Molecular Characterization of Adult-Type Lower-Grade Glioma (WHO Grade 1–3) with Targeted Next-Generation Sequencing: A Retrospective, Single-Institution Experience
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Population
2.2. Next-Generation Sequencing (NGS)
2.3. Treatment Strategy
2.4. Radiological Follow-Up Strategy
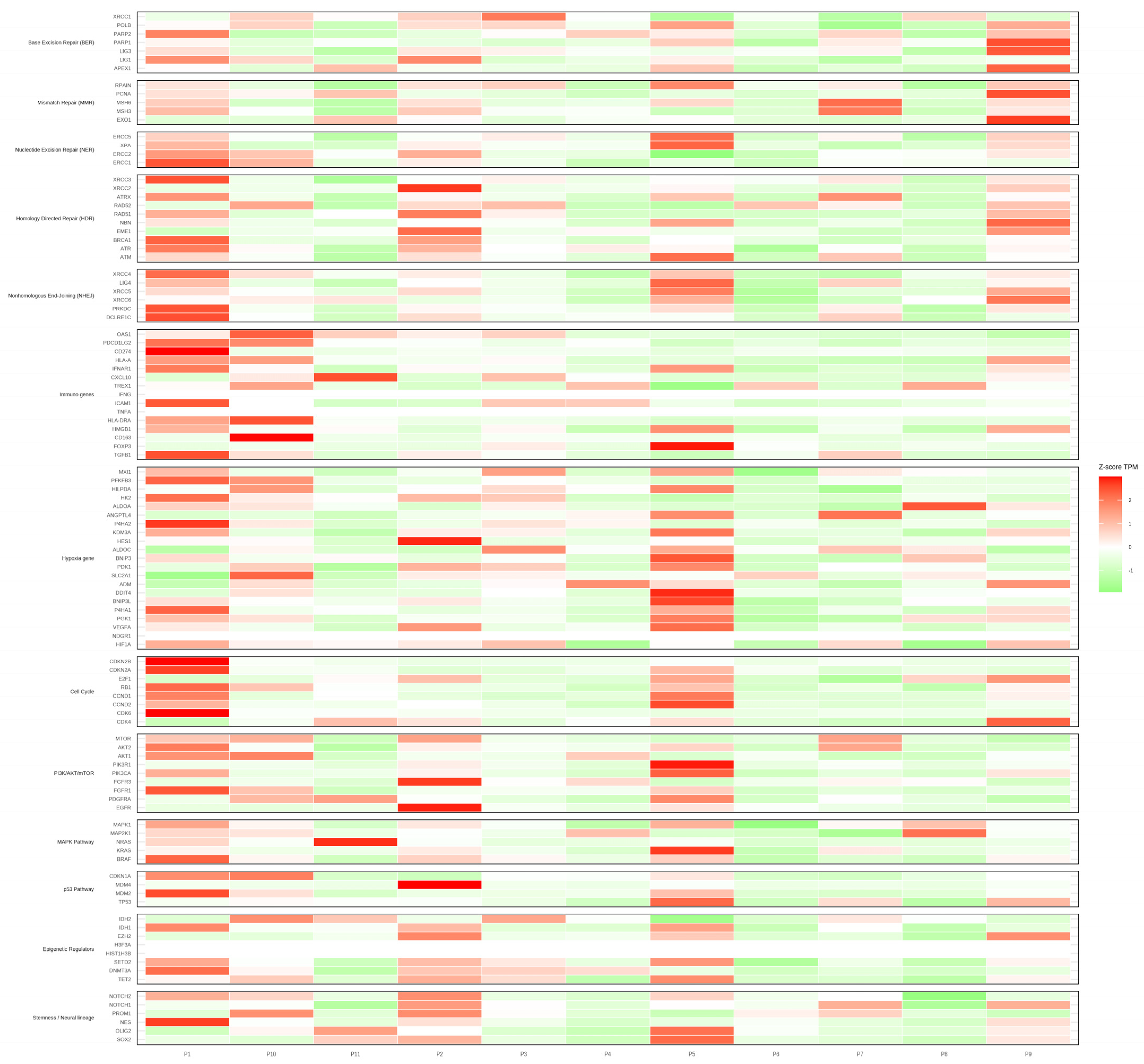
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| BRAF | B-Raf proto-oncogene, serine/threonine kinase |
| CNS | Central nervous system |
| EGFR | Epithelial Growth Factor Receptor |
| FLAIR | Fluid-attenuated inversion recovery |
| LGG | Low-grade glioma |
| MRI | Magnetic resonance imaging |
| NGS | Next-Generation Sequencing |
| OS | Overall survival |
| PFS | Progression-free survival |
| TERT | Telomerase Reverse Transcriptase |
References
- Ostrom, Q.T.; Cioffi, G.; Gittleman, H.; Patil, N.; Waite, K.; Kruchko, C.; Barnholtz-Sloan, J.S. CBTRUS Statistical Report: Primary Brain and Other Central Nervous System Tumors Diagnosed in the United States in 2012–2016. Neuro-Oncology 2019, 21, v1–v100. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Louis, D.N.; von Deimling, A. Grading of diffuse astrocytic gliomas: Broders, Kernohan, Zülch, the WHO… and Shakespeare. Acta Neuropathol. 2017, 134, 517–520. [Google Scholar] [CrossRef] [PubMed]
- Louis, D.N.; Wesseling, P.; Brandner, S.; Brat, D.J.; Ellison, D.W.; Giangaspero, F.; Hattab, E.M.; Hawkins, C.; Judge, M.J.; Kleinschmidt-DeMasters, B.; et al. Data Sets for the Reporting of Tumors of the Central Nervous System: Recommendations From The International Collaboration on Cancer Reporting. Arch. Pathol. Lab. Med. 2020, 144, 196–206. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez Castro, L.N.; Wesseling, P. The cIMPACT-NOW updates and their significance to current neuro-oncology practice. Neuro-Oncol. Pract. 2020, 8, 4–10. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Louis, D.N.; Perry, A.; Wesseling, P.; Brat, D.J.; Cree, I.A.; Figarella-Branger, D.; Hawkins, C.; Ng, H.K.; Pfister, S.M.; Reifenberger, G.; et al. The 2021 WHO Classification of Tumors of the Central Nervous System: A summary. Neuro-Oncology 2021, 23, 1231–1251. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Brat, D.J.; Aldape, K.; Colman, H.; Holland, E.C.; Louis, D.N.; Jenkins, R.B.; Kleinschmidt-DeMasters, B.K.; Perry, A.; Reifenberger, G.; Stupp, R.; et al. cIMPACT-NOW update 3: Recommended diagnostic criteria for “Diffuse astrocytic glioma, IDH-wildtype, with molecular features of glioblastoma, WHO grade IV”. Acta Neuropathol. 2018, 136, 805–810. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Horbinski, C.; Miller, C.R.; Perry, A. Gone FISHing: Clinical lessons learned in brain tumor molecular diagnostics over the last decade. Brain Pathol. 2011, 21, 57–73. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Al-Kateb, H.; Knight, S.M.; Sivasankaran, G.; Voss, J.S.; Pitel, B.A.; Blommel, J.H.; Jerde, C.R.; Rumilla, K.M.; Lee, J.L.; Mattson, N.R.; et al. Clinical Validation of the TruSight Oncology 500 Assay for the Detection and Reporting of Pan-Cancer Biomarkers. J. Mol. Diagn. 2025, 27, 292–305. [Google Scholar] [CrossRef] [PubMed]
- Froyen, G.; Geerdens, E.; Berden, S.; Cruys, B.; Maes, B. Diagnostic Validation of a Comprehensive Targeted Panel for Broad Mutational and Biomarker Analysis in Solid Tumors. Cancers 2022, 14, 2457. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Dal Bo, M.; Polano, M.; Ius, T.; Di Cintio, F.; Mondello, A.; Manini, I.; Pegolo, E.; Cesselli, D.; Di Loreto, C.; Skrap, M.; et al. Machine learning to improve interpretability of clinical, radiological and panel-based genomic data of glioma grade 4 patients undergoing surgical resection. J. Transl. Med. 2023, 21, 450. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Chen, S.; Liu, M.; Huang, T.; Liao, W.; Xu, M.; Gu, J. GeneFuse: Detection and visualization of target gene fusions from DNA sequencing data. Int. J. Biol. Sci. 2018, 14, 843–848. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Stupp, R.; Mason, W.P.; van den Bent, M.J.; Weller, M.; Fisher, B.; Taphoorn, M.J.B.; Belanger, K.; Brandes, A.A.; Marosi, C.; Bogdahn, U.; et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N. Engl. J. Med. 2005, 352, 987–996. [Google Scholar] [CrossRef] [PubMed]
- Weller, M.; van den Bent, M.; Preusser, M.; Le Rhun, E.; Tonn, J.C.; Minniti, G.; Bendszus, M.; Balana, C.; Chinot, O.; Dirven, L.; et al. EANO guidelines on the diagnosis and treatment of diffuse gliomas of adulthood. Nat. Rev. Clin. Oncol. 2021, 18, 170–186, Erratum in Nat. Rev. Clin. Oncol. 2022, 19, 357–358. https://doi.org/10.1038/s41571-022-00623-3. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Wen, P.Y.; Bent, M.v.D.; Youssef, G.; Cloughesy, T.F.; Ellingson, B.M.; Weller, M.; Galanis, E.; Barboriak, D.P.; de Groot, J.; Gilbert, M.R.; et al. RANO 2.0: Update to the Response Assessment in Neuro-Oncology Criteria for High- and Low-Grade Gliomas in Adults. J. Clin. Oncol. 2023, 41, 5187–5199. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Tesileanu, C.M.S.; Dirven, L.; Wijnenga, M.M.J.; Koekkoek, J.A.F.; Vincent, A.J.P.E.; Dubbink, H.J.; Atmodimedjo, P.N.; Kros, J.M.; Van Duinen, S.G.; Smits, M.; et al. Survival of diffuse astrocytic glioma, IDH1/2 wildtype, with molecular features of glioblastoma, WHO grade IV: A confirmation of the cIMPACT-NOW criteria. Neuro-Oncology 2020, 22, 515–523. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Parsons, D.W.; Jones, S.; Zhang, X.; Lin, J.C.-H.; Leary, R.J.; Angenendt, P.; Mankoo, P.; Carter, H.; Siu, I.-M.; Gallia, G.L.; et al. An integrated genomic analysis of human glioblastoma multiforme. Science 2008, 321, 1807–1812. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Balss, J.; Meyer, J.; Mueller, W.; Korshunov, A.; Hartmann, C.; von Deimling, A. Analysis of the IDH1 codon 132 mutation in brain tumors. Acta Neuropathol. 2008, 116, 597–602. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, T.; Nobusawa, S.; Kleihues, P.; Ohgaki, H. IDH1 mutations are early events in the development of astrocytomas and oligodendrogliomas. Am. J. Pathol. 2009, 174, 1149–1153. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Yan, H.; Parsons, D.W.; Jin, G.; McLendon, R.; Rasheed, B.A.; Yuan, W.; Kos, I.; Batinic-Haberle, I.; Jones, S.; Riggins, G.J.; et al. IDH1 and IDH2 mutations in gliomas. N. Engl. J. Med. 2009, 360, 765–773. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Dang, L.; White, D.W.; Gross, S.; Bennett, B.D.; Bittinger, M.A.; Driggers, E.M.; Fantin, V.R.; Jang, H.G.; Jin, S.; Keenan, M.C.; et al. Cancer-associated IDH1 mutations produce 2-hydroxyglutarate. Nature 2009, 462, 739–744. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Turcan, S.; Rohle, D.; Goenka, A.; Walsh, L.A.; Fang, F.; Yilmaz, E.; Campos, C.; Fabius, A.W.M.; Lu, C.; Ward, P.S.; et al. IDH1 mutation is sufficient to establish the glioma hypermethylator phenotype. Nature 2012, 483, 479–483. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Cancer Genome Atlas Research Network; Brat, D.J.; Verhaak, R.G.; Aldape, K.D.; Yung, W.K.; Salama, S.R.; Cooper, L.A.; Rheinbay, E.; Miller, C.R.; Vitucci, M.; et al. Comprehensive, Integrative Genomic Analysis of Diffuse Lower-Grade Gliomas. N. Engl. J. Med. 2015, 372, 2481–2498. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Brat, D.J.; Aldape, K.; Colman, H.; Figrarella-Branger, D.; Fuller, G.N.; Giannini, C.; Holland, E.C.; Jenkins, R.B.; Kleinschmidt-DeMasters, B.; Komori, T.; et al. cIMPACT-NOW update 5: Recommended grading criteria and terminologies for IDH-mutant astrocytomas. Acta Neuropathol. 2020, 139, 603–608. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Taşkan, T.; Noori, F.; Kurukahvecioğlu, O.; Karaman, N.; Gönenç, A. Neurturin gene IVSI-663 polymorphism but not RET variants is associated with increased risk for breast cancer. Lab. Med. 2025, 56, 351–359. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Elisei, R.; Cosci, B.; Romei, C.; Bottici, V.; Sculli, M.; Lari, R.; Barale, R.; Pacini, F.; Pinchera, A. RET exon 11 (G691S) polymorphism is significantly more frequent in sporadic medullary thyroid carcinoma than in the general population. J. Clin. Endocrinol. Metab. 2004, 89, 3579–3584. [Google Scholar] [CrossRef] [PubMed]
- Yu, Z.; Li, H.; Wang, M.; Luo, W.; Xue, Y. GDNF regulates lipid metabolism and glioma growth through RET/ERK/HIF-1/SREBP-1. Int. J. Oncol. 2022, 61, 109. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Gabler, L.; Jaunecker, C.N.; Katz, S.; van Schoonhoven, S.; Englinger, B.; Pirker, C.; Mohr, T.; Vician, P.; Stojanovic, M.; Woitzuck, V.; et al. Fibroblast growth factor receptor 4 promotes glioblastoma progression: A central role of integrin-mediated cell invasiveness. Acta Neuropathol. Commun. 2022, 10, 65. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Colombo, C.; Minna, E.; Rizzetti, M.G.; Romeo, P.; Lecis, D.; Persani, L.; Mondellini, P.; Pierotti, M.A.; Greco, A.; Fugazzola, L.; et al. The modifier role of RET-G691S polymorphism in hereditary medullary thyroid carcinoma: Functional characterization and expression/penetrance studies. Orphanet J. Rare Dis. 2015, 10, 25. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]



| Cohort Size | 11 | |
|---|---|---|
| Gender | Males | 9 (81.81%) |
| Females | 2 (18.18%) | |
| Age at diagnosis | Years | 40 (19–75) |
| Type of diagnosis | Incidental | 3 (27.27%) |
| Symptomatic | 8 (72.73%) | |
| Tumor location | Frontal | 8 (72.73%) |
| Parietal | 2 (18.18%) | |
| Brainstem | 1 (9.09%) | |
| Extent of resection | GTR | 6 (54.55%) |
| STR | 4 (36.36%) | |
| PR (biopsy) | 1 (9.09%) | |
| Adjuvant treatments | Chemoradiotherapy with Temozolomide | 3 (27.27%) |
| PCV protocol | 1 (9.09%) | |
| Radiotherapy alone | 2 (18.18%) | |
| None (or not available) | 5 (45.45%) | |
| Follow-up | Months | 14.67 (4.7–92.07) |
| PFS (6 patients progressed) | Months | 17.5 (1.8–50.3) |
| Case Id | Diagnosis and Grade | Gene | Mutation |
|---|---|---|---|
| 1 | Astrocytoma IDH-mutant, G2 | RET | G691S |
| IDH1 | R132H | ||
| CHEK2 | R474H | ||
| ALK | A1601T | ||
| BLM | S1209T | ||
| IGF1R | R511Q | ||
| SLX4 | R669H | ||
| 2 | Astrocytoma IDH-mutant, G2 | IRS2 | G1057D |
| IDH1 | R132H | ||
| FGFR4 | G388R | ||
| APC | No match | ||
| TP53 | C238R | ||
| 3 | Oligodendroglioma IDH-mutant and 1p/19q co-deleted, G3 | RET | G691S |
| RET | R982C | ||
| IDH1 | R132H | ||
| FGFR4 | G388R | ||
| RANBP2 | V2787G | ||
| RANBP2 | D2828E | ||
| ERBB3 | K747N | ||
| POLE | No match | ||
| PALB2 | L332H | ||
| TP53 | No match | ||
| JAK3 | V718L | ||
| 4 | Astrocytoma IDH-mutant, G2 | IRS2 | G1057D |
| IDH1 | R132H | ||
| FGFR4 | G388R | ||
| ATM | L1420F | ||
| TP53 | R273C | ||
| AXIN2 | S762N | ||
| 5 | Diffuse astrocytic tumor IDH-wild type, G2 | GATA2 | F270C |
| RET | G691S | ||
| IRS2 | G1057D | ||
| 6 | Astrocytoma IDH-mutant, G2 | RET | G691S |
| IDH1 | R132H | ||
| TP53 | R273C | ||
| ATRX | K1176M | ||
| 7 | Oligodendroglioma IDH-mutant and 1p/19q co-deleted, G3 | RET | G691S |
| IDH1 | R132H | ||
| BLM | P355R | ||
| 8 | Ganglioglioma | RET | G691S |
| IRS2 | G1057D | ||
| SPTA1 | G1497E | ||
| BLM | N1393D | ||
| CHEK2 | R474H | ||
| CHEK2 | L174F | ||
| 9 | Pilocytic astrocytoma | IRS2 | G1057D |
| FGFR4 | G388R | ||
| DDR2 | M441I | ||
| ATM | D1853V | ||
| ZFHX3 | G585S | ||
| AXIN2 | S762N | ||
| 10 | Diffuse astrocytic tumor IDH-wild type, G2 | FGFR4 | G388R |
| SPTA1 | T1522A | ||
| EGFR | G598V | ||
| MEN1 | R176Q | ||
| POLE | G6R | ||
| ERCC4 | P379S | ||
| 11 | Astrocytoma IDH-mutant, G2 | FGFR4 | G388R |
| CDC73 | Q338P | ||
| IDH1 | R132H | ||
| RET | G691S | ||
| RET | R982C |
| Case Id | Diagnosis and Grade | Amplifications |
|---|---|---|
| 1 | Astrocytoma IDH-mutant, G2 | None |
| 2 | Astrocytoma IDH-mutant, G2 | None |
| 3 | Oligodendroglioma IDH-mutant and 1p/19q co-deleted, G3 | MYCN |
| MYC | ||
| CCND1 | ||
| FGF19 | ||
| ATM | ||
| CDK4 | ||
| FGF14 | ||
| 4 | Astrocytoma IDH-mutant, G2 | CDK4 |
| 5 | Diffuse astrocytic tumor IDH-wild type, G2 | None |
| 6 | Astrocytoma IDH-mutant, G2 | None |
| 7 | Oligodendroglioma IDH-mutant and 1p/19q co-deleted, G3 | MDM4 |
| MYC | ||
| 8 | Ganglioglioma | CDK6 |
| MET | ||
| BRAF | ||
| 9 | Pilocytic astrocytoma | FGF1 |
| CDK6 | ||
| BRAF | ||
| 10 | Diffuse astrocytic tumor IDH-wild type, G2 | MDM4 |
| MYCN | ||
| EGFR | ||
| CDK6 | ||
| BRAF | ||
| CDK4 | ||
| 11 | Astrocytoma IDH-mutant, G2 | CDK4 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license.
Share and Cite
Pinamonti, M.; Polano, M.; Cester, G.; Saturno Spurio, F.; Roman-Pognuz, E.; Ukmar, M.; Dal Bo, M.; Zanconati, F.; Tacconi, L.; Meola, A. Molecular Characterization of Adult-Type Lower-Grade Glioma (WHO Grade 1–3) with Targeted Next-Generation Sequencing: A Retrospective, Single-Institution Experience. J. Clin. Med. 2026, 15, 53. https://doi.org/10.3390/jcm15010053
Pinamonti M, Polano M, Cester G, Saturno Spurio F, Roman-Pognuz E, Ukmar M, Dal Bo M, Zanconati F, Tacconi L, Meola A. Molecular Characterization of Adult-Type Lower-Grade Glioma (WHO Grade 1–3) with Targeted Next-Generation Sequencing: A Retrospective, Single-Institution Experience. Journal of Clinical Medicine. 2026; 15(1):53. https://doi.org/10.3390/jcm15010053
Chicago/Turabian StylePinamonti, Maurizio, Maurizio Polano, Giacomo Cester, Federico Saturno Spurio, Erik Roman-Pognuz, Maja Ukmar, Michele Dal Bo, Fabrizio Zanconati, Leonello Tacconi, and Antonio Meola. 2026. "Molecular Characterization of Adult-Type Lower-Grade Glioma (WHO Grade 1–3) with Targeted Next-Generation Sequencing: A Retrospective, Single-Institution Experience" Journal of Clinical Medicine 15, no. 1: 53. https://doi.org/10.3390/jcm15010053
APA StylePinamonti, M., Polano, M., Cester, G., Saturno Spurio, F., Roman-Pognuz, E., Ukmar, M., Dal Bo, M., Zanconati, F., Tacconi, L., & Meola, A. (2026). Molecular Characterization of Adult-Type Lower-Grade Glioma (WHO Grade 1–3) with Targeted Next-Generation Sequencing: A Retrospective, Single-Institution Experience. Journal of Clinical Medicine, 15(1), 53. https://doi.org/10.3390/jcm15010053






