7 | Mean ± SD | 3.6 ± 0.6 | 4.5 ± 1.2 | 0.0279 | 3.6±0.8 | 5.0 ± 0.8 | 0.0124 | |
| E6^E7 | Mean ± SD | 3.4 ± 0.6 | 4.4 ± 1.1 | 0.0380 | 3.5±0.8 | 4.8 ± 0.9 | 0.0177 | |
| HPV16 Viral Load Type | CIN2+ Histology Threshold | CIN3+ Histology Threshold | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| TV | YI | NPV. % (95%CI) | PPV. % (95%CI) | Se.% (95%CI) | Spe. % (95%CI) | AUC (95%CI) | TV | YI | NPV. % (95%CI) | PPV. % (95%CI) | Se.% (95%CI) | Spe. % (95%CI) | AUC (95%CI) | ||
| DNA | Total | 6.96 | 0.60 | 80 (44–97) | 80 (44–97) | 80 (44–97) | 80 (44–97) | 0.76 (0.52–1.00) | 7.19 | 0.62 | 92 (36–100) | 63 (24–91) | 83 (36–100) | 79 (49–95) | 0.81 (0.53–1.00) |
| Integrated | 6.79 | 0.50 | 78 (40–97) | 73 (39–94) | 80 (44–97) | 70 (35–93) | 0.77 (0.55–0.99) | 7.08 | 0.62 | 92 (62–100) | 63 (24–91) | 83 (36–100) | 79 (49–95) | 0.79 (0.54–1.00) | |
| mRNA | FL-E6/E7 | 2.54 | 0.60 | 88 (47–100) | 75 (43–95) | 90 (56–100) | 70 (35–99) | 0.84 (0.65–1.00) | 2.54 | 0.57 | 100 (63–100) | 50 (21–79) | 100 (54–100) | 57 (29–82) | 0.92 (0.77–1.00) |
| SP-E6/E7 | 3.16 | 0.80 | 90 (56–100) | 90 (56–100) | 90 (56–100) | 90 (56–100) | 0.88 (0.69–1.00) | 3.17 | 0.79 | 100 (72–100) | 67 (30–93) | 100 (54–100) | 79 (49–95) | 0.97 (0.89–1.00) | |
| T-E6/E7 | 3.51 | 0.50 | 86 (42–100) | 69 (39–91) | 90 (56–100) | 60 (26–88) | 0.82 (0.60–1.00) | 4.72 | 0.76 | 93 (66–100) | 83 (36–100) | 83 (36–100) | 93 (66–100) | 0.91 (0.73–1.00) | |
| E6^E7 | 3.23 | 0.40 | 83 (36–100) | 64 (35–87) | 90 (56–100) | 50 (19–81) | 0.80 (0.58–1.00) | 3.23 | 0.43 | 100 (54–100) | 43 (18–71) | 100 (54–100) | 43 (18–71) | 0.88 (0.69–1.00) | |
| Pap test | NA | NA | 83 (52–98) | 100 (63–100) | 80 (44–97) | 100 (69–100) | NA | NA | NA | 100 (74–100) | 75 (35–97) | 100 (54–100) | 86 (57–98) | NA | |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Camus, C.; Vitale, S.; Loubatier, C.; Pénaranda, G.; Khiri, H.; Plauzolles, A.; Carcopino, X.; Halfon, P.; Giordanengo, V. Quantification of HPV16 E6/E7 mRNA Spliced Isoforms Viral Load as a Novel Diagnostic Tool for Improving Cervical Cancer Screening. J. Clin. Med. 2018, 7, 530. https://doi.org/10.3390/jcm7120530
Camus C, Vitale S, Loubatier C, Pénaranda G, Khiri H, Plauzolles A, Carcopino X, Halfon P, Giordanengo V. Quantification of HPV16 E6/E7 mRNA Spliced Isoforms Viral Load as a Novel Diagnostic Tool for Improving Cervical Cancer Screening. Journal of Clinical Medicine. 2018; 7(12):530. https://doi.org/10.3390/jcm7120530
Chicago/Turabian StyleCamus, Claire, Sébastien Vitale, Céline Loubatier, Guillaume Pénaranda, Hacène Khiri, Anne Plauzolles, Xavier Carcopino, Philippe Halfon, and Valérie Giordanengo. 2018. "Quantification of HPV16 E6/E7 mRNA Spliced Isoforms Viral Load as a Novel Diagnostic Tool for Improving Cervical Cancer Screening" Journal of Clinical Medicine 7, no. 12: 530. https://doi.org/10.3390/jcm7120530
APA StyleCamus, C., Vitale, S., Loubatier, C., Pénaranda, G., Khiri, H., Plauzolles, A., Carcopino, X., Halfon, P., & Giordanengo, V. (2018). Quantification of HPV16 E6/E7 mRNA Spliced Isoforms Viral Load as a Novel Diagnostic Tool for Improving Cervical Cancer Screening. Journal of Clinical Medicine, 7(12), 530. https://doi.org/10.3390/jcm7120530





