Fetal Aneuploidy Detection by Cell-Free DNA Sequencing for Multiple Pregnancies and Quality Issues with Vanishing Twins
Abstract
:1. Introduction
2. Materials and Methods
2.1. Subject Enrollment
2.2. Sample Processing and DNA Extraction
2.3. Quality Control of cffDNA (QuantYfeX)
2.4. Maternal Plasma DNA Sequencing and Data Analysis
3. Results and Discussion
3.1. Retrospective Study on Stored DNA Libraries
| Sample | Chr13 z-score | Chr18 z-score | Chr21 z-score | Fetal fraction (%) | Gestational age (p.m.) | NIPT result | Fetus A karyotype | Fetus B karyotype | Invasive method |
|---|---|---|---|---|---|---|---|---|---|
| RDLN015823 | 0.0 | 0.0 | −0.4 | 37 | 10 + 6 | negative | 46,XX | 46,XY | CVS |
| RDLN015835 | −0.6 | 0.9 | 0.7 | 35 | 12 + 6 | negative | 46,XY | 46,XX | CVS |
| RDLN015916 | 1.8 | 1.9 | 0.8 | 24 | 16 + 2 | negative | 46,XY | 46,XY | AC |
| RDLN016042 | 0.9 | 0.5 | 1.0 | 23 | 17 + 4 | negative | 46,XX | 46,XY | AC |
| RDLN016047 | 1.3 | 0.7 | −1.6 | 45 | 13 + 5 | negative | 46,XY | 46,XY | CVS |
| RDLN016114 | −0.9 | −0.2 | 8.4 | 29 | 14 + 4 | T21 positive | 47,XY,+21 | 46,XX | CVS |
| RDLN016116 | −0.4 | 0.7 | 4.4 | 20 | 13 + 4 | T21 positive | 47,XX,+21 | 46,XX | CVS |
| RDLN016450 | −0.1 | 1.4 | 8.4 | 31 | 16 + 0 | T21 positive | 47,XX,+21 | 46,XX | AC |
| RDLN016457 | 1.0 | 0.9 | −0.3 | 22 | 17 + 5 | negative | 46,XX | 46,XY | AC |
| RDLN016474 | 0.8 | 0.3 | 5.4 | 16 | 18 + 4 | T21 positive | 47,XX,+21 | 46,XX | AC |
| RDLN016519 | 0.2 | −1.0 | 0.2 | 20 | 15 + 0 | negative | 46,XX | 46,XX | AC |
| RDLN016778 | 0.2 | −0.1 | −0.1 | 12 | 16 + 0 | negative | 46,XX | 46,XX | AC |
| RDLN017192 | −1.2 | −0.4 | −1.0 | 13 | 16 + 1 | negative | 46,XY | 46,XY | AC |
| RDLN017624 | 1.0 | 0.9 | 0.7 | 15 | 17 + 0 | negative | 46,XY | 46,XY | AC |
| RDLN017641 | −1.0 | 0.3 | 0.7 | 8 | 15 + 2 | negative | 46,XY | 46,XY | AC |
| RDLN017670 | 0.8 | 0.1 | −0.9 | 24 | 17 + 1 | negative | 46,XX | 46,XX | AC |
3.2. Prospective Study on Blood Samples from Multiple Pregnancies Collected during Laboratory Routine
| Sample | Chr13 z-score | Chr18 z-score | Chr21 z-score | Fetal fraction (%) | Gestational age (p.m.) | No. of fetuses, chorionicity, amnionicity | NIPT result |
|---|---|---|---|---|---|---|---|
| LCMPC01 | 0.8 | −0.4 | 1.0 | n.a. | 11 + 0 | 2, monochorionic, n.a. | Negative |
| LCMPC02 | 0.0 | 0.3 | 0.2 | n.a. | 21 + 0 | 2, dichorionic, diamniotic | Negative |
| LCMPC03 | 0.4 | 1.0 | 0.1 | n.a. | 22 + 0 | 2, dichorionic, diamniotic | negative |
| LCMPC04 | −0.3 | −0.6 | 0.0 | n.a. | n.a. | 3, n.a., n.a. | negative |
| LCMPC05 | 1.3 | −1.0 | −0.8 | 16.7 | 11 + 5 | 3, trichorionic, triamniotic | negative |
| LCMPC06 | −0.4 | 1.1 | 8.5 | 18.0 | 13 + 2 | 2, monochorionic, n.a. | T21 positive |
| LCMPC07 | −1.0 | 0.3 | 0.9 | 7.9 | 19 + 0 | 2, dichorionic, diamniotic | negative |
| LCMPC08 | 0.7 | 1.2 | 0.0 | 16.5 | 18 + 1 | 2, dichorionic, diamniotic | negative |
| LCMPC09 | 0.6 | −0.8 | 0.7 | 8.9 | 11 + 5 | 2, monochorionic, diamniotic | negative |
| LCMPC10 | 0.3 | 0.7 | −0.7 | 17.6 | 20 + 4 | 2, dichorionic, diamniotic | negative |
| LCMPC11 | −0.9 | −0.8 | 0.7 | 11.5 | 23 + 0 | 2, dichorionic, diamniotic | negative |
| LCMPC12 | −0.9 | −0.7 | −2.0 | 13.3 | 11 + 1 | 2, monochorionic, diamniotic | negative |
| LCMPC13 | 1.3 | 0.1 | 0.3 | 21.4 | 16 + 0 | 2, dichorionic, diamniotic | negative |
| LCMPC14 | 0.2 | −0.3 | 0.0 | 6.8 | 12 + 5 | 2, n.a., n.a. | negative |
| LCMPC15 | 2.2 | 0.1 | 14.7 | 24.8 | 16 + 0 | 2, dichorionic, diamniotic | T21 positive |
| LCMPC16 | 1.1 | 1.7 | 0.5 | 5.4 | 12 + 5 | 2, n.a., n.a. | negative |
| LCMPC17 | 0.7 | 1.4 | 0.5 | 16.5 | 14 + 2 | 2, n.a., n.a. | negative |
| LCMPC18 | 0.3 | 2.6 | 0.0 | 18.5 | 18 + 3 | 2, n.a., n.a. | negative |
| LCMPC19 | −0.2 | 0.8 | 0.3 | 16.6 | 14 + 0 | 2, dichorionic, diamniotic | negative |
| LCMPC20 | −0.7 | −0.9 | 0.1 | 13.1 | 15 + 4 | 2, dichorionic, diamniotic | negative |
| LCMPC21 | 1.0 | −0.7 | 1.2 | 8.4 | 9 + 3 | 2, dichorionic, diamniotic | negative |
| LCMPC22 | −1.1 | −0.2 | 0.3 | 5.6 | 16 + 2 | 2, monochorionic, n.a. | negative |
| LCMPC23 | −2.2 | 2.2 | −0.8 | 20.6 | 19 + 5 | 2, monochorionic, n.a. | negative |
| LCMPC24 | −1.6 | −0.4 | −0.5 | 14.7 | 22 + 2 | 2, monochorionic, diamniotic | negative |
| LCMPC25 | −0.8 | −0.2 | −1.5 | 12.1 | 11 + 5 | 2, n.a., n.a. | negative |
| LCMPC26 | −0.4 | −0.6 | −1.3 | 7.5 | 13 + 0 | 2, dichorionic, diamniotic | negative |
| LCMPC27 | 0.5 | −0.8 | −0.4 | 16.3 | 12 + 6 | 2, n.a., n.a. | negative |
| LCMPC28 | −1.2 | −0.3 | −0.7 | 19.4 | 10 + 1 | 2, dichorionic, diamniotic | negative |
| LCMPC29 | −0.8 | 0.7 | −0.4 | 14.2 | 13 + 2 | 2, monochorionic, n.a. | negative |
| LCMPC30 | 0.7 | 0.3 | 0.9 | 14.9 | 12 + 2 | 2, monochorionic, monoamniotic | negative |
| LCMPC31 | −0.2 | 0.3 | −0.9 | 19.3 | 19 + 1 | 2, dichorionic, diamniotic | negative |
| LCMPC32 | −1.1 | 2.5 | −2.2 | 11.6 | 20 + 0 | 2, dichorionic, diamniotic | negative |
| LCMPC33 | 0.2 | 2.2 | −1.6 | 8.6 | 11 + 0 | 2, dichorionic, diamniotic | negative |
| LCMPC34 | −1.0 | 1.2 | 0.0 | 15.1 | 15 + 4 | 2, dichorionic, diamniotic | negative |
| LCMPC35 | −0.3 | −0.8 | −0.3 | 19.2 | 12 + 0 | 2, dichorionic, diamniotic | negative |
| LCMPC36 | −1.4 | −0.5 | −0.8 | 13.9 | 12 + 0 | 2, dichorionic, diamniotic | negative |
| LCMPC37 | 1.8 | −0.7 | 0.1 | 13.8 | 17 + 6 | 2, dichorionic, diamniotic | negative |
| LCMPC38 | −0.1 | 1.1 | −0.7 | 13.4 | 13 + 1 | 2, dichorionic, diamniotic | negative |
| LCMPC39 | −1.9 | 0.2 | −2.2 | 15.0 | 17 + 0 | 2, dichorionic, diamniotic | negative |
| LCMPC40 | 0.6 | −0.4 | 0.8 | 16.2 | 18 + 3 | 2, dichorionic, diamniotic | negative |
3.3. Minimum Fetal Fraction Needed for the Detection of Aneuploidies in Multiple Pregnancies
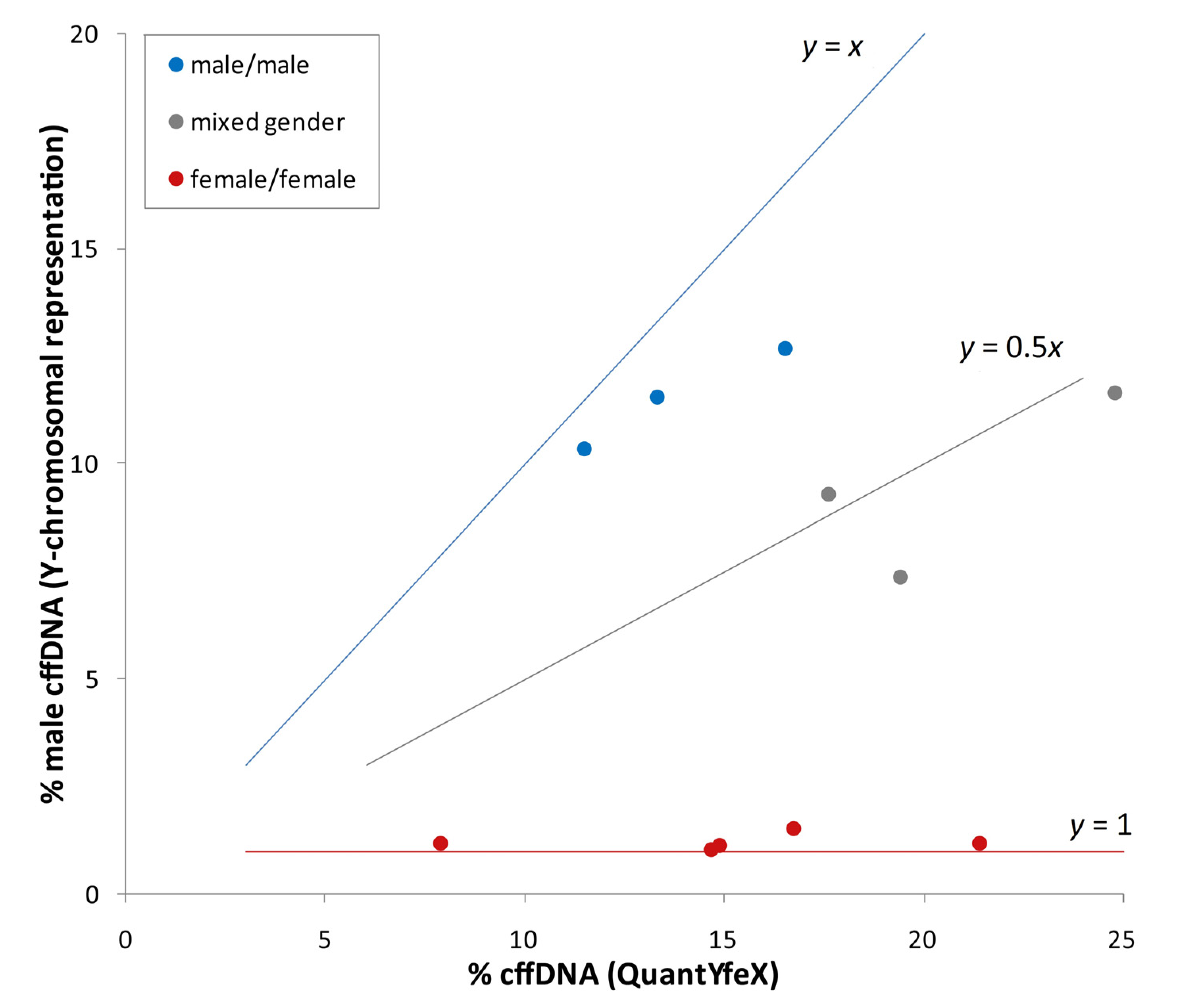
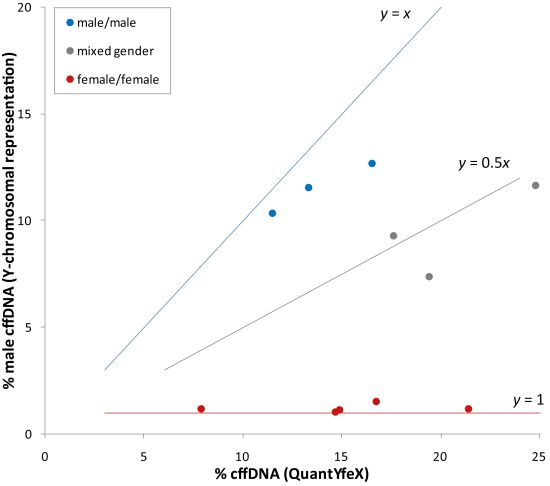
3.4. Case Reports of Discordant NIPT Results Due to Vanishing Twins
| Sample code | Sample type | Gestational age | Total reads (×106) | Chr13 z-score | Chr18 z-score | Chr21 z-score | % cffDNA calculation by ChromRep Y | Fetal fraction (QuantYfeX) |
|---|---|---|---|---|---|---|---|---|
| VTA01 | maternal plasma sample | 17 + 2 | 14.94 | 1.3 | −1.5 | 13.5 | 9.2 | 20.7 |
| VTA02 | Back-up sample | 17 + 2 | 17.08 | 0.4 | −1.7 | 11.1 | 9.3 | 24.8 |
| VTA03 | maternal plasma sample collected prior birth | 38 + 2 | 17.76 | 0.8 | 1.9 | −0.3 | 21.7 | 21.4 |
| VTB01 | initial maternal plasma sample | 13 + 2 | 16.79 | −0.2 | 0.1 | 3.4 | 3.0 | 13.4 |
| VTB02 | Back-up sample | 13 + 2 | 11.97 | −0.1 | 0.3 | 2.6 | 2.7 | 10.0 |
4. Conclusions
Appendix
| Component | Sequence | Concentration | Final concentration (μM) | 1× μL | |||||
|---|---|---|---|---|---|---|---|---|---|
| master mix | 2× | 1× | 12.5 | ||||||
| RASSF1 sens/insens | primer for p0009 | ATT GAG CTG CGG GAG CTG GC | 100 μM | 1.4 | 0.35 | ||||
| primer re p0010 | TGC CGT GTG GGG TTG CAC | 100 μM | 1.4 | 0.35 | |||||
| probe s0001 | FAM-ACC CGG CTG GAG CGT-MGB | 100 μM | 0.14 | 0.035 | |||||
| primer for p0003 | GGT CAT CCA CCA CCA AGA AC | 100 μM | 1.4 | 0.35 | |||||
| primer re p0004 | TGC CCA AGG ATG CTG TCA AG | 100 μM | 1.4 | 0.35 | |||||
| probe s0002 | VIC-GGG CCT CAA TGA CTT CAC GT-MGB | 100 μM | 0.14 | 0.035 | |||||
| TBX3 sens/insens | primer for p0011 | GGT GCG AAC TCC TCT TTG TC | 100 μM | 1.4 | 0.35 | ||||
| primer re p0012 | TTA ATC ACC CAG CGC ATG GC | 100 μM | 1.4 | 0.35 | |||||
| probe s0010 | 6FAM-CCC TCC CGG TGG GTG ATA AA-MGBNFQ | 100 μM | 0.14 | 0.035 | |||||
| Primer for p0021 | TGT TCA CTG GAG GAC TCA TC | 100 μM | 1.4 | 0.35 | |||||
| primer re p0022 | CAG TCC ATG AGG GTG TTT G | 100 μM | 1.4 | 0.35 | |||||
| probe s0011 | VIC-GAG GTC CCA TTC TCC TTT-MGBNFQ | 100 μM | 0.14 | 0.035 | |||||
| general reagents | DMSO | 100% | 0.025 | 0.625 | |||||
| MgCl2 | 50 mM | 2 | 1 | ||||||
| template | 10 | ||||||||
| water | - | ||||||||
| summary | 25 | ||||||||
| Temperature | Time | Cycles | Analysis mode | |
|---|---|---|---|---|
| pre-incubation | 95 °C | 5 min | 1 | none |
| denaturation | 95 °C | 10 s | 45 | quantification |
| annealing | 60 °C | 10 s | none | |
| elongation | 72 °C | 8 s | single | |
| cooling | 40 °C | none |
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Benn, P.; Borell, A.; Chiu, R.; Cuckle, H.; Dugoff, L.; Faas, B.; Gross, S.; Johnson, J.; Maymon, R.; Norton, M.; et al. Position statement from the Aneuploidy Screening Committee on behalf of the Board of the International Society for Prenatal Diagnosis. Prena. Diagn. 2013, 33, 622–629. [Google Scholar] [CrossRef]
- Nikolaides, K.H. The 11–13 + 6 weeks scan. Available online: http://www.fetalmedicine.com/fmf/FMF-English.pdf (accessed on 20 March 2014).
- Agarwal, K.; Alfirevic, Z. Pregnancy loss after chorionic villus sampling and genetic amniocentesis in twin pregnancies: A systematic review. Ultrasound Obstet. Gynecol. 2012, 40, 128–134. [Google Scholar] [CrossRef]
- Chiu, R.W.; Akolekar, R.; Zheng, Y.W.; Leung, T.Y.; Sun, H.; Chan, K.C.; Lun, F.M.; Go, A.T.; Lau, E.T.; To, W.W.; et al. Non-invasive prenatal assessment of trisomy 21 by multiplexed maternal plasma DNA sequencing: Large-scale validity study. BMJ 2011, 342. [Google Scholar] [CrossRef]
- Ehrich, M.; Deciu, C.; Zwiefelhofer, T.; Tynan, J.A.; Cagasan, L.; Tim, R.; Lu, V.; McCullough, R.; Mc-Carthy, E.; Nygren, A.O.; et al. Noninvasive detection of fetal tri-somy 21 by sequencing of DNA in maternal blood: A study in a clinical setting. Am. J. Obstet. Gynecol. 2011, 204. [Google Scholar] [CrossRef]
- Palomaki, G.E.; Kloza, E.M.; Lambert-Messerlian, G.M.; Haddow, J.E.; Neveux, L.M.; Ehrich, M.; van den Boom, D.; Bombard, A.T.; Deciu, C.; Grody, W.W.; et al. DNA sequencing of maternal plasma to detect Down syndrome: An international clinical validation study. Genet. Med. 2011, 13, 913–920. [Google Scholar] [CrossRef]
- Sehnert, A.J.; Rhees, B.; Comstock, D.; de Feo, E.; Heilek, G.; Burke, J.; Rava, R.P. Optimal detec-tion of fetal chromosomal abnormalities by massively parallel DNA sequencing of cell-free fetal DNA from maternal blood. Clin. Chem. 2011, 57, 1042–1049. [Google Scholar] [CrossRef]
- Canick, J.A.; Kloza, E.M.; Lambert-Messerlian, G.M.; Haddow, J.E.; Ehrich, M.; van den Boom, D.; Bombard, A.T.; Deciu, C.; Palomaki, G.E. DNA sequencing of maternal plasma to identify Down syndrome and other trisomies in multiple gestations. Prenat. Diagn. 2012, 32, 730–734. [Google Scholar] [CrossRef]
- Gil, M.M.; Quezada, M.S.; Bregant, B.; Syngelaki, A.; Nicolaides, K.H. Cell-Free DNA Analysis for Trisomy Risk Assessment in First-Trimester Twin Pregnancies. Fetal. Diagn. Ther. 2014, 35, 204–211. [Google Scholar] [CrossRef]
- Leung, T.Y.; Qu, J.Z.; Liao, G.J.; Jiang, P.; Cheng, Y.K.; Chan, K.C.; Chiu, R.W.; Lo, Y.M. Noninvasive twin zygosity assessment and aneuploidy detection by maternal plasma DNA sequencing. Prenat. Diagn. 2013, 33, 675–681. [Google Scholar] [CrossRef]
- Nygren, A.O.; Dean, J.; Jensen, T.J.; Kruse, S.; Kwong, W.; van den Boom, D.; Ehrich, M. Quantification of fetal DNA by use of methylation-based DNA discrimination. Clin. Chem. 2010, 56, 1627–1635. [Google Scholar] [CrossRef]
- Chan, K.C.A.; Ding, C.; Gerovassili, A.; Yeung, S.W.; Chiu, R.W.K.; Leung, T.N.; Chim, S.S.; Chung, G.T.; Nicolaides, K.H.; Lo, Y.M. Hypermethylated RASSF1A in maternal plasma: A universal fetal DNA marker that improves the reliability of noninvasive prenatal diagnosis. Clin. Chem. 2006, 52, 2211–2218. [Google Scholar] [CrossRef]
- Chiu, R.W.; Chim, S.S.; Wong, I.H.; Wong, C.S.; Lee, W.S.; To, K.F.; Tong, J.H.; Yuen, R.K.; Shum, A.S.; Chan, J.K.; et al. Hypermethylation of RASSF1A in human and rhesus placentas. Am. J. Pathol. 2007, 170, 941–950. [Google Scholar] [CrossRef]
- Stumm, M.; Entezami, M.; Haug, K.; Blank, C.; Wüstemann, M.; Schulze, B.; Raabe-Meyer, G.; Hempel, M.; Schelling, M.; Ostermayer, E.; et al. Diagnostic accuracy of random massively parallel sequencing for non-invasive prenatal detection of common autosomal aneuploidies: A collaborative study in Europe. Prenat. Diagn. 2014, 34, 185–191. [Google Scholar] [CrossRef]
- Qu, J.Z.; Leung, T.Y.; Jiang, P.; Liao, G.J.; Cheng, Y.K.; Sun, H.; Chiu, R.W.; Chan, K.C.; Lo, Y.M. Noninvasive prenatal determination of twin zygosity by maternal plasma DNA analysis. Clin. Chem. 2013, 59, 427–435. [Google Scholar] [CrossRef]
- Struble, C.A.; Syngelaki, A.; Oliphant, A.; Song, K.; Nicolaides, K.H. Fetal Fraction Estimate in Twin Pregnancies Using Directed Cell-Free DNA Analysis. Fetal. Diagn. Ther. 2014, 35, 199–203. [Google Scholar] [CrossRef]
- Chen, K.; Chmait, R.H.; Vanderbilt, D.; Wu, S.; Randolph, L. Chimerism in monochorionic dizygotic twins: Case study and review. Am. J. Med. Genet. A 2013, 161A, 1817–1824. [Google Scholar]
- Sperling, L.; Tabor, A. Twin pregnancy: The role of ultrasound in management. Acta Obstet. Gynecol. Scand. 2001, 80, 287–299. [Google Scholar] [CrossRef]
- Zheng, J.; Xu, C.; Guo, J.; Wei, Y.; Ge, H.; Li, X.; Zhang, C.; Jiang, H.; Pan, L.; Tang, W.; et al. Effective Noninvasive Zygosity Determination by Maternal Plasma Target Region Sequencing. PLoS One 2013, 8, e65050. [Google Scholar]
- Futch, T.; Spinosa, J.; Bhatt, S.; de Feo, E.; Rava, R.P.; Sehnert, A.J. Initial clinical laboratory experience in noninvasive prenatal testing for fetal aneuploidy from maternal plasma DNA samples. Prenat. Diagn. 2013, 33, 569–574. [Google Scholar] [CrossRef]
- Hofmann, W.; LifeCodexx AG, Konstanz, Germany. Unpublished work. 2014.
© 2014 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Grömminger, S.; Yagmur, E.; Erkan, S.; Nagy, S.; Schöck, U.; Bonnet, J.; Smerdka, P.; Ehrich, M.; Wegner, R.-D.; Hofmann, W.; et al. Fetal Aneuploidy Detection by Cell-Free DNA Sequencing for Multiple Pregnancies and Quality Issues with Vanishing Twins. J. Clin. Med. 2014, 3, 679-692. https://doi.org/10.3390/jcm3030679
Grömminger S, Yagmur E, Erkan S, Nagy S, Schöck U, Bonnet J, Smerdka P, Ehrich M, Wegner R-D, Hofmann W, et al. Fetal Aneuploidy Detection by Cell-Free DNA Sequencing for Multiple Pregnancies and Quality Issues with Vanishing Twins. Journal of Clinical Medicine. 2014; 3(3):679-692. https://doi.org/10.3390/jcm3030679
Chicago/Turabian StyleGrömminger, Sebastian, Erbil Yagmur, Sanli Erkan, Sándor Nagy, Ulrike Schöck, Joachim Bonnet, Patricia Smerdka, Mathias Ehrich, Rolf-Dieter Wegner, Wera Hofmann, and et al. 2014. "Fetal Aneuploidy Detection by Cell-Free DNA Sequencing for Multiple Pregnancies and Quality Issues with Vanishing Twins" Journal of Clinical Medicine 3, no. 3: 679-692. https://doi.org/10.3390/jcm3030679
APA StyleGrömminger, S., Yagmur, E., Erkan, S., Nagy, S., Schöck, U., Bonnet, J., Smerdka, P., Ehrich, M., Wegner, R.-D., Hofmann, W., & Stumm, M. (2014). Fetal Aneuploidy Detection by Cell-Free DNA Sequencing for Multiple Pregnancies and Quality Issues with Vanishing Twins. Journal of Clinical Medicine, 3(3), 679-692. https://doi.org/10.3390/jcm3030679