4.1. Summary of Main Results
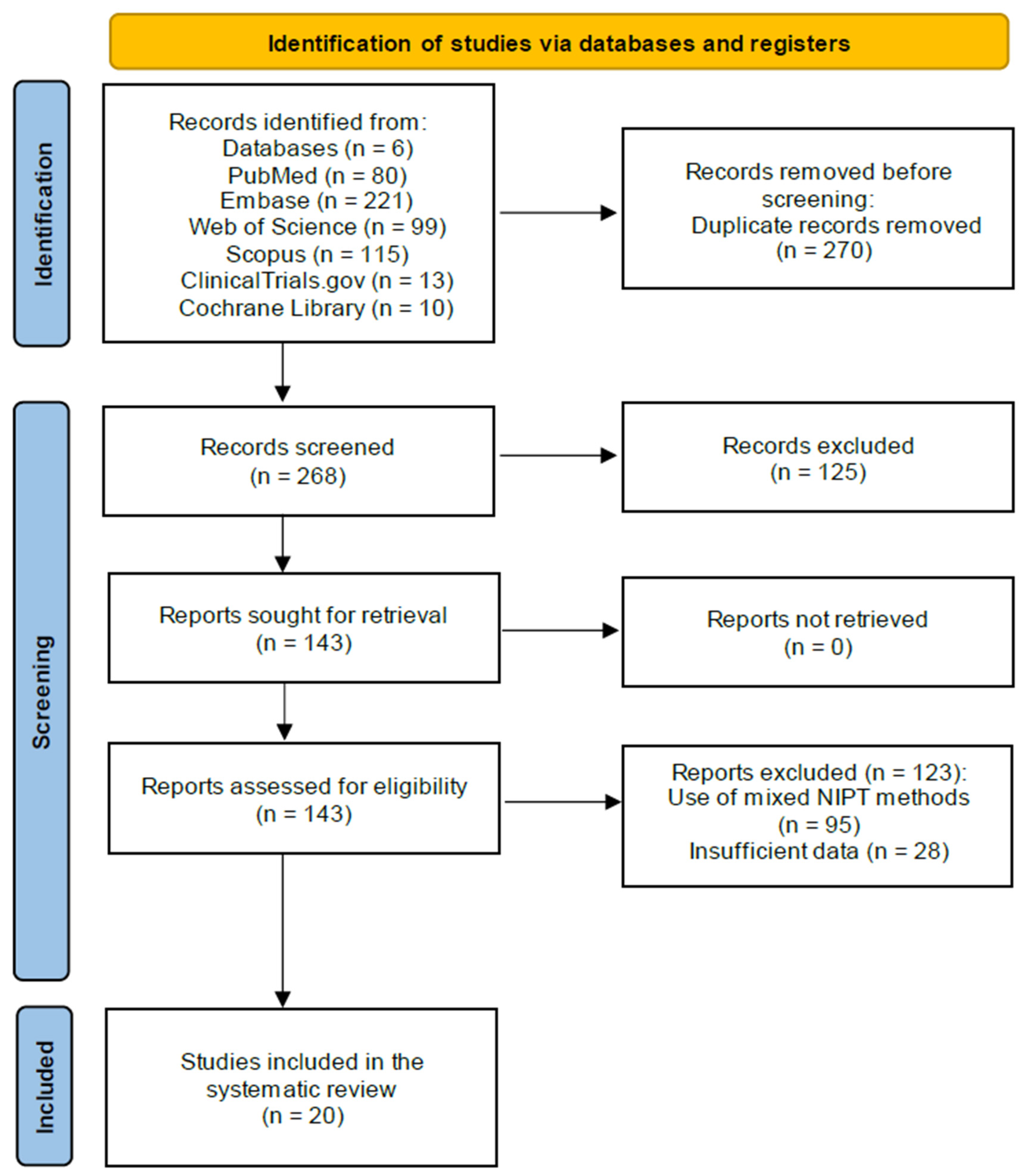
This review includes data from 21 studies involving 92,164 pregnant women, of whom 1245 were affected by T21, tested using genomics-based non-invasive prenatal testing (NIPT).
Out of the included studies, eleven were conducted in unselected populations, two in high-risk cohorts, and eight studies did not specify how the participants were recruited. In the two studies involving high-risk participants, two NIPT methods were evaluated—the WGS method [
29] and the RCA method [
37]. Both studies showed high accuracy in detecting the three major trisomies (T21, T18 and T13), with sensitivities ranging from 95.0% to 100% depending on the trisomy, and specificity above 99%.
Because of the different sample sizes, we calculated the weighted average for the different NIPT methods. This was done by multiplying each data point by its sample size, summing the products, and then dividing it by the total number of samples.
We compared the averages of sensitivity, specificity, PPV, and NPV for each NIPT method used for T21 detection (
Table 2 and
Table 3). Values lower than 95% and higher than 90% were colored blue, while values lower than 90% were highlighted red. As for sensitivity, the RCA test showed slightly lower values (97.63%), while all the other tests, such as the WGS-based tests, the SNP test, and the microarray test, showed high sensitivity (above 98.9%). This difference might be the result of the slight variations and limited number of studies. Therefore, it cannot be concluded that the RCA method has significantly lower sensitivity than the other methods. In contrast, all tests showed similarly high specificity values exceeding 99%.
Regarding the positive predictive value, NIPT cannot reach 100%, as the cfDNA used is of placental origin, and may harbor confined placental mosaicism. Further, 100% PPV was not reached for T21 as the SNP and RCA tests had 92.97% and 75.98% PPVs, respectively. Two publications were outliers, with PPVs of 80.0% in one of the SNP tests [
33] and 64.44% in one the RCA tests reported [
19], partly explaining the relatively lower PPVs for these tests. It is worth noting that when we excluded these two studies, the average PPVs of the SNP and RCA tests still remained lower than that of other tests (93.83% and 93.25%), supporting the previous result that the SNP and RCA tests have lower PPVs for T21 detection. In terms of NPV, all NIPT methods performed well, with values above 99%.
In conclusion, the figures show that all tests utilizing WGS or targeted methodologies such as SNPs, microarrays, and RCA perform similarly in terms of clinical sensitivity, specificity and NPV for the detection of fetal T21. However, two targeted methods—using SNPs and RCA—demonstrated lower PPV values compared to other NIPT methods.
When we compared the weighted averages of sensitivity, specificity, PPV and NPV of each NIPT method used for detecting T18 (
Table 4 and
Table 5), the microarray-based test showed lower sensitivity, at 77.22%. This was primarily due to the low sensitivity scores of all but one report on the microarray-based methods, with sensitivities of 73.33%, 71.43%, 100%, and 80%, respectively [
18,
34,
35,
36]. Based on these results, the microarray-based (Harmony) test showed lower sensitivity for the detection of T18. The WGS-based methods performed well in terms of sensitivity, nearing 100%. One product showed slightly lower sensitivity, at 96.40%, which can be explained by one outlier in a single study reporting 95% sensitivity, while the rest of the tests reported 100% sensitivity [
27,
28,
29,
30]. For this reason, we cannot conclude that this particular WGS-using product has lower sensitivity than the other tests. All NIPT tests proved to be highly specific for detecting T18, with specificity above 99%, regardless of the method or product. In terms of PPV, the values varied significantly, with two WGS tests scoring only 77.56% and 84.74%, and the RCA test scored only 66.98%. One of the WGS products did particularly poorly, with PPV for T18 ranging between 66.67% and 83.33% [
27,
28,
29,
30].
Interestingly, when we looked more closely at the performances of the other WGS-based test, the PPV for T18 was 100% and 77.27% [
31,
32]. But the sample size for the Stumm study [
32] was approximately one-tenth of that in the Hofmann study [
31], suggesting that the PPV for this product is clearly lower than those of other methods. Based on the published figures, the PPV of T18 detection using the RCA test was lower, with the two lowest scores—60% and 65.12%—associated with the studies with the largest sample (
Table 4) [
19,
34,
37,
38,
39]. While two of the WGS-based methods and the RCA test had lower PPVs for T18 detection, another WGS-based NIPT had only a slightly lower PPV of 92.56% based on one study, but with a relatively high number of tested cases. There was no significant difference for the PPV between one of the WGS tests, the SNP, and the microarray-based test (98.05%, 95.31%, and 100%, respectively). Each NIPT method had a high NPV (all above 99%).
In summary, all types of non-invasive prenatal tests, utilizing WGS, SNPs, microarray or RCA, perform similarly in terms of clinical specificity and NPV for the detection of fetal T18. The microarray method, however, had lower sensitivity compared to other tests. All tests demonstrated relatively lower PPV, except for one test utilizing WGS, as well as the microarray-based method.
In terms of sensitivity, there was only one outlier in the RCA group [
39], with 62.5% sensitivity, compared to all the other tests, which had 100% sensitivity (
Table 6). All tests are found to be similarly efficient when comparing the weighted averages of sensitivity, specificity, PPV and NPV of each NIPT method for T13 detection (
Table 7). For specificity, all NIPT methods performed well, with values above 99%, thus we did not detect any differences in specificity between the tests. There was a relatively high PPV in all tests, at above 99%, except for one of the WGS methods, which had a PPV of only 84.74%. Similarly to the PPV in T18, the same authors reported a low PPV figure for T13 testing in a large study population, which lowered the overall PPV for this product (100% and 83.33%, respectively) [
31,
32]. Every NIPT method had a high NPV, above 99%, for T13.
As a conclusion, these results show that nearly all tests utilizing whole-genome sequencing (WGS) or targeted methods such as single nucleotide polymorphisms (SNPs), microarrays, and rolling circle amplification (RCA) perform similarly in terms of clinical sensitivity, specificity, PPV, and NPV for the detection of fetal T13.
4.2. Comparison with Classic Screening Methods and Health Economical Considerations
In the 21 selected studies, all NIPT methods showed greater sensitivity for the detection of T21, above 98%, compared to traditional screening tests; the detection rate of the combined test is 82–87%, while NT alone could detect 70% of Down syndrome (DS) cases. The detection rate of the quadruple test is 81%, and the detection rates of the full integrated test, serum integrated test, and stepwise sequential testing are 96%, 88% and 95%, respectively [
13].
It is not our aim to “compare” the traditional methods with the new cfDNA-based technique. Nevertheless, in the recent era, considerations of cost-effectiveness are becoming increasingly important, and the issue is far more complex than simply comparing the cost of the tests themselves. In the past, the use of NIPT as an alternative to current first-tier screening tests, which include both biochemical and ultrasound methods, has been deemed non-cost-ineffective [
10,
40].
Changes in the cost of NIPT have led to its adoption as a first-tier screening test in several countries [
41,
42].
Looking at long-term health and economic considerations, it transpired that including NIPT in existing prenatal screening for DS was beneficial over conventional testing [
43]. The authors also stated that, based on the prices at the time, using NIPT as a second-tier screening method is more cost-effective than NIPT as the first choice. But long term costs were considered by Xiao et al., who compared the costs of different diagnostic strategies for Down syndrome in a large population (17,363 patients). Down syndrome seems to have the largest impact on society, since it is compatible with life; the overall cost also includes long-term direct medical and indirect, non-medical health and social costs, such as the development of services and education for individuals with Down syndrome. They observed three groups: The first group used classical serum screening, and subsequent invasive testing if the Down syndrome risk was ≥1/270. The second group employed serum screening, and those with a T21 risk ≥ 1/270 underwent invasive testing, while those with a risk between 1/270 and 1/1000 had NIPT, followed by invasive testing based on the NIPT result. The third method applied NIPT to all patients with a risk ≥ 1/1000, and if indicated, this was followed by invasive tests. In the fourth method, all patients underwent NIPT [
44]. In summary, serum screening alone would have missed 14 cases of Down syndrome, while the 2nd and 3rd approaches would have resulted in eight missed cases, and none would have resulted with the fourth method (NIPT for all). When they compared the lifetime health economic costs of the different strategies, group 1 was the most expensive and resulted in the most “missed” Down syndrome cases. The “lifetime” costs, for groups 1, 2, 3 and 4 were 100%, 63%, 61%, and 23%, respectively.
A recent publication concluded that the application of ultrasound combined with NIPT in prenatal testing as a complimentary method seems to be the most effective approach, providing the most accurate data for obstetricians and resulting in the best health and economic outcomes [
45].
We would like to conclude our considerations with the 2023 consensus statement of the board of the International Society for Prenatal Diagnosis on the use of non-invasive prenatal testing for the detection of fetal chromosomal conditions in singleton pregnancies, as follows: [
46] (1) NIPT is the most accurate screening test for common autosomal aneuploidies (trisomies 21, 13 and 18) in unselected singleton populations, and those at known increased probability. (2) False positive results occur with NIPT. Therefore, ISPD strongly recommends that all pregnant individuals with a high chance of an NIPT result undergo genetic counseling and diagnostic testing if they are considering termination of pregnancy.
4.6. Further Considerations
In the past, the use of NIPT as an alternative to current first-tier screening tests, which include both biochemical and ultrasound methods, was deemed cost-ineffective [
10,
47]. More recently, a number of countries have adopted cfDNA-based NIPT as the single first-tier screening method for trisomies [
41].
It is worth noting that, compared to targeted methods such as SNPs, microarrays, and RCA, WGS have the potential to screen for a variety of chromosomal abnormalities. There is insufficient data on the clinical sensitivity of detecting rare abnormalities, and it is beyond the scope of this paper to comment on microdeletion detection or single-gene NIPT. It is not documented in this study, but needs to be mentioned, that triploidy can only be diagnosed with the SNP method [
48,
49]. WGS benefits only a very small number of patients who actually have rare abnormalities, but it also increases false positive rates, identifying patients as high-risk pregnancies, resulting in heightened surveillance and unnecessary invasive diagnostics. Currently, WGS is more expensive than other targeted methods such as SNPs, microarrays, or RCA. More data are required, and financial feasibility issues should be addressed in order to prove the potential of WGS-based whole-chromosome screening [
50,
51,
52].
We did not specifically address the failure rates of NIPT. Gil et al. (2017) [
53], in their metanalysis, addressed this problem, referring to the “no result” of cfDNA-based tests. The three reasons they quote are as follows: Problems with blood collection and the transportation of the sample, and issues with hemolysis, etc. (0.03–11.1%). The second issue is the low fetal fraction, below 4% (0.1–6.1%), and the third reason is assay failure (failed DNA extraction, amplification or sequencing). They also state that it was not possible to draw a correlation between the failure rates and the method used for the analysis, but it seemed that the failure rate for sex chromosome aneuploidies was higher than for trisomies [
53]. A look at the failure rate of the different tests gave the following result: for the WGS-based method, failure rates ranged from 0.24% to 1.46%; with the microarray, the highest failure rate was 3.2% (0.04–3.2%), whilst the RCA failure rate ranged from 0.07 to 0.93%. Considering the above-mentioned reasons for the “no result”, comparing failure rates seems unreliable due to the heterogeneity of the methods and the bias caused by human factors.
With regard to the versatility, WGS has the most benefits. Apart from trisomies, it can detect sex chromosome abnormalities, and also microdeletions. Its technology requires a simpler and faster laboratory pipeline. Targeted methods, such as the SNP and microarray, are similarly adaptable. SNP requires the multiplex amplification of SNP sequencing in a single PCR reaction, followed by next-generation sequencing. SNP can detect common aneuploidies, sex chromosome alterations, microdeletions and also monogenic diseases. Microarray-based technology is currently available for the detection of common trisomies, sex chromosome aneuploidy and DiGeorge microdeletion. RCA, which is an easy test, will give fewer answers (only trisomies), but the method offers a simpler and more cost-effective pipeline [
54].